Abstract
The axe–txe operon encodes a toxin–antitoxin (TA) pair, Axe–Txe, that was initially identified on the multidrug-resistance plasmid pRUM in Enterococcus faecium. In Escherichia coli, expression of the Txe toxin is known to inhibit cell growth, and co-expression of the antitoxin, Axe, counteracts the toxic effect of Txe. Here, we report the nucleotide sequence of pS177, a 39 kb multidrug-resistant plasmid isolated from vancomycin-resistant Ent. faecium, which harbours the axe–txe operon and the vanA gene cluster. RT-PCR analysis revealed that the axe–txe transcript is produced by strain S177 as well as by other vancomycin-resistant enteroccoci. Moreover, we determine the mechanism by which the Txe protein exerts its toxic activity. Txe inhibits protein synthesis in E. coli without affecting DNA or RNA synthesis, and inhibits protein synthesis in a cell-free system. Using in vivo primer extension analysis, we demonstrate that Txe preferentially cleaves single-stranded mRNA at the first base after an AUG start codon. We conclude that Txe is an endoribonuclease which cleaves mRNA and inhibits protein synthesis.
INTRODUCTION
Enterococcal species such as Enterococcus faecium and Enterococcus faecalis have emerged as significant nosocomial pathogens throughout Europe and the United States. These bacteria are increasingly responsible for a variety of illnesses, particularly in immunocompromised patients, and are one of the main causes of surgical-site infections (Hidron et al., 2008; Owens & Stoessel, 2008; Richards et al., 2000). Clinical isolates of enterococci are often resistant to multiple antibiotics either intrinsically or through the lateral gene transfer of antibiotic resistance determinants, the most problematic of which are the genetic elements that encode vancomycin resistance (Baldassarri et al., 2005; Deshpande et al., 2007; Dowzicky & Park, 2008; Guardabassi & Dalsgaard, 2004; Low et al., 2001; Sader & Jones, 2009; Werner et al., 2008). A recent survey of Gram-positive pathogens isolated from hospitals in the United States found that, for between 2002 and 2008, up to 33 % of enterococcal species tested as part of the surveillance network were vancomycin-resistant enterococci (VRE) (JMI, 2009). As vancomycin is a preferred antimicrobial treatment for enterococcal infections, the increasing occurrence of multidrug-resistant VRE severely limits therapeutic options and highlights the need for new antibiotics.
In the search for novel antimicrobial agents, one intriguing approach involves the explicit targeting of toxin–antitoxin (TA) systems to exploit their innate toxicity (DeNap & Hergenrother, 2005; Engelberg-Kulka et al., 2004; Williams & Hergenrother, 2008). TA genes were originally discovered on plasmids, where they function as plasmid maintenance systems (Ogura & Hiraga, 1983). In plasmids containing the genes for proteic (type II) TA systems, a stable toxin and a labile antitoxin are encoded in a single operon. In a cell carrying the plasmid, both proteins are expressed and the antitoxin binds to and inhibits the toxin. However, if during cell division a plasmid-free segregant arises, the labile antitoxin is rapidly degraded, freeing the toxin to kill the cell (Aizenman et al., 1996; Hayes, 2003). This post-segregational killing has led TA systems to be dubbed ‘plasmid addiction' systems, as the cell will die if the plasmid is lost (Gerdes et al., 1986). The first TA system identified was ccdAB, on the F plasmid of Escherichia coli (Ogura & Hiraga, 1983), and since its discovery, a number of TA systems have been identified on a variety of plasmids and bacterial chromosomes (Moritz & Hergenrother, 2007a; Pandey & Gerdes, 2005; Sletvold et al., 2007). Among the type II TA systems, the toxins can be grouped into several superfamilies based on sequence and structural homology, including the RelE/ParE, MazF/CcdB, Doc, HipA, ζ and PIN domain superfamilies, and most have been shown to act as either endoribonucleases (also termed mRNA interferases) or inhibitors of DNA gyrase (Anantharaman & Aravind, 2003; Bernard & Couturier, 1992; Christensen-Dalsgaard & Gerdes, 2008; Christensen-Dalsgaard et al., 2008; Fico & Mahillon, 2006; Francuski & Saenger, 2009; Jiang et al., 2002; Makarova et al., 2009; Miallau et al., 2009; Van Melderen, 2002).
Reports of TA systems in Gram-positive bacteria have been much rarer compared with the numerous studies of TA systems in Gram-negative bacteria. One of the first proteic TA systems described in a Gram-positive organism was the Axe–Txe system, which was identified on the 24.8 kb non-conjugative plasmid pRUM, obtained from a multidrug-resistant clinical isolate of Ent. faecium (Grady & Hayes, 2003). pRUM confers resistance to chloramphenicol, erythromycin, streptomycin and streptothricin, and coexists in its host with a 60 kb conjugative vancomycin resistance plasmid. Sequence analysis of pRUM suggested it arose from a variety of mobile genetic elements, recombination events and smaller plasmids (Grady & Hayes, 2003). Although it was originally described in 2003, there have been no further reports on the biochemical characterization of Axe–Txe.
We have previously shown that in a collection of 75 VRE clinical isolates, 56 contained the genes for axe–txe, as determined by PCR analysis (Moritz & Hergenrother, 2007a). Of those 56 isolates, gel extractions and conjugative matings suggested a physical linkage between the vancomycin-resistance genes and the axe–txe genes in 44 isolates (Moritz & Hergenrother, 2007a). Recently, a separate analysis of a collection of Ent. faecium isolates found that of 42 strains positive by PCR for the pRUM replicon type, 90 % were also PCR-positive for axe–txe (Rosvoll et al., 2010). Additionally, co-hybridization studies suggested that a genetic linkage existed between the pRUM replicon and the axe–txe genes (Rosvoll et al., 2010). Thus, the axe–txe genes appear to be common features of plasmids in enterococci where they may play a role in the persistence and stability of plasmid-encoded antibiotic resistance, including vancomycin resistance.
Although several TA systems have been well characterized biochemically, many of these were discovered on the chromosomes of Gram-negative bacteria (Christensen et al., 2001; Engelberg-Kulka & Glaser, 1999). In contrast, there is a relative paucity of studies on TA systems of Gram-positive origin, and TA systems that were discovered on multidrug resistance plasmids. As the dissemination of such plasmids enables widespread resistance to antibiotics, the mechanisms by which these plasmids maintain themselves in their hosts are of critical importance. The development of toxin activation strategies for Gram-positive bacteria requires the identification of a suitable TA system that is functional in these hosts, and information about the mechanism of action of the toxin.
In this study, we report the nucleotide sequence of plasmid pS177 (39 023 bp), isolated from the vancomycin-resistant Ent. faecium clinical isolate S177. pS177 confers resistance to kanamycin, streptothricin, streptomycin, erythromycin and vancomcyin and harbours the genes for axe–txe and a relBE homologue, relBEEf. Additionally, we confirmed by RT-PCR analysis that the axe–txe transcript is synthesized in six VRE clinical isolates, including strain S177. Although axe–txe has been shown to function as a plasmid stabilization system in Ent. faecium, Bacillus thuringiensis and E. coli (Grady & Hayes, 2003), the mechanism by which Txe elicits its toxic effect on the cell is unknown. Here, we demonstrate that expression of the toxin inhibits protein synthesis in the cell but does not affect DNA or RNA synthesis. Primer extension analysis reveals that Txe is an endoribonuclease that cleaves cellular RNA three bases downstream of an AUG start codon. This is the first report, to our knowledge, of the biochemical mode of action of Txe, and one of the few studies on TA systems of Gram-positive origin.
METHODS
Plasmid DNA isolation.
Plasmid DNA was isolated from Ent. faecium by using a modified alkaline lysis midiprep protocol (Sambrook & Russell, 2001). A 50 ml bacterial culture grown in brain heart infusion broth was harvested after 12–14 h growth and the pellet was resuspended in 2 ml solution I (25 mM Tris, pH 8.0, 50 mM glucose, 10 mM EDTA) and 200 μl lysozyme (100 mg ml−1 in 25 mM Tris, pH 8.0). The suspension was incubated at 37 °C for 1 h. Solution II (3 ml; 0.2 M NaOH, 1 % SDS) was added and the tube was inverted gently six times, followed by 4.5 min incubation on ice. Finally, 3 ml solution III (5 M potassium acetate, 11.5 % glacial acetic acid) was added and the tube was inverted eight times, followed by 5 min incubation on ice. Cell debris was collected by centrifuging for 20 min at 20 000 g at 4 °C. To extract the nucleic acid, 7 ml supernatant was transferred to a new tube and an equal volume of phenol/chloroform/isoamyl alcohol (25 : 24 : 1, w/v/v) was added and the tube was shaken vigorously, followed by centrifugation as described previously. Nucleic acid was precipitated from 6 ml of the aqueous layer by adding an equal volume of 2-propanol and incubating at room temperature for 2 min, followed by centrifuging for 30 min at 27 000 g. The nucleic acid pellet was washed with 5 ml 70 % ethanol and centrifuged for 10 min at 27 000 g at 4 °C. All ethanol was removed and the pellet was allowed to air-dry, then dissolved in 100 μl 10 mM Tris, pH 8.0, overnight at 4 °C. The RNA was digested by incubation with 5 μl 10 mg RNase A ml−1 for 30 min at 37 °C.
Plasmid DNA isolated from Ent. faecium S177 was separated by electrophoresis in a 0.65 % agarose gel, containing 0.5 μg ethidium bromide ml−1. The dominant supercoiled plasmid band was excised from the gel and the DNA was recovered by electroelution, following a modification of the protocol described by Strong et al. (1997). Dialysis tubing with a 12–14 kDa nominal molecular mass cut-off (Spectra/Por) was prepared by boiling in 25 mM EDTA, pH 8.0, for 15 min, rinsed thoroughly in dH2O, and rinsed in 1× Tris-acetate-EDTA (TAE) at 4 °C. The gel slice and approximately 2 ml 1× TAE were secured in the dialysis tubing and electrophoresed in 1× TAE at 70 V for 2 h, at which point the polarity was reversed for 2 min. Without removing the gel slice, the solution was subjected to dialysis against 10 mM Tris, pH 8.0, overnight at 4 °C. The electroeluted plasmid DNA was concentrated by centrifuging at 500 g in a 0.5 ml 30 kDa molecular mass cut-off tube (Microcon) until the volume was reduced to 100 μl.
Plasmid DNA sequencing.
Shotgun cloning, sequencing and assembly of plasmid pS177 was performed by the University of Illinois W. M. Keck Center for Comparative and Functional Genomics. Briefly, DNA was sheared with a nebulizer, end-repaired and dephosphorylated. The DNA was separated by electrophoresis in a 0.8 % low-melting-point agarose gel, from which DNA ranging from 1.5 to 5.0 kb was purified and cloned into the pSMART-HCKan vector (Lucigen) according to the manufacturer's instructions. Sequencing was performed on 480 subclones from both the 5′ and the 3′ ends of the insert by using ABI Big-Dye terminator chemistry (Applied Biosystems). Custom primers were used in PCRs and sequencing reactions to close gaps and to ensure at least 3× coverage at each nucleotide (average coverage was 12.4×). Sequence data were assembled by using the Phrap (http://www.phrap.org) and Sequencher (GeneCodes) software programs. Analysis of the assembled plasmid via the blast program (Altschul et al., 1990) was used to determine sequence similarity, and annotations were made accordingly.
Plasmid construction.
To construct plasmid pET21a-txe-His6, the txe gene was amplified by PCR with primers txe-NheI-F, 5′-CATCCCGCTAGCATGATTAAGG-3′, and txe-XhoI-R, 5′-CGCCCTCGAGTTCATAGTG-3′. The PCR product was digested with NheI and XhoI and inserted into pET21a (Novagen) digested with NheI and XhoI. His6–Txe is expressed from pET21a-txe-His6 upon addition of IPTG to E. coli Rosetta BL21(DE3).
To construct plasmid pACYCDuet1-His6-axe, the axe gene was amplified by PCR with primers axe-start-EcoRI, 5′-GGGTGAATTCAATGGAAGCA-3′, and axe-stop-SalI, 5′-TCAGACTGCGACTTAATCATC-3′. The PCR product was digested with EcoRI and SalI and inserted into pACYCDuet1 (Novagen) digested with EcoRI and SalI. His6–Axe is expressed from pACYCDuet1-His6-axe upon addition of IPTG to E. coli BL21(DE3) or Rosetta BL21(DE3).
To construct plasmid pET21a-axe-His6, the axe gene was amplified by PCR with primers axe-cterm-F1, 5′-CCAGGATCCGCATATGATGGAAGC-3′, and axe-XhoI-R, 5′-CAAGCTCGAGGACTTCATCATC-3′. The PCR product was digested with NdeI and XhoI and inserted into pET21a (Novagen) digested with NdeI and XhoI. Axe–His6 is expressed from pET21a-axe-His6 upon addition of IPTG to E. coli BL21(DE3) or Rosetta BL21(DE3).
To construct plasmid pET28a-relB-His6, relB was amplified by PCR with primers relB-NcoI-F2, 5′-GGAGATATACCATGGGAACAATGG-3′, and relB-HindIII-R, 5′-CCAGAAAAGCTTCCATCCGAGTTC-3′. The PCR product was digested with NcoI and HindIII and inserted into pET28a treated with NcoI and HindIII. pET28a-relBE-His6 produces RelB–His6 upon addition of IPTG to E. coli BL21(DE3).
To construct plasmid pRSF-1b-His6-lpp, the lpp gene was amplified by PCR with primers lpp-1-BamHI, 5′-CCCCGGATCCGGAGATTAACTCAATCTAGAGG-3′, and lpp-2-SalI, 5′-CCCCGTCGACGCGCCATTTTTCACTTCACAG-3′. The PCR product was digested with BamHI and SalI and inserted into pRSF-1b (Novagen) digested with BamHI and SalI. His6–Lpp is expressed from pRSF-1b-His6-lpp upon addition of IPTG to E. coli Rosetta BL21(DE3) or E. coli BL21(DE3) cells.
Macromolecular synthesis assays.
For each strain tested, an overnight culture grown in Luria–Bertani (LB) with 100 μg ampicillin ml−1 was used to seed 10 ml filter-sterilized M9 broth (42 mM Na2HPO4, 24 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 0.5 % glucose, 1 % Casamino acids) and shaken at 37 °C until an OD600 of 0.3 was reached. To measure protein synthesis, [3H]leucine (l-[3, 4, 5-3H(N)]leucine; PerkinElmer) was added to the culture at a final concentration of 2 μCi ml−1 (74 kBq ml−1) and shaken at 37 °C for 5 min. To measure DNA synthesis, [3H]thymidine ([6-3H]thymidine; PerkinElmer) was added to the culture at a final concentration of 0.1 μCi ml−1 (3.7 kBq ml−1) and shaken at 37 °C for 1 min. To measure RNA synthesis, [3H]uridine ([5,6-3H]uridine; PerkinElmer) was added to the culture at a final concentration of 0.1 μCi ml−1 (3.7 kBq ml−1) and shaken at 37 °C for 1 min.
Before inducing expression, 0.5 ml was taken from the culture and added to 1 ml cold 15 % trichloroacetic acid (TCA), inverted once and placed on ice for at least 30 min to precipitate (via TCA) all macromolecules in the cell. The remaining culture was split in two; one half received 0.05 mM IPTG and the other received no IPTG. The cultures were shaken again at 37 °C and aliquots were TCA-precipitated, as above, 60, 120 and 180 min after IPTG was added. Precipitates were filtered on a glass microfibre disc (Whatman or Fisher Scientific), washed twice with cold 100 % ethanol and allowed to dry overnight. Discs were placed in scintillation fluid and c.p.m. was measured.
Cell-free protein synthesis assays.
The S30 T7 high-yield protein expression system (Promega) was used for cell-free protein synthesis according to the manufacturer's protocol with some modifications. The construct encoding the protein of interest, namely pET21-txe-His6, pET21a-axe-His6 or pET21a-His6-axe, was incubated with 10 μl S30 Premix Plus and 9 μl T7 S30 extract (at 37 °C for 20 min) to allow synthesis of Txe–His6, Axe–His6 or His6–Axe. After the initial 20 min of incubation, 300 ng DNA (10 mM Tris/HCl, pH 8.0) encoding the reporter protein (pET28a-relB-His6 or pRSF1b-His6-lpp) was added and shaken at 37 °C for 1 h. Reactions were stopped by placing tubes on ice for 5 min and by adding 1 μg RNase A to each tube and incubating at room temperature for 5 min.
Protein was isolated from each reaction by precipitation with cold 10 % TCA, followed by centrifugation at 21 130 g for 15 min at room temperature. Pellets were washed with cold 100 % ethanol and allowed to air-dry. Pellets were resuspended in 30 μl 50 mM Tris/HCl, pH 8.0, 300 mM NaCl and 20 μl 6× SDS loading dye and heated at 95 °C for 5 min. For each sample, the entire volume was run on a 15 % polyacrylamide Tris/HCl precast gel (Bio-Rad). After SDS-PAGE, proteins were transferred to an Immobilon-P PVDF membrane (Millipore) in cold Towbin transfer buffer. Membranes were blocked with PBS containing 5 % BSA and 0.05 % Tween-20 overnight with gentle shaking at 4 °C. Membranes were probed with HisProbe–HRP (Pierce) at 1 μg ml−1 in PBS with 0.05 % Tween-20 for 1 h at room temperature. Antibody binding was detected with Supersignal West Pico chemiluminescent substrate (Pierce), followed by exposure to autoradiography film.
RNA extraction and RT-PCR analysis.
Total RNA from enterococci was isolated in a manner identical to the extraction method described previously (Moritz & Hergenrother, 2007a). Intragenic primers used for axe–txe RT-PCR analysis were designed from the sequence of the plasmid pRUM. The primers used were axe-txe-RT-F, 5′-CAGTAGCTTATTCAAATTTCCGCC-3′, and axe-txe-RT-R, 5′-GTTCATCTGTAATTCTTCTGGACC-3′. Control primers were based on the tuf gene specific to enterococcal species (Garcia-Migura et al., 2007). The primers used were Ent-tuf F, 5′-TACTGACAAACCATTCATGATG-3′, and Ent-tuf R, 5-′AACTTCGTCACCAACGCGAAC-3′. RT-PCR and PCR for DNA contamination were performed by using the SuperScript One-Step RT-PCR with Platinum Taq kit (Invitrogen) as described previously (Moritz & Hergenrother, 2007a). RT-PCR amplification products were analysed by agarose gel electrophoresis in 1 % agarose and were stained with ethidium bromide.
Primer extension analysis.
Primer lpp 21 (5′-CTGAACGTCAGAAGACAGCTGATCG-3′) (Christensen et al., 2003) was 5′-end-labelled with [γ-32P]ATP according to a modification of the protocol reported by Kitami & Hiwada (1999). In a 10 μl reaction volume, 10 pmol of the lpp 21 primer was mixed with 1 μl 10× T4 polynucleotide kinase (PNK) reaction buffer (NEB), 1 μl 10 U T4 PNK (NEB) ml−1 and 3 μl [γ-32P]ATP [6 mCi mmol−1 (222 MBq mmol−1), 10 mCi ml−1 (370 MBq ml−1) or adjusted appropriately for decay]. The reaction mixture was incubated at 37 °C for 30 min followed by 2 min at 90 °C to inactivate the enzyme. Unincorporated nucleotides were removed with a NucAway Spin column according to the manufacturer's protocol (Ambion). The final concentration of the end-labelled primer was adjusted to 100 fmol μl−1 with RNase-free dH2O.
The DNA sequencing ladder was prepared with the primer lpp 21 and pRSF-1b-His6-lpp as template to map the 5′-end of the lpp mRNA. Sequencing reactions were prepared by using the Sequenase version 2.0 DNA sequencing kit and [α-32P]dATP according to the manufacturer's protocol (USB).
E. coli Rosetta BL21(DE3) carrying pET21-txe-His6 was grown in LB containing 100 μg ampicillin ml−1 to an OD600 of 0.3. Before adding 0.5 mM IPTG, a 1 ml aliquot was centrifuged for 5 min at room temperature and the pellet was frozen at −80 °C. Following addition of IPTG, aliquots were also taken at 30, 60 and 120 min post-induction. RNA was extracted by using the RNeasy mini kit for isolation of total RNA from bacteria and treated with RNase-free DNase (Qiagen). The 32P-labelled lpp 21 primer was hybridized with 10 μg total RNA from each time point according to a modification of the protocol reported by Kitami & Hiwada (1999). In a 40 μl reaction volume, 10 μg total RNA, 200 fmol 32P-labelled lpp 21 primer, 4 μl 10× avian Myeloblastosis virus reverse transcriptase (AMV RT) reaction buffer (NEB) and 4 μl 10× dNTP solution (10 mM each dNTP) were added together. The reaction mixture was heated at 90 °C for 2 min and then incubated at 54 °C overnight (18–20 h).
The next day, 20 μl of a primer extension mastermix (6 μl 10× AMV RT reaction buffer, 6 μl 10× dNTP solution, 4.2 μl 40 mM sodium pyrophosphate, 1.8 μl dH2O and 2 μl 10 U AMV RT μl−1) was added to each hybridization reaction, mixed gently and incubated at 42 °C for 1 h. Reactions were then treated with 2 μl 10 mg RNase A ml−1 at 37 °C for 30 min and extracted twice with phenol/chloroform/isoamyl alcohol (25 : 24 : 1, w/v/v). DNA was precipitated by adding 0.1 vols 3 M sodium acetate, pH 5.5, and 2.5 vols 70 % ethanol and storing at −80 °C for 30 min. Precipitates were centrifuged at room temperature for 15 min, washed with 70 % ethanol and allowed to air-dry before resuspending in 10 μl stop solution (USB).
Gel analysis was performed by using a 6 % polyacrylamide TBE–urea gel prepared with 6 % PAGE/urea and complete buffer solutions (Ambion). The gel was pre-electrophoresed at 45 W in 1× Tris-borate-EDTA (TBE) buffer for 30 min. Primer extension samples (10 μl) were heated at 90 °C for 5 min before loading onto the gel, and the 32P-labelled sequencing reactions (2 μl) were heated at 75 °C for 2 min before loading onto the gel. After running the gel at 45 W in 1× TBE buffer, it was dried under vacuum at 80 °C for 1 h. The gel was cooled before exposing to a storage phosphor screen cassette (Molecular Dynamics) at room temperature overnight. Primer extension products and the DNA sequencing ladder were detected by using the Storm PhosphorImager 840 (Molecular Dyanmics/GE Lifesciences).
RESULTS
Although it has been suggested that activation of toxins of TA pairs could be a viable antibacterial strategy, the success of such an approach is dependent on the TA system being present and functional in the bacteria of interest (Williams & Hergenrother, 2008). The axe–txe operon has been shown to reside on plasmids within VRE (Moritz & Hergenrother, 2007a; Rosvoll et al., 2010), although PCR alone was used to characterize axe–txe in these studies. In an effort to link axe–txe definitively to plasmid-encoded vancomycin resistance genes and to establish the functionality of axe–txe within VRE, we first sought to characterize fully an enterococcal plasmid harbouring axe–txe, and to determine if VRE clinical isolates synthesize the axe–txe mRNA transcript. For this, a collection of VRE clinical isolates previously shown to harbour a plasmid with axe–txe was used (Moritz & Hergenrother, 2007a).
Sequence analysis of pS177
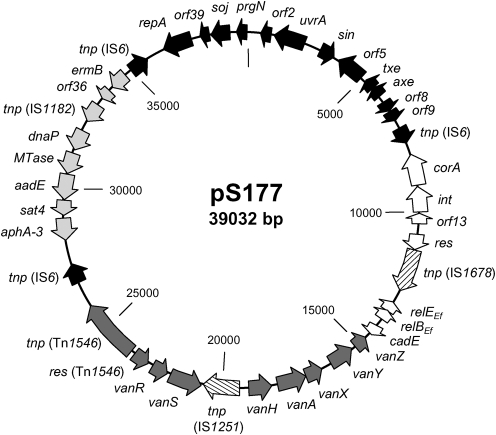
The nucleotide sequence of plasmid pS177 was determined by shotgun cloning and sequencing, with average coverage of 12.4×. pS177 is a 39 032 bp non-conjugative plasmid isolated from Ent. faecium clinical strain S177. Its DNA G+C content is 35.5 mol%, consistent with that of other enterococcal plasmids and genomes (Sletvold et al., 2007). Nucleotide blast analysis of pS177 revealed that 37 358 bp (95.7 %) shared significant (99–100 %) similarity with sequences deposited in the GenBank database (Altschul et al., 1990). Forty ORFs were identified and annotated based on these known sequences. For ORFs annotated in the database as hypothetical proteins, blastp searches were performed to assign a putative function for that potential protein based on amino acid similarity (Altschul et al., 1990). pS177 has a mosaic structure comprising a pRUM backbone, two resistance gene cassettes and five insertion elements (Fig. 1, Table 1). Interestingly, pS177 has 12 748 bp (32.6 %) and 18 930 bp (48.4 %) in common with plasmids p5753cA (GenBank accession no. GQ900435) and p5753cB (GenBank accession no. GQ900487), respectively, both isolated from Ent. faecium.
Fig. 1.
Genetic organization and mosaic structure of multidrug-resistant plasmid pS177. Coding sequences are indicated by arrows showing the predicted direction of transcription. Position 1 of pS177 was assigned as the same nucleotide position 1 of pRUM. The origins of the components of pS177 are shown by black, white or grey arrows: genes from pRUM, black; genes from p5753cA, white; solitary insertion elements, hatched; VanA-type glycopeptide resistance gene cassette, dark grey; S. intermedius resistance gene cassette, light grey.
Table 1.
ORFs of plasmid pS177
| ORF* | Gene | Nucleotide position | Protein length (aa) | Database match (GenBank accession no.) | Amino acid identity (%) | |
|---|---|---|---|---|---|---|
| 5′ | 3′ | |||||
| 1c | prgN | 1 | 291 | 97 | pRUM PrgN (AAO52827) | 100 |
| 2c | orf2 | 603 | 980 | 126 | pRUM conserved hypothetical protein, Ent. faecium (AAO52828) | 100 |
| 3c | uvrA | 946 | 2271 | 442 | pRUM UvrA, structural gene for UV resistance (AAO52829) | 100 |
| 4 | sin | 2784 | 3335 | 184 | pRUM Sin recombinase (AAO52830) | 100 |
| 5c | orf5 | 3500 | 4504 | 335 | pRUM hypothetical protein (AAO52831) | 100 |
| 6c | txe | 4963 | 5220 | 86 | pRUM Txe, toxin component of TA system (AAO52832) | 100 |
| 7c | axe | 5213 | 5482 | 90 | pRUM Axe, antitoxin component of TA system (AAO52833) | 100 |
| 8 | orf8 | 5824 | 6126 | 101 | pRUM conserved hypothetical protein (AAO52834) | 100 |
| 9 | orf9 | 6169 | 6387 | 73 | pRUM conserved hypothetical protein (AAO52835) | 100 |
| 10 | tnp (IS6) | 6559 | 7245 | 229 | pRUM IS6 transposase (AAO52848) | 100 |
| 11c | corA | 7536 | 8924 | 463 | Ent. faecium magnesium/nickel/cobalt transporter CorA (ZP_05660433) | 100 |
| 12c | int | 8989 | 9942 | 318 | Ent. faecium integrase, catalytic region (ZP_00603158) | 100 |
| 13c | orf13 | 10 026 | 10 156 | 43 | p5753cA hypothetical protein (ADA62235) | 100 |
| 14 | res | 10 585 | 11 148 | 188 | p5753cA resolvase, N-terminal domain (ADA62236) | 100 |
| 15 | tnp (IS1678) | 11 336 | 12 655 | 440 | Ent. faecium transposase, IS1678 (AAW32124) | 99 |
| 16c | relEEf | 13 111 | 13 398 | 96 | Lactobacillus antri RelE-like toxin component of TA system (ZP_05745775) | 53 |
| 17c | relBEf | 13 388 | 13 717 | 110 | Ent. faecalis Abr-family antitoxin component of TA system (ZP_06623423) | 79 |
| 18 | cadE | 14 022 | 14 351 | 110 | p5753cA cadmium efflux system accessory protein (ADA62239) | 100 |
| 19c | vanZ | 14 638 | 15 123 | 162 | Tn1546 VanZ, teicoplanin resistance protein (AAA65959) | 100 |
| 20c | vanY | 15 276 | 16 187 | 304 | Tn1546 VanY, truncated carboxypeptidase (AAA65958) | 99 |
| 21c | vanX | 16 615 | 17 223 | 203 | Tn1546 VanX, d-Ala–d-Ala dipeptidase (AAA65957) | 99 |
| 22c | vanA | 17 229 | 18 260 | 344 | Tn1546 VanA, d-Ala–d-Lac ligase (AAA65956) | 99 |
| 23c | vanH | 18 253 | 19 221 | 323 | Tn1546 VanH, pyruvate dehydrogenase (AAA65955) | 100 |
| 24 | tnp (IS1251) | 19 553 | 20 845 | 431 | Ent. faecium IS1251-like transposase (AAF73111) | 100 |
| 25c | vanS | 20 943 | 22 097 | 385 | Tn1546 VanS, truncated histidine kinase (AAA65954) | 100 |
| 26c | vanR | 22 075 | 22 770 | 232 | Tn1546 VanR, regulator of two-component regulatory system (AAA65953) | 100 |
| 27c | res | 22 984 | 23 559 | 192 | Tn1546 resolvase (AAA65952) | 100 |
| 28 | tnp (Tn1546) | 23 705 | 25 856 | 717 | Tn1546 transposase (AAA65951) | 100 |
| 29 | tnp (IS6) | 26 661 | 27 347 | 229 | pRUM IS6 transposase (AAO52854) | 100 |
| 30c | aph-3 | 28 195 | 28 980 | 262 | S. intermedius Aph-3, aminoglycoside phosphotransferase (AAG42234) | 100 |
| 31c | sat4 | 29 082 | 29 612 | 177 | S. intermedius Sat4, streptothricin acetyltransferase (AAG42233) | 100 |
| 32c | aadE | 29 621 | 30 529 | 303 | S. intermedius AadE, streptomycin adenyltransferase (AAG42232) | 100 |
| 33c | MTase | 30 562 | 31 296 | 245 | Ent. faecium methyltransferase (ZP_00603123) | 100 |
| 34c | dnaP | 31 277 | 32 146 | 290 | Ent. faecium DNA polymerase, beta-like region (ZP_00603124) | 100 |
| 35c | tnp (IS1182) | 32 521 | 33 195 | 225 | S. intermedius transposase, IS1182 (AAG42229) | 99 |
| 36c | orf36 | 33 652 | 33 783 | 44 | S. intermedius orf3 (AAG42228) | 100 |
| 37c | ermB | 33 788 | 34 525 | 246 | S. intermedius ErmB, erythromycin resistance methylase (AAG42227) | 100 |
| 38 | tnp (IS6) | 34 873 | 35 559 | 229 | pRUM IS6 transposase (AAO52854) | 100 |
| 39c | repA | 36 175 | 37 215 | 347 | pRUM putative RepA replication protein (AAO52855) | 100 |
| 40c | orf40 | 37 564 | 37 890 | 109 | pRUM hypothetical protein (AAO52856) | 100 |
| 41c | soj | 37 877 | 38 680 | 268 | pRUM Soj partitioning protein (AAO52857) | 100 |
*ORFs predicted to be transcribed on the complementary strand are denoted with ‘c’.
pS177 shares 12 341 bp (31.6 %) of its sequence with pRUM and contains 75 % of the total pRUM plasmid (GenBank accession no. AF50797). It harbours a pRUM-like replicon consisting of the putative RepA replication protein observed in various enterococcal plasmids (Grady & Hayes, 2003; Rosvoll et al., 2010). In addition to genes encoding stability mechanisms such as plasmid replication and partitioning, pS177 carries the TA system axe–txe (Grady & Hayes, 2003). The axe–txe genes and upstream promoter region share 100 % homology with the pRUM sequence, suggesting that this TA system is functional (Grady & Hayes, 2003). Plasmid pS177 harbours a gene cassette from Staphylococcus intermedius (GenBank accession no. AF299292), which confers resistance to streptothricin, streptomycin, kanamycin and erythromycin (Boerlin et al., 2001). pRUM carries a nearly identical gene cassette in which the aphA-3 gene encoding kanamycin resistance is truncated (Grady & Hayes, 2003). The full-length S. intermedius resistance gene cassette is also carried on the enterococcal plasmids pRE25 (Schwarz et al., 2001) and p5753cB.
Additionally, vancomycin resistance is encoded by the VanA-type glycopeptide resistance determinant Tn1546, observed on many plasmids identified in VRE, including plasmids pVEF3 (Sletvold et al., 2008), p5753cA and pIP816 (GenBank accession no. AM932524). However, the Tn1546 cassette on pS177 contains the insertion element IS1251 between vanS and vanH. This unusual vanA cluster type has been observed previously (Camargo et al., 2004; Handwerger & Skoble, 1995; Handwerger et al., 1995) and is also present on the enterococcal plasmid p5753cA, which carries a truncated vanA-type gene cassette with IS1251 in the same position as observed in pS177.
Plasmid pS177 shares 12 748 bp (32.6 %) with p5753cA, including the putative RelBE TA system homologue from Ent. faecium, which we designate relBEEf. The ORFs encoding RelBEf and RelEEf were identified based on their homology with p5753cA, in which their putative gene products were annotated as hypothetical proteins. A blastp search of the amino acids encoded by relEEf showed up to 100 % identity with a variety of Ent. faecium hypothetical proteins, as well as 53 % amino acid identity with the RelE protein from Lactobacillus antri (Altschul et al., 1990). The blastp search of the amino acids encoded by relBEf resulted in hits sharing up to 100 % homology with other Ent. faecium hypothetical proteins, as well as 79 % amino acid identity with the Ent. faecalis AbrB family antitoxin component.
Transcription of axe–txe in VRE clinical isolates
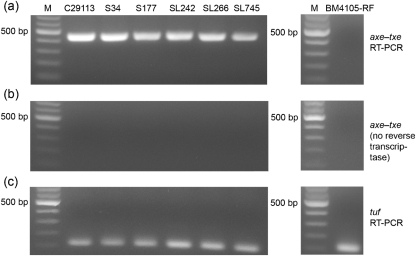
To determine whether the mRNA transcript coding for the Axe–Txe proteins is produced in VRE strains previously shown by PCR to carry axe–txe (Moritz & Hergenrother, 2007a), RT-PCR was performed from total mRNA isolated from six VRE clinical isolates. As shown in Fig. 2(a), all six isolates (C29113, S34, S177, SL242, SL266 and SL745) produce the axe–txe transcript. RT-PCR analysis was also performed on total RNA isolated from the plasmid-free Ent. faecium strain BM4105-RF, which does not contain the axe–txe genes, and no axe–txe transcript was detected (Fig. 2a). To ensure that the observed RT-PCR products were not due to DNA contamination, controls were performed in which the reverse transcriptase enzyme was not added, but all other components (including thermostable DNA polymerase and primers for axe–txe) were included. As shown in Fig. 2(b), no amplification product is observed under these conditions, confirming that the products seen by RT-PCR were due to the presence of transcript and not DNA contamination. Finally, RT-PCR with primers for the enterococcal elongation factor tuf gave the expected positive result for all seven strains examined (Fig. 2c) (Garcia-Migura et al., 2007). Thus, RT-PCR analysis demonstrated that the axe–txe transcript is synthesized in VRE clinical isolates carrying the axe–txe genes.
Fig. 2.
RT-PCR analysis of six VRE isolates and a plasmid-free enterococcal strain (BM4105-RF). The VRE isolates tested were C29113, S34, S177, SL242, SL266 and SL745. (a) RT-PCR with primers complementary to axe–txe (product size=447 bp). (b) Controls for DNA contamination, in which reverse transcriptase was omitted from the reaction mix. (c) RT-PCR with primers complementary to the enterococci tuf gene (product size=112 bp). Lane M, DNA molecular mass markers.
Txe expression inhibits global protein synthesis
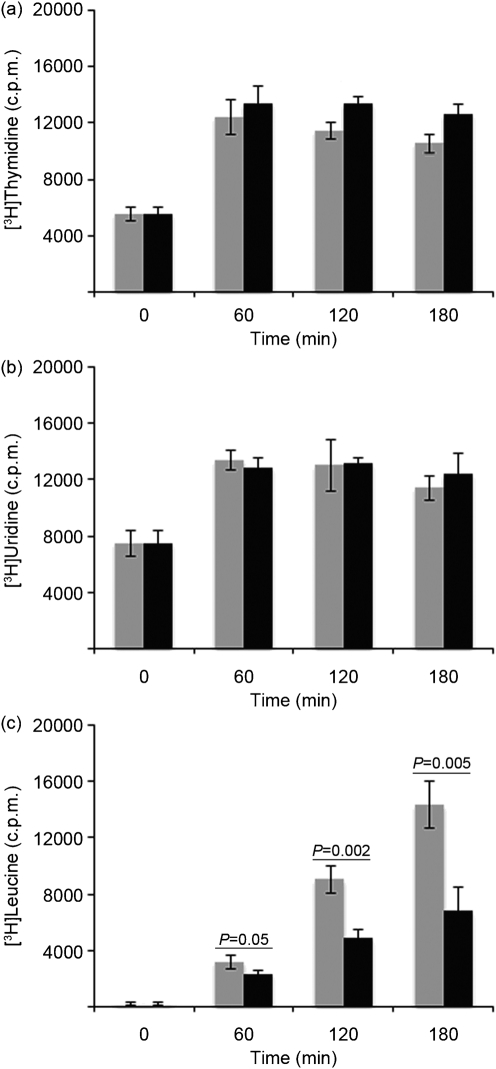
As sequencing of pS177 and RT-PCR analysis of six VRE isolates revealed that axe–txe is present and transcribed in VRE, we sought to determine the mechanism by which Txe exerts its toxic activity. Such information could be exploited in the development of TA-specific antimicrobial therapies. Txe mechanistic experiments began by assessing the toxin's effect on global biosynthetic pathways in the cell. Radiolabel incorporation assays were carried out with [3H]thymidine to measure DNA synthesis, [3H]uridine to measure RNA synthesis and [3H]leucine to measure protein synthesis upon induction of Txe–His6 expression with IPTG in E. coli Rosetta BL21(DE3). As shown in Fig. 3(a, b), levels of DNA and RNA synthesis were not inhibited by the expression of Txe–His6 in E. coli. In contrast, the level of protein synthesis was significantly inhibited by the expression of Txe–His6 in E. coli (Fig. 3c). By 2 h post-induction, the amount of [3H]leucine incorporation measured was greatly reduced in cells receiving IPTG compared with cells that did not receive IPTG. Shorter time points (5, 10, 15 and 30 min) were investigated to determine how quickly expression of Txe–His6 inhibited protein synthesis; however, differences in [3H]leucine incorporation were not observed until 60 min post-induction (Supplementary Fig. S1, available with the online version of this paper). Growth of E. coli harbouring the pET21a-txe-His6 plasmid in liquid culture was also examined; induction of Txe–His6 expression with IPTG in E. coli resulted in a decrease in the viable cell count (Supplementary Fig. S2).
Fig. 3.
DNA, RNA and protein synthesis following IPTG induction of Txe–His6, as assessed immediately before and following addition of 0 (grey bars) or 0.05 (black bars) mM IPTG to E. coli Rosetta BL21(DE3) carrying the pET21a-txe-His6 plasmid. Incorporation of [3H]thymidine (a), [3H]uridine (b) and [3H]leucine (c). Results are the mean of three independent experiments. Error bars, sd; P, statistical significance between designated groups.
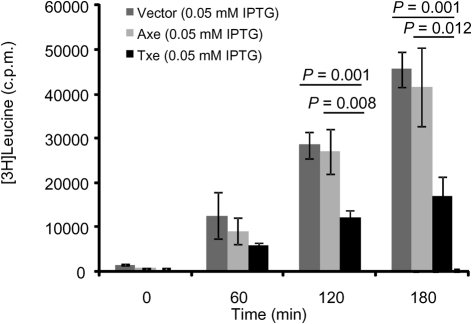
To determine whether the inhibition of protein synthesis observed was specific for the expression of Txe–His6, radiolabel incorporation assays were performed with [3H]leucine and E. coli carrying the pET21a-axe-His6 plasmid or the empty pET21a vector. As shown in Supplementary Fig. S3(a, b), the level of protein synthesis was not inhibited by the expression of Axe–His6 in E. coli or when IPTG was added to cells carrying the empty vector. A direct comparison of levels of [3H]leucine incorporation following addition of IPTG to E. coli with pET21a, pET21a-axe-His6 and pET21a-txe-His6 at the various time points examined indicated that there is considerable inhibition of protein synthesis by Txe–His6 (Fig. 4).
Fig. 4.
Protein synthesis following IPTG induction of Txe–His6, Axe–His6 and empty pET21a vector. Incorporation of [3H]leucine immediately before and following addition of 0.05 mM IPTG to E. coli Rosetta BL21(DE3) carrying pET21a, pET21a-txe-His6 or pET21a-axe-His6. Results are the mean of three independent experiments. Error bars, sd; P, statistical significance between designated groups.
Txe inhibits protein synthesis in a cell-free system
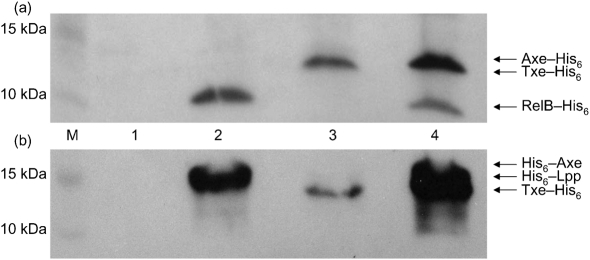
A cell-free system was used to examine further the inhibitory effect of Txe–His6 on protein synthesis. An E. coli T7 S30 extract system was preincubated with the pET21a-txe-His6 plasmid to begin synthesis of the Txe–His6 protein, followed by the addition of DNA encoding the reporter (pET28a-relB-His6 or pRSF1b-His6-lpp) to allow synthesis of RelB–His6 (the antitoxin of the RelBE TA system) or the major lipoprotein, Lpp, of the E. coli outer membrane (Christensen & Gerdes, 2003; Gotfredsen & Gerdes, 1998). Txe–His6 inhibited the synthesis of RelB–His6 (Fig. 5a, lane 3) and His6–Lpp (Fig. 5b, lane 3). When His6–Axe or Axe–His6 was synthesized instead of Txe–His6, protein synthesis of RelB–His6 and His6–Lpp was not inhibited (Fig. 5a, b, lane 4). Interestingly, both tagged versions of the antitoxin were unable to significantly prevent protein synthesis inhibition by Txe–His6 (data not shown). This may be the result of Txe–His6 inhibiting protein synthesis of the antitoxin as well as RelB–His6 and His6–Lpp, or interference of the His6-tag on Axe with its ability to bind the toxin.
Fig. 5.
Effect of Txe–His6, Axe–His6 and His6–Axe on cell-free protein synthesis. (a) Western blot with HisProbe–HRP of cell-free protein synthesis reactions with no DNA added (lane 1), pET28a-relB-His6 DNA added (2), pET28a-relB-His6 and pET21a-txe-His6 DNA added (3), and pET28a-relB-His6 and pET21a-axe-His6 DNA added (4). (b) Western blot with HisProbe-HRP of cell-free protein synthesis reactions with no DNA added (lane 1), pRSF1b-His6-lpp DNA added (2), pRSF1b-His6-lpp and pET21a-txe-His6 DNA added (3), and pRSF1b-His6-lpp and pACYCDuet1-His6-axe DNA added (4). M, Marker lane.
Txe cleaves mRNA in a sequence-specific manner
In vivo primer extension analysis was used to determine if Txe-mediated protein synthesis inhibition is achieved via mRNA cleavage in the cell. Cleavage of the 5′ end of lpp mRNA was chosen for analysis given its frequent use as an RNA substrate for both Northern blot analysis and primer extension analysis in mechanistic investigations of other TA system toxins (Christensen-Dalsgaard & Gerdes, 2008; Christensen & Gerdes, 2003; Christensen et al., 2003; Zhang et al., 2004).
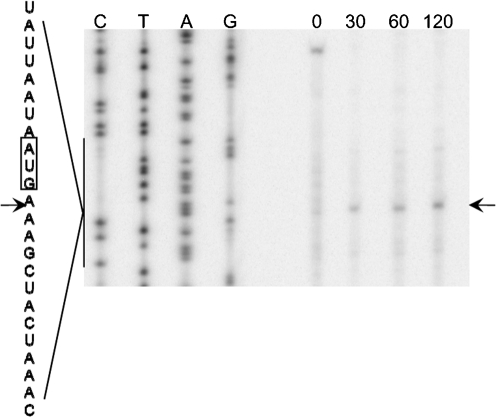
To perform primer extension analysis in cell culture, RNA was isolated from E. coli Rosetta BL21(DE3) cultures before and after induction of txe expression by IPTG. As shown in Fig. 6, induction of Txe–His6 expression led to the disappearance of the full-length lpp mRNA band and the appearance of a major cleavage band as early as 30 min post-induction (Fig. 6). The major cleavage band appeared three bases downstream of the A of the AUG start codon. These results suggest that Txe–His6 cleaves mRNA and that this RNase activity may lead to the protein synthesis inhibition observed upon induction of Txe–His6 expression in the macromolecular synthesis assays and cell-free protein synthesis assays.
Fig. 6.
Txe-induced cleavage of lpp mRNA. The 5′ end of lpp mRNA was mapped by using the primer lpp 21. Numbers indicate times (min) at which mRNA was harvested after the addition of IPTG. The major cleavage site is indicated by an arrow. The sequence around the major cleavage site is shown to the side, with the cleavage site again indicated by an arrow.
DISCUSSION
The Axe–Txe TA system was first discovered on a multidrug-resistant plasmid, pRUM, in a vancomycin-resistant Ent. faecium clinical isolate (Grady & Hayes, 2003). Axe–Txe was one of the first proteic TA systems to be characterized from Gram-positive bacteria; it was also notable in that axe–txe was discovered on a bacterial plasmid, whereas most TA systems were discovered on bacterial chromosomes. Preliminary analysis of Axe–Txe demonstrated that it functions as a characteristic TA system: expression of Txe is toxic to cells, expression of Axe alleviates Txe-induced toxicity and Axe–Txe increases the segregational stability of plasmids (Grady & Hayes, 2003). Due to the prevalence of the axe–txe genes encoded on plasmids in enterococcal isolates (Moritz & Hergenrother, 2007a; Rosvoll et al., 2010), activation of Txe presents an attractive antimicrobial strategy. However, a lack of biochemical characterization of the Axe–Txe proteins, including knowledge of the biological target or mechanism of action of Txe, hampers exploration of Axe–Txe as an antimicrobial target.
In this study, the complete nucleotide sequence of the 39 kb non-conjugative plasmid pS177 isolated from a VRE clinical strain S177 was determined. A blast analysis revealed extensive homology to known sequences, including the enterococcal plasmid pRUM, the vanA-type glycopeptide resistance determinant Tn1546 and the S. intermedius resistance gene cassette (Altschul et al., 1990). Thus, pS177 appears to have arisen from multiple recombination events between smaller plasmids and mobile genetic elements. This is the first report, to our knowledge, of a completely sequenced VRE plasmid that harbours a full-length vanA cassette containing the insertion element IS1251 between vanS and vanH. Additionally, plasmid pS177 confers resistance to vancomycin, kanamycin, streptomycin, streptothricin and erythromycin and, indeed, VRE strain S177 was resistant to gentamicin, erythromycin and vancomycin (Moritz & Hergenrother, 2007a). The presence of TA systems on pS177, including axe–txe and relBEEf, probably enhances plasmid stability and enables the persistence of this multidrug-resistant plasmid in clinical isolates of VRE. This is also the first report of a completely sequenced plasmid carrying both the VanA-type resistance determinant and axe–txe. In addition, it is shown here for the first time to our knowledge that the axe–txe transcript is produced in VRE clinical isolates. The conclusive link between the pRUM-like replicon, axe–txe, the vanA-type resistance determinant (as demonstrated from sequencing) and the presence of axe–txe transcripts in VRE clinical isolates further supports the importance of axe–txe in the maintenance of plasmids coding for multidrug resistance, and bolsters the notion of targeting axe–txe for antimicrobial development (Moritz & Hergenrother, 2007b; Rosvoll et al., 2010).
Txe shares significant sequence similarity with the YoeB toxin, which is part of the RelE toxin superfamily, and is structurally related to the archaeal RelE monomer (Francuski & Saenger, 2009; Kamada, 2005). The RelE and YoeB toxins from E. coli and two YoeB homologues in Staphylococcus aureus have been shown to inhibit protein synthesis through mRNA cleavage (Grady & Hayes, 2003; Zhang & Inouye, 2009; Yoshizumi et al., 2009; Kamada, 2005). The results of the macromolecular synthesis assays and cell-free protein synthesis assays described herein indicate that Txe significantly inhibits protein synthesis. The results of primer extension analysis indicate that Txe cleaves mRNA, and this activity is probably the mechanism by which Txe inhibits protein synthesis.
The two major mechanisms of action for toxins of TA systems are the inhibition of DNA gyrase and the cleavage of cellular RNA. There is reasonable amino acid sequence similarity between Txe and the YoeB RNase (Grady & Hayes, 2003); however, it has been noted that these two toxins have very different effects on cells, with yoeB expression dramatically reducing viable cell numbers, whereas txe expression halts growth but does not induce death (Grady & Hayes, 2003). Although structural and sequence similarity to other toxins can be informative in discerning mode of action, it can be misleading as well. For example, significant structural similarity exists between the gyrase poison CcdB and the RNase MazF (Buts et al., 2005). Thus, it was not obvious a priori under which category Txe would fall.
With the completion of the experiments described herein, it is now apparent that Txe is an RNase harbouring similarities to YoeB and sharing some, but not all, characteristics with other members of the RelE superfamily. Txe cleaved lpp mRNA three bases downstream of the A of the AUG start codon, similar to what has been observed with the YoeB toxin (Yoshizumi et al., 2009; Zhang & Inouye, 2009). In contrast, the RelE toxin has been shown to cleave mRNA at both sense and stop codons, with a strong preference for cleavage of the stop codon UAG between the second and third bases (Christensen & Gerdes, 2003; Pedersen et al., 2003).
The data presented here indicate that the Txe toxin is an RNase and inhibits protein synthesis, activities which may play a role in the ability of axe–txe to stabilize plasmids in enterococci, including plasmids harbouring the genes for antibiotic resistance (Grady & Hayes, 2003). As axe–txe appears to be widespread in VRE clinical isolates (Moritz & Hergenrother, 2007a; Rosvoll et al., 2010), the artificial activation of Txe could be an effective antibacterial strategy.
Acknowledgments
This work was supported by the National Institutes of Health (2R01-GM068385). E. M. H. and J. J. W. were partially supported by a National Institutes of Health Cell and Molecular Biology Training grant (T32 GM007283). We thank Laura Guest for plasmid sequencing and assembly.
Abbreviations
PNK, polynucleotide kinase
TA, toxin–antitoxin
TCA, trichloroacetic acid
VRE, vancomycin-resistant enterococci
Footnotes
The GenBank/EMBL/DDBJ accession number for the sequence of plasmid pS177 reported in this paper is HQ115078.
Three supplementary figures, showing protein synthesis following IPTG induction of Txe–His6, Axe–His6 and empty pET21a vector, and the effect of Txe–His6 expression in Escherichia coli Rosetta BL21(DE3), are available with the online version of this paper.
References
- Aizenman, E., Engelberg-Kulka, H. & Glaser, G. (1996). An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci U S A 93, 6059–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Anantharaman, V. & Aravind, L. (2003). New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol 4, R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarri, L., Bertuccini, L., Creti, R., Orefici, G., Dicuonzo, G., Gherardi, G., Venditti, M. & Di Rosa, R. (2005). Clonality among Enterococcus faecium clinical isolates. Microb Drug Resist 11, 141–145. [DOI] [PubMed] [Google Scholar]
- Bernard, P. & Couturier, M. (1992). Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol 226, 735–745. [DOI] [PubMed] [Google Scholar]
- Boerlin, P., Burnens, A. P., Frey, J., Kuhnert, P. & Nicolet, J. (2001). Molecular epidemiology and genetic linkage of macrolide and aminoglycoside resistance in Staphylococcus intermedius of canine origin. Vet Microbiol 79, 155–169. [DOI] [PubMed] [Google Scholar]
- Buts, L., Lah, J., Dao-Thi, M. H., Wyns, L. & Loris, R. (2005). Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci 30, 672–679. [DOI] [PubMed] [Google Scholar]
- Camargo, I. L., Del Peloso, P. F., Da Costa Leite, C. F., Goldman, G. H. & Darini, A. L. (2004). Identification of an unusual VanA element in glycopeptide-resistant Enterococcus faecium in Brazil following international transfer of a bone marrow transplant patient. Can J Microbiol 50, 767–770. [DOI] [PubMed] [Google Scholar]
- Christensen, S. K. & Gerdes, K. (2003). RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol 48, 1389–1400. [DOI] [PubMed] [Google Scholar]
- Christensen, S. K., Mikkelsen, M., Pedersen, K. & Gerdes, K. (2001). RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci U S A 98, 14328–14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S. K., Pedersen, K., Hansen, F. G. & Gerdes, K. (2003). Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol 332, 809–819. [DOI] [PubMed] [Google Scholar]
- Christensen-Dalsgaard, M. & Gerdes, K. (2008). Translation affects YoeB and MazF messenger RNA interferase activities by different mechanisms. Nucleic Acids Res 36, 6472–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen-Dalsgaard, M., Overgaard, M., Winther, K. S. & Gerdes, K. (2008). RNA decay by messenger RNA interferases. Methods Enzymol 447, 521–535. [DOI] [PubMed] [Google Scholar]
- DeNap, J. C. & Hergenrother, P. J. (2005). Bacterial death comes full circle: targeting plasmid replication in drug-resistant bacteria. Org Biomol Chem 3, 959–966. [DOI] [PubMed] [Google Scholar]
- Deshpande, L. M., Fritsche, T. R., Moet, G. J., Biedenbach, D. J. & Jones, R. N. (2007). Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn Microbiol Infect Dis 58, 163–170. [DOI] [PubMed] [Google Scholar]
- Dowzicky, M. J. & Park, C. H. (2008). Update on antimicrobial susceptibility rates among gram-negative and Gram-positive organisms in the United States: results from the Tigecycline Evaluation and Surveillance Trial (TEST) 2005 to 2007. Clin Ther 30, 2040–2050. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka, H. & Glaser, G. (1999). Addiction modules and programmed cell death and antideath in bacterial cultures. Annu Rev Microbiol 53, 43–70. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka, H., Sat, B., Reches, M., Amitai, S. & Hazan, R. (2004). Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol 12, 66–71. [DOI] [PubMed] [Google Scholar]
- Fico, S. & Mahillon, J. (2006). TasA-tasB, a new putative toxin-antitoxin (TA) system from Bacillus thuringiensis pGI1 plasmid is a widely distributed composite mazE-doc TA system. BMC Genomics 7, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francuski, D. & Saenger, W. (2009). Crystal structure of the antitoxin-toxin protein complex RelB-RelE from Methanococcus jannaschii. J Mol Biol 393, 898–908. [DOI] [PubMed] [Google Scholar]
- Garcia-Migura, L., Liebana, E., Jensen, L. B., Barnes, S. & Pleydell, E. (2007). A longitudinal study to assess the persistence of vancomycin-resistant Enterococcus faecium (VREF) on an intensive broiler farm in the United Kingdom. FEMS Microbiol Lett 275, 319–325. [DOI] [PubMed] [Google Scholar]
- Gerdes, K., Rasmussen, P. B. & Molin, S. (1986). Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A 83, 3116–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotfredsen, M. & Gerdes, K. (1998). The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol 29, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Grady, R. & Hayes, F. (2003). Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol Microbiol 47, 1419–1432. [DOI] [PubMed] [Google Scholar]
- Guardabassi, L. & Dalsgaard, A. (2004). Occurrence, structure, and mobility of Tn1546-like elements in environmental isolates of vancomycin-resistant enterococci. Appl Environ Microbiol 70, 984–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger, S. & Skoble, J. (1995). Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob Agents Chemother 39, 2446–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger, S., Skoble, J., Discotto, L. F. & Pucci, M. J. (1995). Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob Agents Chemother 39, 362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, F. (2003). Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301, 1496–1499. [DOI] [PubMed] [Google Scholar]
- Hidron, A. I., Edwards, J. R., Patel, J., Horan, T. C., Sievert, D. M., Pollock, D. A. & Fridkin, S. K. (2008). NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol 29, 996–1011. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., Pogliano, J., Helinski, D. R. & Konieczny, I. (2002). ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol 44, 971–979. [DOI] [PubMed] [Google Scholar]
- JMI (2009). Susceptibility of Gram-positive pathogens. http://www.gp-pathogens.com/data/default.cfm.
- Kamada, K. & Hanaoka, F. (2005). Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol Cell 19, 497–510. [DOI] [PubMed] [Google Scholar]
- Kitami, Y. & Hiwada, K. (1999). Vascular disease: molecular biology and gene therapy protocols. In Methods in Molecular Medicine, pp. 133–142. Edited by Baker, A. H.. Bristol. : Bristol Heart Institute, University of Bristol.
- Low, D. E., Keller, N., Barth, A. & Jones, R. N. (2001). Clinical prevalence, antimicrobial susceptibility, and geographic resistance patterns of enterococci: results from the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis 32 (Suppl. 2), S133–S145. [DOI] [PubMed] [Google Scholar]
- Makarova, K. S., Wolf, Y. I. & Koonin, E. V. (2009). Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct 4, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miallau, L., Faller, M., Chiang, J., Arbing, M., Guo, F., Cascio, D. & Eisenberg, D. (2009). Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems. VapBC-5 from Mycobacterium tuberculosis. J Biol Chem 284, 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, E. M. & Hergenrother, P. J. (2007a). Toxin-antitoxin systems are ubiquitous and plasmid-encoded in vancomycin-resistant enterococci. Proc Natl Acad Sci U S A 104, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, E. M. & Hergenrother, P. J. (2007b). The prevalence of plasmids and other mobile genetic elements in clinically important drug-resistant bacteria. In Antimicrobial Resistance in Bacteria, pp. 25–53. Edited by Amabile-Ceuvas, C. F.. Norwich. : Horizon Scientific Press.
- Ogura, T. & Hiraga, S. (1983). Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A 80, 4784–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens, C. D. & Stoessel, K. (2008). Surgical site infections: epidemiology, microbiology and prevention. J Hosp Infect 70 (Suppl. 2), 3–10. [DOI] [PubMed] [Google Scholar]
- Pandey, D. P. & Gerdes, K. (2005). Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33, 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, K., Zavialov, A. V., Pavlov, M. Y., Elf, J., Gerdes, K. & Ehrenberg, M. (2003). The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140. [DOI] [PubMed] [Google Scholar]
- Richards, M. J., Edwards, J. R., Culver, D. H. & Gaynes, R. P. (2000). Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect Control Hosp Epidemiol 21, 510–515. [DOI] [PubMed] [Google Scholar]
- Rosvoll, T. C., Pedersen, T., Sletvold, H., Johnsen, P. J., Sollid, J. E., Simonsen, G. S., Jensen, L. B., Nielsen, K. M. & Sundsfjord, A. (2010). PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTbeta-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol Med Microbiol 58, 254–268. [DOI] [PubMed] [Google Scholar]
- Sader, H. S. & Jones, R. N. (2009). Antimicrobial susceptibility of Gram-positive bacteria isolated from US medical centers: results of the Daptomycin Surveillance Program (2007–2008). Diagn Microbiol Infect Dis 65, 158–162. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D. W. (2001). Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY. : Cold Spring Harbor Laboratory.
- Schwarz, F. V., Perreten, V. & Teuber, M. (2001). Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46, 170–187. [DOI] [PubMed] [Google Scholar]
- Sletvold, H., Johnsen, P. J., Simonsen, G. S., Aasnaes, B., Sundsfjord, A. & Nielsen, K. M. (2007). Comparative DNA analysis of two vanA plasmids from Enterococcus faecium strains isolated from poultry and a poultry farmer in Norway. Antimicrob Agents Chemother 51, 736–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletvold, H., Johnsen, P. J., Hamre, I., Simonsen, G. S., Sundsfjord, A. & Nielsen, K. M. (2008). Complete sequence of Enterococcus faecium pVEF3 and the detection of an omega-epsilon-zeta toxin-antitoxin module and an ABC transporter. Plasmid 60, 75–85. [DOI] [PubMed] [Google Scholar]
- Strong, S. J., Ohta, Y., Litman, G. W. & Amemiya, C. T. (1997). Marked improvement of PAC and BAC cloning is achieved using electroelution of pulsed-field gel-separated partial digests of genomic DNA. Nucleic Acids Res 25, 3959–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen, L. (2002). Molecular interactions of the CcdB poison with its bacterial target, the DNA gyrase. Int J Med Microbiol 291, 537–544. [DOI] [PubMed] [Google Scholar]
- Werner, G., Coque, T. M., Hammerum, A. M., Hope, R., Hryniewicz, W., Johnson, A., Klare, I., Kristinsson, K. G., Leclercq, R. & other authors (2008). Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill 13, 19046. [PubMed] [Google Scholar]
- Williams, J. J. & Hergenrother, P. J. (2008). Exposing plasmids as the Achilles’ heel of drug-resistant bacteria. Curr Opin Chem Biol 12, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizumi, S., Zhang, Y., Yamaguchi, Y., Chen, L., Kreiswirth, B. N. & Inouye, M. (2009). Staphylococcus aureus YoeB homologues inhibit translation initiation. J Bacteriol 191, 5868–5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. & Inouye, M. (2009). The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J Biol Chem 284, 6627–6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Zhang, Y., Zhu, L., Suzuki, M. & Inouye, M. (2004). Interference of mRNA function by sequence-specific endoribonuclease PemK. J Biol Chem 279, 20678–20684. [DOI] [PubMed] [Google Scholar]