Abstract
Vibrio cholerae is a human diarrhoeal pathogen that is a major cause of gastrointestinal disease and death worldwide. Pathogenic V. cholerae strains are characterized by the presence of a Vibrio pathogenicity island (VPI) that encodes virulence factors, including the toxin co-regulated pilus (TCP). TagA is encoded within the VPI and is positively co-regulated with cholera toxin and TCP. TagA is a sequelogue of the StcE mucinase of Escherichia coli O157 : H7. We investigated whether this sequence homology reflected a conserved enzymic substrate profile. TagA exhibited metalloprotease activity toward crude purified mucins, salivary mucin and LS174T goblet cell surface mucin. Like StcE, TagA did not cleave general protease substrates, but unlike StcE, TagA did not cleave the mucin-like serpin C1 esterase inhibitor. Both proteins cleaved the immune cell surface mucin CD43, but TagA demonstrated reduced enzymic efficiency relative to StcE. TagA was expressed and secreted by V. cholerae under ToxR-dependent conditions. A tagA-deficient V. cholerae strain showed no defect in a model of in vitro attachment to the HEp-2 cell line; however, overexpression of a proteolytically inactive mutant, TagA(E433D), caused a significant increase in attachment. The increased attachment was reduced by pretreatment of epithelial monolayers with active TagA. Our results indicate that TagA is a mucinase and suggest that TagA may directly modify host cell surface molecules during V. cholerae infection.
INTRODUCTION
The Gram-negative bacterium Vibrio cholerae causes a secretory diarrhoeal disease in humans that has been the cause of at least seven worldwide pandemics since the 19th century (Sack et al., 2004). Both pathogenic and non-pathogenic V. cholerae are found in the environment, and the vast majority of disease is caused by V. cholerae of the O1 serotype. Disease-causing strains of V. cholerae possess two distinct pathogenic elements, the Vibrio pathogenicity island (VPI) and the cholera toxin phage (ctxΦ) (Kaper et al., 1995). The VPI is a 40 kb pathogenicity island that encodes several potential virulence factors and the toxin co-regulated pilus (TCP) (Karaolis et al., 1998, 2001). TCP is a type IV pilus that is required for colonization of the human intestine (Herrington et al., 1988; Taylor et al., 1987). Cholera toxin (CT) is transcriptionally co-regulated with TCP by the ToxR/S/T regulon, which activates numerous genes of the VPI (Champion et al., 1997; Matson et al., 2007). CT is the major cause of secretory diarrhoea induced by V. cholerae and can recapitulate disease when given alone (Levine et al., 1983). However, vaccine strains lacking CT can cause diarrhoea in human volunteers, suggesting that other virulence factors, including some VPI genes, may play a role in disease (Silva et al., 1996; Harrison et al., 2008; Rui et al., 2010).
One potential VPI-encoded virulence factor is TagA (ToxR-activated gene A), a homologue of the StcE mucinase of enterohaemorrhagic E. coli (EHEC). The small intestinal niche of V. cholerae is coated with a protective layer of thick mucus to which the bacteria preferentially adhere (Yamamoto et al., 1988). Mucinase activity of V. cholerae was first described over 50 years ago as a complex with both neuraminidase and protease activities that were eventually attributed to the 32 kDa haemagglutinin/protease (HA/P) (Finkelstein et al., 1983; Stewart-Tull et al., 1986; Burnet, 1949; Schneider & Parker, 1982). HA/P is an elastase-like general metalloprotease that cleaves a wide variety of substrates, including ovomucin, BSA, casein, elastin and CT (Finkelstein et al., 1983; Häse & Finkelstein, 1990; Booth et al., 1983, 1984). Although HA/P promotes mucin gel penetration (Silva et al., 2003), it is not required for virulence (Silva et al., 2006; Finkelstein et al., 1992; Häse & Finkelstein, 1991). Rather, it provides detachase activity that may allow bacteria to disengage from the intestinal epithelium and disseminate (Finkelstein et al., 1992). Expression profiling suggests that HA/P is active late in infection and regulated oppositely to virulence genes (Zhu et al., 2002). Penetration of the mucus layer is presumably important during initial colonization, and a role for a specific mucinase early in infection remains to be described.
TagA is encoded by the VPI and is part of the ToxR/S/T regulon (Withey & Dirita, 2005). Microarray analysis of stool samples from patients with cholera detected tagA transcript, suggesting that it is expressed during infection (Bina et al., 2003). TagA is a putative 115 kDa secreted lipoprotein that is homologous to StcE (Secreted protease of C1 esterase inhibitor), a metalloprotease secreted by E. coli O157 : H7. StcE is the prototypic member of the SLiMe (StcE-like metalloprotease) family, a group of large proteins of Gram-negative bacteria that share a consensus protease domain with a conserved zinc metalloprotease active site. StcE cleaves specific mucin-type O-glycosylated proteins, including C1 esterase inhibitor (C1-INH), CD43 and mucin 7 (MUC7), and increases the ability of E. coli O157 : H7 to intimately adhere to epithelial cells (Grys et al., 2005; Lathem et al., 2002; Szabady et al., 2009). The StcE homologue of Aeromonas hydrophila contributes to virulence in a mouse peritonitis model (Pillai et al., 2006), and evidence suggests that StcE may be involved in colonization of the rabbit intestine by EHEC (Ho et al., 2008). Here we investigate the hypothesis that TagA, similar to StcE, is a secreted mucinase and contributes to colonization of the intestinal epithelium.
METHODS
Bacterial strains and plasmids.
Bacterial strains were maintained in LB medium with 50 % glycerol at −80 °C. Bacteria were grown under ToxR-inducing conditions at 30 °C in LB medium pH 6.5 (Miller & Mekalanos, 1988). Ampicillin was used at 100 μg ml–1. V. cholerae O1 El tor strain N16961 was obtained from ATCC (Manassas, VA, USA). Classical biotype V. cholerae O395 SmR and JJM43 (V. cholerae O395 SmR toxR43) were kind gifts from R. K. Taylor (Dartmouth College, Hanover, NH, USA). MC4100 λpir containing pKAS32 was a kind gift from V. J. DiRita (University of Michigan, Ann Arbor, MI, USA).
A tagA deletion in V. cholerae O395 SmR was constructed using allelic exchange as described by Skorupski & Taylor (1996). Briefly, DNA flanking tagA was PCR amplified from the VPI (GenBank accession no. AF325733) using the oligos A (5′-CGGGTACCCGGAAGCAGCACCTTCTTGTG-3′) and B (5′-CCACAGTCATCAAAAAAAGTTCTATAGTTTAACTCCTCGTAAACATTTTTCGA-3′) for nt 3355–3988, and oligos C (5′-TCGAAAAATGTTTACGAGGAGTTAAACTATAGAACTTTTTTTGATGACTGTGG-3′) and D (5′-CGGAGCTCGGAATCGCTGTATGCAGGTATC-3′) for nt 6998–8093, using genomic DNA as template and TripleMaster DNA polymerase (Eppendorf). The resultant PCR fragments were then ligated by PCR with primers A and D. The resultant product was ligated into pKAS32, an R6K suicide vector that expresses rpsL (Skorupski & Taylor, 1996), transformed into CC118 λpir (Herrero et al., 1990) and mated into V. cholerae O395 SmR. Transconjugants were isolated on TCBS agar with 250 μg carbenicillin ml–1, recovered on LB agar and counterselected on LB agar with 1 mg streptomycin ml–1. Deletion of tagA (ΔtagA) was confirmed by PCR.
pBAD24 AR (Guzman et al., 1995) was a gift from Charles Cowles (University of Wisconsin, Madison, WI, USA). The entire tagA gene, including the signal sequence, was PCR-amplified from V. cholerae O1 genomic DNA using oligos E (5′-CGGGCTAGCTACGAGGAGTTAGTGGTGG-3′) and F (5′-GCGGCATGCTCTATAGTTTTAATGGTAGGTCATCG-3′) and cloned into pBAD24 using NheI and SphI (NEB) to create pTagA. The proteolytically inactive TagA(E433D) mutant was created by PCR mutagenesis using primer G (5′-CGAATGGTCTCATGATTTGGGACATAACTATGGATTGGGAC-3′) and its complementary oligonucleotide to create pE433D. pTagA and pE433D were confirmed by DNA sequencing. As a control for attachment experiments, pBAD24 was transformed into V. cholerae O395 wt and ΔtagA.
For analysis of TagA expression, V. cholerae O395 wt, ΔtagA or the toxR null mutant (JJM43) were grown under ToxR-inducing conditions overnight (Taylor et al., 1987). Cells were pelleted and proteins in culture supernatants were precipitated with 10 % trichloroacetic acid on ice. Samples were separated by SDS-PAGE and transferred to nitrocellulose. Protein expression was detected with polyclonal antiserum to a TagA peptide (aa 44–57, KKPSRPIIDEKNKG) raised in rabbits (Open Biosystems) and used at 1 : 2000 dilution.
Protein purification.
pTagA and pE433D were transformed into V. cholerae O395 ΔtagA. Bacteria were grown to mid-exponential phase in LB (pH 6.5) with ampicillin, and protein expression was induced with 0.1 % arabinose for 24 h at 30 °C. Culture supernatants were filtered and precipitated with ammonium sulfate (AMS). The 35–55 % AMS pellet was resuspended in Buffer A (20 mM Tris, pH 8.0, 50 mM NaCl). Soluble protein was applied to a Mono Q anion exchange column (GE Healthcare) equilibrated in Buffer A, and protein was eluted with a linear gradient with 20 mM Tris, pH 8.0, 1 M NaCl. Peak fractions were further purified by gel filtration using an S200 16/60 column (GE Healthcare) in 20 mM Tris, pH 8.0, 150 mM NaCl, and glycerol was added to 20 % prior to storage at –20 °C.
Mucinase assays.
Human saliva (10 %), bovine submaxillary mucin (0.04 %, BSM; Sigma) and porcine gastric mucin type II or III (0.04 %, PGM; Sigma) in PBS were treated with 2 μg enzyme or equivalent volume PBS vehicle control overnight in a final volume of 100 μl at room temperature. EDTA (50 mM) was used where indicated to inhibit metalloprotease activity. Samples were separated by electrophoresis through 1 % agarose with 0.1 % SDS at 23 V overnight in TAE buffer. Proteins were transferred to polyvinylidene difluoride (Amersham) and probed with 1 : 10 000 biotinylated wheatgerm agglutinin (WGA; US Biological) followed by horseradish peroxidase (HRP)-conjugated streptavidin (Bio-Rad), and detected by enhanced chemiluminescence (Thermo Fisher). Collection of human saliva samples was in accordance with a protocol approved by the University of Wisconsin Institutional Review Board. For experiments with cell-bound mucins, goblet-like LS174T cells (ATCC) were grown in Eagle's Minimum Essential Medium (Mediatech) with 10 % fetal bovine serum (Atlanta Biologicals). Confluent monolayers in 24-well plates were treated with 5 μg enzyme for 3 h at 37 °C, 5 % CO2. Cells were lysed and samples analysed as above.
Protease assays.
For MUC7 cleavage, 30 μl whole human saliva was incubated with 2 μg enzyme or PBS vehicle control for 90 min at 37 °C. Samples were separated by SDS-PAGE, transferred to nitrocellulose and probed with 1 : 1000 rabbit anti-MUC7 (kind gift from J. G. M. Bolscher, Academic Centre for Dentistry Amsterdam, The Netherlands) and anti-rabbit-HRP. For C1-INH cleavage, 1 μg C1-INH (Comptech) was treated with 1 μg enzyme in a final volume of 30 μl for 3 h at 37 °C. Samples were analysed as above using 1 : 2000 goat anti-C1-INH and anti-goat-HRP. For casein cleavage, 5 or 10 μg enzyme was assayed using the EnzChek green fluorescence protease assay kit (Molecular Probes/Invitrogen), according to the manufacturer's instructions.
Flow cytometry.
The human T-lymphocyte Jurkat cell line was obtained from ATCC and maintained as per the supplier's instructions. Cells were suspended to 5×105 ml–1 in RPMI with 5 % fetal bovine serum and treated with 1 μg enzyme in a final volume of 500 μl for the indicated times at 37 °C, 5 % CO2. Cells were pelleted at 400 g for 5 min and blocked by addition of 0.05 % BSA (Sigma) in Dulbecco's PBS with Ca2+/Mg2+. Samples were stained with phycoerythrin-conjugated anti-CD43 (clone L10; Invitrogen) and analysed by flow cytometry on an LSR II (Becton Dickinson).
Attachment assays.
The HEp-2 human epithelial-like cell line was acquired from ATCC and maintained as per the supplier's instructions. Confluent monolayers (5×105) in 24-well tissue culture plates were used for attachment experiments and washed twice before use. Bacteria were grown to an OD600 of 0.2–0.3, tagA expression was induced with 0.1 % arabinose for 2 h, and bacteria were applied to monolayers for 30 min at 37 °C, 5 % CO2. Non-attached bacteria were removed by three PBS washes, HEp-2 cells were lysed in 1 % Triton X-100, and recovered bacteria were enumerated by serial dilution and plating on LB agar with ampicillin. Percentage attachment was calculated as output c.f.u./input c.f.u.×100 %. Statistical analyses were performed using GraphPad Prism. For immunofluorescence analysis, HEp-2 cells were grown to confluent monolayers on Lab-Tek II chamber slides (Nalge Nunc International) and incubated with V. cholerae as above for 30 min, followed by washing to remove non-attached bacteria. Samples were fixed with 2 % paraformaldehyde and permeabilized with 0.1 % Triton X-100 (Sigma), stained with 1 : 5000 anti-TcpA antibody (a kind gift of V. J. DiRita) followed by 1 : 200 Alexa-488 anti-rabbit IgG and 1 : 50 Alexa-594 phalloidin, and mounted in ProLong Gold antifade reagent with DAPI (Molecular Probes). Coverslips were imaged using a ×100 oil immersion lens on a Zeiss Axioplan IIi epifluorescence microscope.
RESULTS
Identification of TagA as a SLiMe
We previously identified and characterized StcE, a secreted protein of E. coli O157 : H7, that has been identified as the prototype member of family M66 within the merops protease database (http://merops.sanger.ac.uk; Rawlings et al., 2010). Members of this family, which we refer to as the SLiMe family, share a conserved ∼300 aa metalloprotease domain with a predicted zinc-dependent active site sequence of HEXGHXXXGXGH. SLiMe family members are found in several species of the Gammaproteobacteria in the families Vibrionaceae, Enterobacteriaceae, Aeromonadaceae and Shewanellaceae. blast analysis and clustal w alignment confirmed that V. cholerae TagA (RefSeq NP_230468.1) is a homologue of StcE. The two proteins share amino acid sequences with 37 % similarity throughout the total protein and 41 % identity within the central, 300 aa metalloprotease domain. In addition to TagA, V. cholerae encodes another SLiMe, TagA-related protein (TARP; RefSeq NP_232548.1), on chromosome II. Vibrio vulnificus also encodes a TARP homologue on chromosome II, but lacks a tagA gene on chromosome I. Several other pathogenic and environmental vibrios, including Vibrio parahaemolyticus and Vibrio fischeri, do not encode any SLiMes. All currently sequenced V. cholerae isolates encode TARP, whereas only those possessing the VPI encode TagA. Because there is evidence that TagA is expressed during human infection (Bina et al., 2003), we chose to focus our studies solely on the functions of this SLiMe family member in V. cholerae.
TagA is a secreted, ToxR-regulated protein
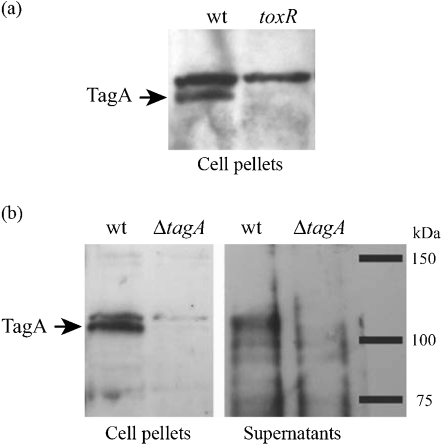
The tagA gene has a ToxT-dependent promoter (Withey & Dirita, 2005) and encodes a 981 aa polypeptide with a predicted molecular mass of 115 kDa. TagA has a leader peptidase II-dependent signal sequence, which predicts that the N-terminal cysteine of the processed protein will be lipidated. Following secretion across the inner membrane, TagA may be secreted extracellularly by a type II secretion system, similar to StcE or the lipoprotein pullulanase (d'Enfert et al., 1987; Stathopoulos et al., 2000), or may be associated with the outer membrane via the attached N-terminal lipid moiety (Bos et al., 2007). We therefore examined TagA expression and secretion in vitro under ToxR-inducing conditions. A polyclonal antibody was produced to a peptide within the N-terminal region of TagA. Immunoblot analysis demonstrated that TagA was expressed by V. cholerae O395 under ToxR-inducing conditions and not in the ToxR null mutant strain JJM43 (Fig. 1a). An isogenic TagA deletion strain (ΔtagA) was created using allelic exchange and used to confirm that the antibody specifically recognized TagA. Owing to a cross-reactive band of a very similar size, samples were electrophoresed until the 75 kDa marker was at the bottom of the acrylamide gel in order to resolve TagA. Natively expressed TagA was detectable in both the supernatant and cell pellet fractions (Fig. 1b). Overexpression of TagA in V. cholerae (see below) resulted in large amounts of TagA in the supernatant, further confirming that it is a secreted protein. The possibility remains that TagA is both secreted and cell-associated, and that some protein may bind back to the cell surface via its lipid moiety.
Fig. 1.
TagA is secreted by V. cholerae in a ToxR-dependent manner. (a) V. cholerae O395 and the isogenic ToxR null mutant strain JJM43 were grown under ToxR-inducing conditions until late-exponential phase. Cells were pelleted, supernatants precipitated with TCA, and samples were analysed by SDS-PAGE followed by immunoblotting with a polyclonal antibody raised to an N-terminal peptide of TagA. (b) Experiments were performed as described for (a), comparing V. cholerae O395 wt with the ΔtagA strain.
TagA is a metal-dependent mucinase
In order to conduct functional protease assays, we attempted to purify TagA. For unknown reasons, the protein was not expressed well in numerous strains of E. coli, with or without a variety of epitope tags. We were able to overexpress TagA with arabinose induction from a tagA recombinant-based pBAD24 vector plasmid in the V. cholerae O395 background. The majority of overexpressed, recombinant TagA was present as a soluble form in the culture supernatant. Ammonium sulfate precipitation of filtered supernatants, followed by ion exchange and subsequent size exclusion chromatography, yielded highly purified TagA protein (data not shown).
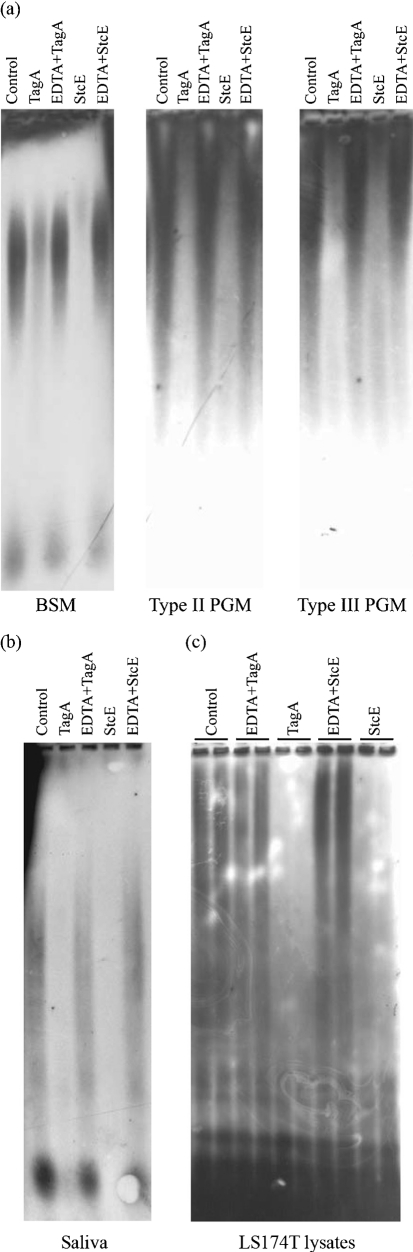
StcE recognizes glycan-induced protein conformations in order to specifically cleave mucin-type glycoproteins. We hypothesize that SLiMe family members share a similar mucin-specific substrate profile. We therefore investigated potential mucinase activity of TagA. Solutions of crude purified BSM or PGM type II or III were treated with TagA. StcE served as a positive control, and EDTA was used to inhibit metalloprotease activity. Mucins were separated by agarose gel electrophoresis and probed with WGA, a lectin that recognizes the N-acetylglucosamine sugar linkage common to many glycoproteins. Mucins run as large, diffuse bands using this method as a result of their heterogeneity in size and glycosylations. Mucin cleavage, detected as clearance of these lectin-reactive bands, was evident in TagA- and StcE-treated samples, and was prevented by incubation with EDTA (Fig. 2a). In addition to reconstituted crude animal mucin preparations, we investigated cleavage of human-derived mucins ex vivo and in vitro. TagA cleaved mucins present in human saliva (Fig. 2b) and mucins that were attached to the LS174T human goblet-like cell line (Fig. 2c).
Fig. 2.
Cleavage of mucins by TagA. (a) Crude purified BSM or PGM type II and III (0.04 % in PBS) were treated with 2 μg TagA or StcE or buffer control overnight at room temperature. EDTA (50 mM) was included as a negative control to inhibit metalloprotease activity where indicated. Samples were separated by agarose gel electrophoresis, transferred to polyvinylidene difluoride and probed with WGA. (b) Saliva (10 %) was treated and probed as described for (a). (c) Differentiated goblet-like LS174T cells were treated with 5 μg enzyme with or without EDTA for 3 h at 37 °C, 5 % CO2. Cells were lysed, and samples were analysed as described for (a).
TagA is not a general protease
StcE uniquely cleaves proteins with mucin-type O-glycosylations (Grys et al., 2006). With the demonstration that TagA cleaved mucins, we sought to confirm that it did not exhibit general proteolytic activity toward non-mucin substrates. Gelatin zymography was performed with purified TagA, using trypsin as a positive control for proteolysis. Like StcE, TagA did not exhibit any proteolytic activity toward gelatin (data not shown). Furthermore, TagA did not cleave hide powder azure and casein, two common substrates used for measuring proteolytic activity (Fig. 2d and data not shown).
Enzymic activities of TagA and StcE are not identical
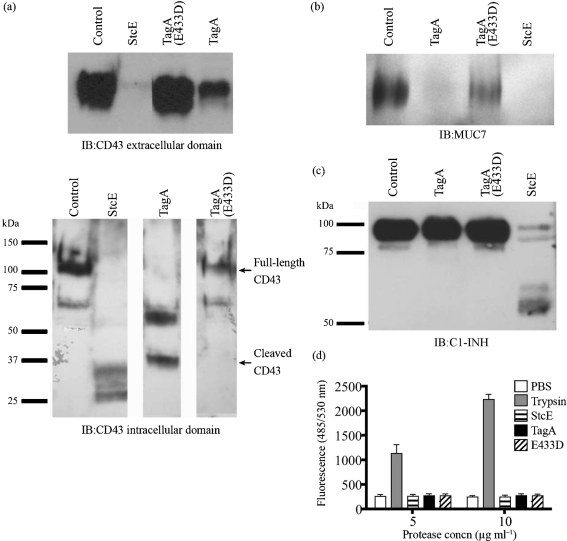
Our data suggest that TagA is a metal-dependent protease that shares a similar mucin substrate profile to StcE. We sought to further delineate the specific substrate profile by examining cleavage of CD43, MUC7 and C1-INH, known substrates of StcE. The cleavage of these substrates may contribute to the role of StcE in EHEC infection (Grys et al., 2005; Lathem et al., 2002; Szabady et al., 2009). As a negative control, we purified a version of TagA with a single point mutation in the active site (E433D), a mutation that was previously demonstrated to inactivate StcE and other zinc metalloproteases (Lathem et al., 2002). CD43 is a large anti-adhesive glycoprotein found on the surface of immune cells. TagA cleaved CD43 from the surface of Jurkat T cells and, similarly to StcE, degraded the O-glycosylated extracellular domain (Fig. 3a). Immunoblotting with an antibody to the CD43 N terminus revealed that TagA cleaved CD43 with less apparent efficiency than StcE (discussed further below). MUC7 is a 250 kDa glycoprotein found in human saliva. Saliva samples were treated with TagA and probed with a MUC7-specific antibody, demonstrating cleavage of MUC7 (Fig. 3b). Cleavage of MUC7 and CD43 was not observed with the TagA(E433D) mutant, confirming that this mutation inactivated the protease active site.
Fig. 3.
Comparisons of TagA and StcE substrate profiles. (a) Intact Jurkat cells (1×106) were treated with 1 μg TagA, TagA(E433D), StcE or PBS control at 37 °C, 5 % CO2 for 30 min. Samples were separated by SDS-PAGE and immunoblotted (IB) for the CD43 extracellular or intracellular domain. (b) Human saliva was treated with control or 2 μg enzyme at 37 °C for 90 min, separated by SDS-PAGE, and immunoblotted for MUC7. (c) Purified C1-INH (1 μg) was treated with control or 1 μg enzyme at 37 °C for 3 h, separated by SDS-PAGE and immunoblotted for C1-INH. (d) Purified enzymes (5 or 10 μg ) were evaluated for cleavage of fluorescent BODIPY-FL casein substrate using the EnzChek protease assay kit. Data are means±sem of two experiments.
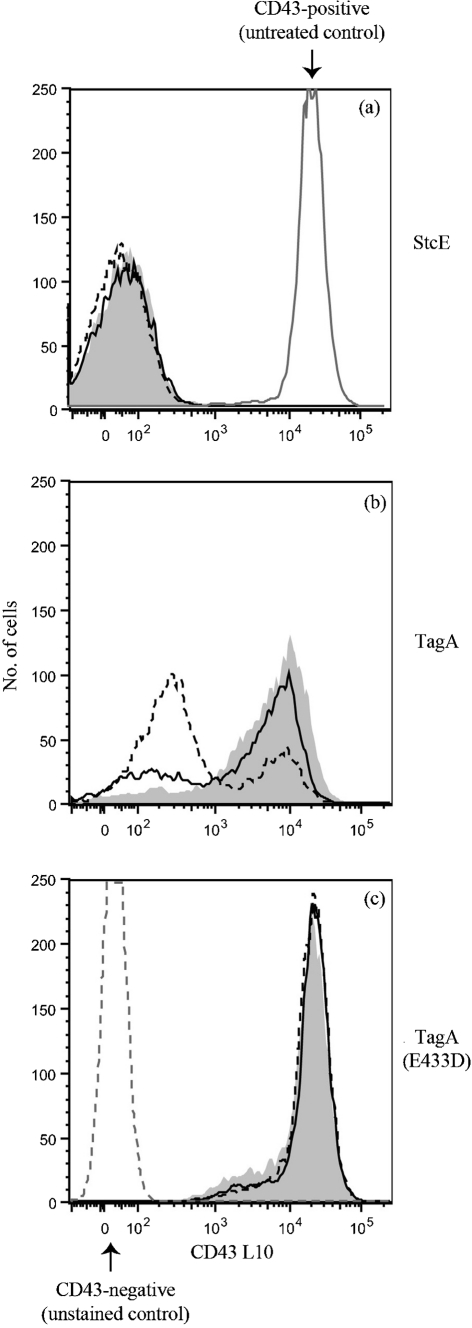
Unlike StcE, TagA was unable to cleave purified C1-INH (Fig. 3c). This finding indicates that despite sharing a conserved metalloprotease domain, the substrate specificities of TagA and StcE are not identical. We examined cleavage efficiency of TagA toward CD43, a substrate that it shares with StcE. Intact Jurkat cells were treated with similar amounts of TagA or StcE for various times, and CD43 on the surface was detected by flow cytometry. StcE cleaved nearly all of the CD43 on the surface after 30 min; in fact, reduction of CD43 on the surface was evident after 5 min of protease treatment (Fig. 4a and data not shown). In contrast, although TagA cleaved some CD43 after 30 min, substantial cleavage was not evident until 1–3 h, and remained incomplete after 3 h (Fig. 4b). TagA(E433D) served as a negative control and did not remove any CD43 from the cell surface (Fig. 4c). Our results indicate that TagA is less efficient at cleaving CD43 than StcE and, combined with the lack of cleavage of C1-INH, suggest that sequence divergence between StcE and TagA might confer differing substrate preferences despite a general similarity in substrate type.
Fig. 4.
TagA and StcE exhibit differing cleavage efficiencies toward CD43. Intact Jurkat cells (5×105) were treated with control or 1 μg enzyme at 37 °C, 5 % CO2 for 30 min (grey shading), 1 h (solid line) or 3 h (dotted line) as indicated. The presence of the CD43 extracellular domain on the cell surface was detected by flow cytometry. (a) StcE, (b) TagA and (c) E433D. Data are from a representative of three independent experiments.
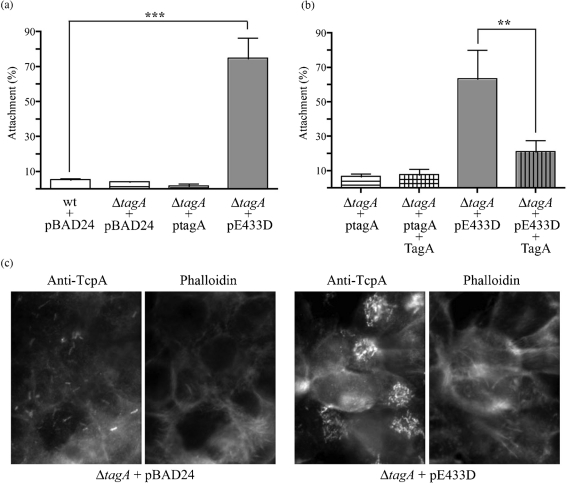
Secretion of a mucinase during infection with V. cholerae could promote intestinal colonization. With the demonstration of TagA mucinase activity, we next evaluated its contribution in an in vitro model of V. cholerae attachment to human epithelial-like cells (HEp-2) that has been used previously to identify V. cholerae adherence factors (Sperandio et al., 1995). Wild-type V. cholerae O395 and the isogenic ΔtagA mutant strain were allowed to attach to HEp-2 monolayers, washed and the attached bacteria enumerated. Of the input V. cholerae cells, 3–15 % adhered to the HEp-2 cells, consistent with previously reported attachment figures in this model (Sperandio et al., 1995). The loss of TagA expression had no significant effect on V. cholerae attachment. However, overexpression of proteolytically inactive TagA(E433D), but not TagA, resulted in a significant 70 % increase in attachment of V. cholerae to the cultured epithelial cells (Fig. 5a). We did not observe any qualitative differences in protein expression or bacterial autoaggregation between the TagA and TagA(E433D) overexpression strains. Immunofluorescence microscopy confirmed the increased attachment of the TagA(E433D) ovexpression strain (Fig. 5c). A possible explanation for the differential attachment observed is that TagA normally binds and cleaves an unidentified epithelial cell surface protein, but in the absence of proteolysis there remains a receptor–ligand interaction that results in increased attachment by the TagA(E433D) overexpression strain. To test this hypothesis, we pretreated HEp-2 cells with purified active TagA. This treatment reduced the hyper-attachment of the TagA(E433D) overexpression strain (Fig. 5b), suggesting that active TagA cleaved the binding partner of the TagA(E433D) mutant protein and eliminated this receptor–ligand interaction. The addition of purified TagA had no effect on the attachment of V. cholerae ΔtagA cells not overexpressing the TagA(E433D) mutant. These assays were repeated in the mucus-producing cell line LS174T with similar results (data not shown). We were unable to identify binding partner(s) for the TagA(E433D) mutant using immunoprecipitation and mass spectrometry, possibly as a result of interference by protein glycosylation with mass spectrometric identification.
Fig. 5.
Attachment of V. cholerae TagA mutants to HEp-2 cells. (a) Exponential-phase V. cholerae O395, ΔtagA or ΔtagA complemented with pBAD24 encoding TagA (ptagA) or TagA(E433D) (pE433D) were incubated with HEp-2 monolayers for 30 min at 37 °C, 5 % CO2. wt and ΔtagA contained pBAD24 alone as a control. Monolayers were washed and attached bacteria were enumerated by serial dilution and plating. Percentage attachment was calculated as output c.f.u./input c.f.u.×100. Data are from a representative of at least three independent experiments, and are presented as means±sd (n=3). Statistical analysis was performed using GraphPad Prism and one-way ANOVA with the Bonferroni post test; ***, P<0.001. (b) Experiments were performed as described for (a), except that HEp-2 monolayers were first treated with 10 μg purified TagA for 2 h and washed prior to adding bacteria. Data were analysed as above; **, P<0.01. (c) HEp-2 monolayers were grown on coverslips and attachment assays were performed as described. Samples were fixed and stained for immunofluorescence with rabbit anti-TcpA to stain V. cholerae, and with phalloidin to label HEp-2 actin.
DISCUSSION
We have demonstrated that TagA, a homologue of E. coli O157 : H7 StcE and a member of the SLiMe family, is expressed and secreted by V. cholerae under ToxR-dependent conditions. Purified TagA cleaves CD43, MUC7, crude BSM and PGM, and LS174T goblet cell surface mucin, but does not digest C1-INH, hide azure powder, casein or gelatin. Substrate cleavage is prevented by EDTA or by a single glutamic acid to aspartic acid substitution in the consensus metalloprotease active site. We have thus shown that TagA is a metalloprotease similar to StcE, which cleaves specific mucins but does not exhibit general proteolysis. TagA activity stands in clear contrast to the classic V. cholerae mucinase, HA/P. HA/P is a broad-spectrum protease that cleaves numerous non-mucin substrates (Finkelstein et al., 1983; Häse & Finkelstein, 1990). HA/P is also regulated by quorum sensing through HapR and is repressed under conditions that induce the ToxR virulence regulon (Zhu et al., 2002), whereas TagA is activated by ToxR (Taylor et al., 1987).
TagA is homologous to StcE, but exhibits a distinct proteolytic activity and is therefore a unique member of the StcE-like metalloprotease family. Aeromonas hydrophila TagA is the only other SLiMe protein whose activity has been characterized. This protein cleaves C1-INH to the same-sized product as the StcE-cleaved product (Pillai et al., 2006), whereas TagA failed to cleave C1-INH. TagA also demonstrated reduced cleavage efficiency toward CD43, a substrate that it shares with StcE. A. hydrophila TagA and EHEC StcE share 68 % sequence similarity, as compared to 35 % for V. cholerae TagA and StcE. This suggests that divergent sequence elements of these proteins might dictate specificity for different substrates.
Our results further suggest that the homologous members of the SLiMe family are restricted to proteolytic cleavage of substrates within the general class of mucin-like O-linked glycoproteins. TagA did not cleave gelatin, which is non-glycosylated, or a fluorogenic casein substrate, which has non-mucin O-glycosylations. Previous research suggests that StcE recognizes an extended random coil structure in the protein backbone that is induced by dense O-glycan attachments, a structure common to mucins and mucin-like glycoproteins (L. Walters & others, unpublished data). We have been unable to identify specific conserved amino acid sequences or glycan linkages that confer recognition and cleavage by StcE. If TagA also recognizes the extended random coil structure, why does StcE cleave C1-INH and TagA does not? We hypothesize that among SLiMe family members with different enzymic efficiencies, their ability to cleave may depend on the length and glycosylation density of the mucin-like region within the substrate. C1-INH has a relatively short mucin-like region that is cleaved by StcE (Lathem et al., 2004), whereas nearly the entire 250 aa extracellular domain of CD43 is O-glycosylated. The intestinal mucins possess variable numbers of tandem O-glycosylated repeats and can form polymers up to 1 MDa in size (Thornton & Sheehan, 2004). It appears that the more efficient StcE mucinase can cleave proteins such as C1-INH with relatively short mucin-like regions, whereas less efficient enzymes like TagA require more extensive mucin-like regions. We have shown that StcE is more efficient at cleaving MUC7 than C1-INH, supporting the hypothesis that the size of the mucin region may contribute to protease activity (Grys et al., 2006). The structural and sequence determinants of enzyme efficiency and specificity of the SLiMe proteins remain to be investigated. Determination of crystal structures and synthesis of recombinant chimeric enzymes could be useful tools to investigate these questions.
Many of the bacteria that encode SLiMes cause disease in mucin-rich environments such as the human intestine, or fish skin in the case of A. hydrophila, and the SLiMe mucinase activity may be useful in promoting bacterial colonization and disease in their respective hosts (Austin et al., 2005; Beaz Hidalgo et al., 2008; Thompson et al., 2004, 2006). A. hydrophila TagA promotes virulence in a mouse peritonitis model (Pillai et al., 2006), and StcE promotes the intimate attachment of E. coli O157 : H7 required for human colonization (Grys et al., 2005). TagA is co-expressed with known virulence factors of V. cholerae and is expressed in vivo during human infection (Bina et al., 2003). V. cholerae colonization of the human intestine occurs in the context of a mucus layer. Although TCP is thought to promote attachment by causing bacterial self-aggregation, a direct receptor–ligand interaction with host cells has not been demonstrated for V. cholerae. We hypothesized that TagA may contribute to V. cholerae attachment by clearing the mucus layer because TagA demonstrably cleaves crude mucin preparations and the surface mucus of goblet-like intestinal cells. However, we were unable to demonstrate in vitro a defect in attachment by bacteria lacking tagA. The attachment of V. cholerae to HEp-2 cells in culture may have insufficient sensitivity to measure the effects of mucin cleavage in an in vivo setting, a more dynamic environment accompanied by peristalsis of the small intestine. Preliminary attachment experiments with the mucus-producing LS174T cell line yielded similar results, further suggesting that more complex assays might be required to fully understand the role of TagA in vivo. StcE does not affect overall adherence of E. coli O157 : H7 to epithelial cells, but specifically increases the ability of the bacteria to attach intimately and form pedestals (Grys et al., 2005).
TagA may play a role in attachment not revealed by our assay, or it may be important for other aspects of V. cholerae colonization and disease. The observation that overexpression of a non-proteolytic mutant of TagA leads to a large increase in V. cholerae attachment suggests that TagA can mediate an interaction with the host cell surface. The location of the tagA gene within the VPI and the fact that TagA is positively co-regulated with TCP and other ToxR targets provide circumstantial evidence that TagA may play a role in V. cholerae disease. The possibility remains that TagA may act similarly to the HA/P mucinase in promoting detachment of V. cholerae cells, therefore aiding transmission to the environment. Alternatively, TagA may modulate the immune response to V. cholerae by interacting with CD43 or other immune cell surface mucins, as we have recently demonstrated for StcE (Szabady et al., 2009). Another possibility is that TagA plays a role promoting survival and growth in the non-human environment for this pathogen. Such a role is a current area of investigation in our laboratory.
Acknowledgments
The authors would like to thank Victor J. DiRita for materials and advice and Joseph Ebinger for experimental assistance. This study was supported by NIH NRSA AI55397 to R. L. S. and NIH AI051735 to R. A. W.
Abbreviations
BSM, bovine submaxillary mucin
C1-INH, C1 esterase inhibitor
CT, cholera toxin
EHEC, enterohaemorrhagic E. coli
HA/P, V. cholerae haemagglutinin/protease
MUC7, mucin 7
PGM, porcine gastric mucin
SLiMe, StcE-like metalloprotease
StcE, secreted protease of C1 esterase inhibitor
TagA, ToxR-activated gene A
TCP, toxin co-regulated pilus
VPI, Vibrio pathogenicity island
WGA, wheatgerm agglutinin
References
- Austin, B., Austin, D., Sutherland, R., Thompson, F. & Swings, J. (2005). Pathogenicity of vibrios to rainbow trout (Oncorhynchus mykiss, Walbaum) and Artemia nauplii. Environ Microbiol 7, 1488–1495. [DOI] [PubMed] [Google Scholar]
- Beaz Hidalgo, R., Cleenwerck, I., Balboa, S., De Wachter, M., Thompson, F. L., Swings, J., De Vos, P. & Romalde, J. L. (2008). Diversity of Vibrios associated with reared clams in Galicia (NW Spain). Syst Appl Microbiol 31, 215–222. [DOI] [PubMed] [Google Scholar]
- Bina, J., Zhu, J., Dziejman, M., Faruque, S., Calderwood, S. & Mekalanos, J. (2003). ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc Natl Acad Sci U S A 100, 2801–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth, B. A., Boesman-Finkelstein, M. & Finkelstein, R. A. (1983). Vibrio cholerae soluble hemagglutinin/protease is a metalloenzyme. Infect Immun 42, 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth, B. A., Boesman-Finkelstein, M. & Finkelstein, R. A. (1984). Vibrio cholerae hemagglutinin/protease nicks cholera enterotoxin. Infect Immun 45, 558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos, M. P., Robert, V. & Tommassen, J. (2007). Biogenesis of the Gram-negative bacterial outer membrane. Annu Rev Microbiol 61, 191–214. [DOI] [PubMed] [Google Scholar]
- Burnet, F. M. (1949). Ovomucin as a substrate for the mucinolytic enzymes of V cholerae filtrates. Aust J Exp Biol Med Sci 27, 245–252. [DOI] [PubMed] [Google Scholar]
- Champion, G. A., Neely, M. N., Brennan, M. A. & DiRita, V. J. (1997). A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol Microbiol 23, 323–331. [DOI] [PubMed] [Google Scholar]
- d'Enfert, C., Chapon, C. & Pugsley, A. P. (1987). Export and secretion of the lipoprotein pullulanase by Klebsiella pneumoniae. Mol Microbiol 1, 107–116. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R. A., Boesman-Finkelstein, M. & Holt, P. (1983). Vibrio cholerae hemagglutinin/lectin/protease hydrolyzes fibronectin and ovomucin: F.M. Burnet revisited. Proc Natl Acad Sci U S A 80, 1092–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R. A., Boesman-Finkelstein, M., Chang, Y. & Häse, C. C. (1992). Vibrio cholerae hemagglutinin/protease, colonial variation, virulence, and detachment. Infect Immun 60, 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grys, T. E., Siegel, M. B., Lathem, W. W. & Welch, R. A. (2005). The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect Immun 73, 1295–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grys, T. E., Walters, L. L. & Welch, R. A. (2006). Characterization of the StcE protease activity of Escherichia coli O157:H7. J Bacteriol 188, 4646–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, L. M., Rallabhandi, P., Michalski, J., Zhou, X., Steyert, S. R., Vogel, S. N. & Kaper, J. B. (2008). Vibrio cholerae flagellins induce Toll-like receptor 5-mediated interleukin-8 production through mitogen-activated protein kinase and NF-kappaB activation. Infect Immun 76, 5524–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häse, C. C. & Finkelstein, R. A. (1990). Comparison of the Vibrio cholerae hemagglutinin/protease and the Pseudomonas aeruginosa elastase. Infect Immun 58, 4011–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häse, C. C. & Finkelstein, R. A. (1991). Cloning and nucleotide sequence of the Vibrio cholerae hemagglutinin/protease (HA/protease) gene and construction of an HA/protease-negative strain. J Bacteriol 173, 3311–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero, M., de Lorenzo, V. & Timmis, K. N. (1990). Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J Bacteriol 172, 6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington, D. A., Hall, R. H., Losonsky, G., Mekalanos, J. J., Taylor, R. K. & Levine, M. M. (1988). Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med 168, 1487–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, T. D., Davis, B. M., Ritchie, J. M. & Waldor, M. K. (2008). Type 2 secretion promotes enterohemorrhagic Escherichia coli adherence and intestinal colonization. Infect Immun 76, 1858–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper, J. B., Morris, J. G. & Levine, M. M. (1995). Cholera. Clin Microbiol Rev 8, 48–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis, D. K., Johnson, J. A., Bailey, C. C., Boedeker, E. C., Kaper, J. B. & Reeves, P. R. (1998). A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci U S A 95, 3134–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis, D. K., Lan, R., Kaper, J. B. & Reeves, P. R. (2001). Comparison of Vibrio cholerae pathogenicity islands in sixth and seventh pandemic strains. Infect Immun 69, 1947–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathem, W. W., Grys, T. E., Witowski, S. E., Torres, A. G., Kaper, J. B., Tarr, P. I. & Welch, R. A. (2002). StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol Microbiol 45, 277–288. [DOI] [PubMed] [Google Scholar]
- Lathem, W. W., Bergsbaken, T. & Welch, R. A. (2004). Potentiation of C1 esterase inhibitor by StcE, a metalloprotease secreted by Escherichia coli O157:H7. J Exp Med 199, 1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, M. M., Kaper, J. B., Black, R. E. & Clements, M. L. (1983). New knowledge on pathogenesis of bacterial enteric infections as applied to vaccine development. Microbiol Rev 47, 510–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson, J. S., Withey, J. H. & DiRita, V. J. (2007). Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect Immun 75, 5542–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, V. L. & Mekalanos, J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170, 2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, L., Sha, J., Erova, T. E., Fadl, A. A., Khajanchi, B. K. & Chopra, A. K. (2006). Molecular and functional characterization of a ToxR-regulated lipoprotein from a clinical isolate of Aeromonas hydrophila. Infect Immun 74, 3742–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings, N. D., Barrett, A. J. & Bateman, A. (2010). merops: the peptidase database. Nucleic Acids Res 38, D227–D233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui, H., Ritchie, J. M., Bronson, R. T., Mekalanos, J. J., Zhang, Y. & Waldor, M. K. (2010). Reactogenicity of live-attenuated Vibrio cholerae vaccines is dependent on flagellins. Proc Natl Acad Sci U S A 107, 4359–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sack, D. A., Sack, R. B., Nair, G. B. & Siddique, A. K. (2004). Cholera. Lancet 363, 223–233. [DOI] [PubMed] [Google Scholar]
- Schneider, D. R. & Parker, C. D. (1982). Purification and characterization of the mucinase of Vibrio cholerae. J Infect Dis 145, 474–482. [DOI] [PubMed] [Google Scholar]
- Silva, T. M., Schleupner, M. A., Tacket, C. O., Steiner, T. S., Kaper, J. B., Edelman, R. & Guerrant, R. (1996). New evidence for an inflammatory component in diarrhea caused by selected new, live attenuated cholera vaccines and by El Tor and Q139 Vibrio cholerae. Infect Immun 64, 2362–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, A. J., Pham, K. & Benitez, J. A. (2003). Haemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology 149, 1883–1891. [DOI] [PubMed] [Google Scholar]
- Silva, A. J., Leitch, G. J., Camilli, A. & Benitez, J. A. (2006). Contribution of hemagglutinin/protease and motility to the pathogenesis of El Tor biotype cholera. Infect Immun 74, 2072–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorupski, K. & Taylor, R. K. (1996). Positive selection vectors for allelic exchange. Gene 169, 47–52. [DOI] [PubMed] [Google Scholar]
- Sperandio, V., Girón, J. A., Silveira, W. D. & Kaper, J. B. (1995). The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect Immun 63, 4433–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos, C., Hendrixson, D. R., Thanassi, D. G., Hultgren, S. J., St Geme, J. W., III & Curtiss, R., III (2000). Secretion of virulence determinants by the general secretory pathway in Gram-negative pathogens: an evolving story. Microbes Infect 2, 1061–1072. [DOI] [PubMed] [Google Scholar]
- Stewart-Tull, D. E., Ollar, R. A. & Scobie, T. S. (1986). Studies on the Vibrio cholerae mucinase complex. I. Enzymic activities associated with the complex. J Med Microbiol 22, 325–333. [DOI] [PubMed] [Google Scholar]
- Szabady, R. L., Lokuta, M. A., Walters, K. B., Huttenlocher, A. & Welch, R. A. (2009). Modulation of neutrophil function by a secreted mucinase of Escherichia coli O157:H7. PLoS Pathog 5, e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, R. K., Miller, V. L., Furlong, D. B. & Mekalanos, J. J. (1987). Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A 84, 2833–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, F. L., Iida, T. & Swings, J. (2004). Biodiversity of vibrios. Microbiol Mol Biol Rev 68, 403–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, F. L. & Klose, K. E., AVIB Group (2006). Vibrio2005: the First International Conference on the Biology of Vibrios. J Bacteriol 188, 4592–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, D. J. & Sheehan, J. K. (2004). From mucins to mucus: toward a more coherent understanding of this essential barrier. Proc Am Thorac Soc 1, 54–61. [DOI] [PubMed] [Google Scholar]
- Withey, J. H. & Dirita, V. J. (2005). Vibrio cholerae ToxT independently activates the divergently transcribed aldA and tagA genes. J Bacteriol 187, 7890–7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T., Kamano, T., Uchimura, M., Iwanaga, M. & Yokota, T. (1988). Vibrio cholerae O1 adherence to villi and lymphoid follicle epithelium: in vitro model using formalin-treated human small intestine and correlation between adherence and cell-associated hemagglutinin levels. Infect Immun 56, 3241–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Miller, M. B., Vance, R. E., Dziejman, M., Bassler, B. L. & Mekalanos, J. J. (2002). Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A 99, 3129–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]