Summary
TGF-β signaling plays a critical role in cartilage and spine tissue development at embryonic stage but its role in postnatal intervertebral disc (IVD) tissue growth and maintenance remain poorly understood. In the present studies, we have deleted the Tgfbr2 gene in inner annulus fibrosus cells of the disc tissue and surrounding growth plate chondrocytes using Col2a1-CreERT2 transgenic mice. We found that TGF-β signaling is required for normal growth plate cartilage and endplate cartilage growth at postnatal stage. The expression of Mmp13 gene is significantly up-regulated in primary disc cells of Tgfbr2 conditional knockout mice. Deletion of the Mmp13 gene under Tgfbr2 null background completely reverses the abnormal disc phenotype found in Tgfbr2 knockout mice.
Keywords: TGF-β, Col2a1-CreERT2, Intervertebral disc, Conditional knockout, MMP-13
Introduction
Low back pain, which is often related to intervertebral disc degeneration, affects approximately 80% of the aging population and causes a significant socio-economic burden [1]. In an autopsy study, 97% of individuals age 50 years or older showed disc degeneration [2]. The intervertebral disc (IVD) is a specialized tissue that permits rotation as well as flexion and extension of the human spine. It consists of an annulus fibrosus that encloses the nucleus pulposus, a highly viscous gel-like tissue that produces aggrecan and other proteoglycans, providing the disc with its critical water-retaining characteristics. The superior and inferior boundaries of the IVD are formed by the growth plate and cartilage endplate [3]. Genetic mouse models are important tools to target specific genes in specific cell populations such as IVD and surrounding cells at different developmental and postnatal stages. Since the cells of annulus fibrosus, nucleus pulposus, growth plate cartilage, and endplate tissues express type II collagen [4], we have generated Col2a1-CreERT2 transgenic mice [5, 6]. We bred these transgenic mice with Rosa26 reporter mice and analyzed the cell population(s) targeted in Col2a1-CreERT2 transgenic mice. In addition, we investigated the Cre-recombination efficiency of the targeted disc cells. Taking advantage of the inducible manner of Cre expression in Col2a1-CreERT2 transgenic mice, we also performed a time-course experiment to investigate the fate of targeted disc cells.
Similar to long bone, the development of IVD tissue starts from mesenchymal cell condensation at around E10.5 in mice. At approximately E12.5, the condensed mesenchyme forms the IVD, and the loose mesenchyme undergoes chondrogenesis to form the vertebral body. Notochord cells derived from the mesoderm are then removed from the vertebral region and expand into the IVD region to form the nucleus pulposus. The region around the nucleus pulposus forms the annulus fibrosus. The inner annulus fibrosus contains hyaline cartilage and the outer annulus fibrosus is more fibrous. The nucleus pulposus is formed in the middle of the IVD region at E15.5–E17.5 stage in mice [7]. It has been demonstrated that defects in the development of IVD tissue was detected in Tgfbr2 cKO embryos targeted by Col2a1-Cre transgenic mice (hereafter referred to as Tgfbr2Col2). The development of annulus fibrosus and the expansion of nucleus pulposus tissue to the IVD region were delayed in E15.5 to E17.5 Tgfbr2Col2 embryos [7]. In addition, it has also been reported that spinal column of E18.5 Tgfbr2Col2 embryos show the neural arch defect from a dorsal-lateral aspect in addition to the craniofacial defects [8]. The TGFβ-2 knockout phenotype has no overlap with the phenotypes observed in the TGFβ-1 and TGFβ-3 null mice [9–12].
Although TGF-β signaling plays a critical role in axial skeletal development [7, 8, 13], its roles in postnatal growth and maintenance of disc tissue remain poorly understood. In the present studies, we have bred Col2a1-CreERT2 transgenic mice with Tgfbr2fx/fx mice [13] and generated inducible Tgfbr2 cKO mice (hereafter referred to as Tgfbr2Col2ER). We specifically deleted the Tgfbr2 gene in inner annulus fibrosus cells and growth plate chondrocytes at the postnatal stage. Defects in disc tissue growth and maintenance were observed in Tgfbr2Col2ER mice. Our studies demonstrate that Col2a1-CreERT2 transgenic mice are an important tool to target cells of IVD and surrounding tissues and TGF-β signaling plays an essential role in the development of IVD and surrounding tissues.
Materials and Methods
Characterization of Col2a1-CreERT2 transgenic mice
Col2a1-CreERT2 transgenic mice were previously generated in our laboratory [5, 6] and these mice were crossed with Rosa26 reporter mice in which the expression of Escherichia coli β-galactosidase can be induced by Cre-mediated recombination [14]. Offspring were genotyped by polymerase chain reaction (PCR) using specific primers for detecting Cre insertion and R26R alleles. The sequences of PCR primers for genotyping Col2a1-CreERT2 mice are: 5′-CCTGGAAAATGCTTCTGTCCGTTTGCC-3′ (forward primer) and 5′-GAGTTGATAGCTGGCTGGTGGCAGAG-3′ (reverse primer) and the size of the PCR product is 600-bp. The sequences of PCR primers for genotyping Rosa26 reporter mice are: R1295: 5′-GCGAAGAGTTTGTCCTCAACC-3′; R523: 5′-GGAGCGGGAGAAATGGATATG-3′ and R26F2: 5-AAAGTCGCTCTGAGTTGTTAT-3′. The 600-bp PCR product was detected in wild-type mice and the 325-bp PCR product was detected in homozygous Rosa26 mice. Both 600- and 325-bp PCR products were detected in heterozygous Rosa26 reporter mice.
Tamoxifen was administered to 2-week-old Col2a1-CreERT2;R26R mice (1 mg/10g body weight, x5 days, i.p. injection). Mice were sacrificed 1, 2, 3, and 4 months after tamoxifen injection. The disc tissues were harvested and fixed in 0.2% glutaraldehyde, decalcified and processed for frozen sectioning followed by X-Gal staining. Nuclear Fast Red staining was performed as a counter stain. The Cre recombination efficiency was determined by counting the lacZ positive cells divided by the total number of cells in growth plate, cartilage endplate, inner and outer annulus fibrosus and nucleus pulposus regions (n=4).
Generation of Tgfbr2 conditional knockout mice
Col2a1-CreERT2 transgenic mice were bred with Tgfbr2fx/fx mice (obtained from Jackson lab) [15]. Tamoxifen was administered to P1 pups (1 mg/10g body weight, single dose, i.p. injection). Mice were sacrificed when they were 2 weeks old. Tamoxifen was also administered to P14 Tgfbr2Col2ER mice (1 mg/10g body weight, x 5 days, i.p. injection). Mice were sacrificed at 3 months old.
Histological analysis
The disc tissues were dissected from Col2a1-CreERT2;R26R mice, Tgfbr2Col2ER mice mice, Tgfbr2Col2ER;Mmp13Col2ER mice, and their corresponding Cre-negative control mice. Samples were fixed in 10% formalin, decalcified in 14% EDTA, and embedded in paraffin. Serial sections were taken at 3 levels with 15 μm apart within the midsagittal region of the intervetebral bodies. Sections were cut at 3 μm thickness. The sections were stained with Alcian blue/H&E (AB/HE) and Safranin O/Fast green (SO/FG).
Disc cells culture and real-time PCR analysis
14-day-old mice were genotyped and then sacrificed. Primary mouse disc cells were isolated from the intervertebral disc tissues of Cre-negative and Tgfbr2-flox mice through digestion with 1 mg/ml Collagenase A (Roche). The cells were then counted and plated in DMEM containing 10% FBS. 12 hours after the seeding, the cells were infected with Ad-Cre (Vector Biolabs) or an Ad-GFP (Vector Biolabs) (control), at 100 MOI for 24 hours. The cells were allowed to recover for 48 hours before harvest. The cells were then subjected to RNA isolation.
Total RNA was prepared using PureLink™ RNA Mini Kit (Invitrogen) according to the manufacturer’s protocol. 1 μg of total RNA was used to synthesize cDNA using iScripts cDNA Synthesis Kit (Bio-Rad). Real-time PCR amplifications were performed using gene specific primers and SYBR green real-time PCR kit. The primers and sequences used for the real-time PCR analysis are shown in Table 1. The thermal cycling conditions were as follows: an initial denaturation at 95°C for 3 min, followed by 40 cycles of 20 seconds of denaturing at 95°C, 20 seconds of annealing at 58°C, and 20 seconds of extension at 72°C. The expression levels of the target genes were normalized to that of β-actin in cDNA samples.
Table 1.
Primers and sequences for real-time PCR analysis
| Upper Primers | Lower Primers | |
|---|---|---|
| Tgfbr2 | 5′-AGATGGCTCGCTGAACACTACCAA-3′ | 5′-AGAATCCTGCTGCCTCTGGTCTTT-3′ |
| Col2a1 | 5′-TGGTCCTCT GGGCATCTCAGG C-3′ | 5′-GGTGAACCTGCTGTTGCCCTC A-3′ |
| Col10a1 | 5′-ACCCCAAGGACCTAAAGGAA-3 | 5′-CCCCAGGATACCCTGTTTTT-3′ |
| Mmp13 | 5′-TTTGAGAACACGGGGAAGA-3′ | 5′-ACTTTGTTGCCAATTCCAGG-3′ |
| Adamts4 | 5′-GGCAGATGACAAGATGGCAGCATT-3′ | 5′-AGACGAGTCACCACCAAGTTGACA-3′ |
| Adamts5 | 5′-CCAAATGCACTTCAGCCACGATCA-3′ | 5′-AATGTCAAGTTGCACTGCTGGGTG-3′ |
| Ptc1 | 5′-TTCTGCTGCCTGTCCTCTTA-3′ | 5′-GCAAACCGGACGACACTT-3′ |
| Pthr1 | 5′-ACTCCTTCCAGGGATTTTTTGTT-3′ | 5′-GAAGTCCAATGCCAGTGTCCA-3′ |
| β-Actin | 5′-TCAGGTTACTGGTTCGGTCTG-3′ | 5′-ACCAGAGGCATACAGGGACAG-3′ |
Results
Administration of tamoxifen to Col2a1-CreERT2;R26R mice induced efficient Cre-recombination in IVD cells up to 4 months after tamoxifen induction (Fig. 1A). High Cre-recombination efficiency was observed in growth plate chondrocytes. It reached 80% in 1-month-old mice and remained high (70%) in 4-month-old mice (Table 2). These results suggest that the cell turnover rates are very low in growth plate chondrocytes of the disc surrounding tissue. Although Col2a1-CreERT2 mice can target cartilage endplate cells, the efficiency is relatively low. While only 10% of endplate cartilage cells were X-Gal positive by 1 month after tamoxifen induction, the efficiency increased to 37% by 4 months after tamoxifen induction (Fig. 1B and Table 1). Col2a1-CreERT2 mice can also efficiently target inner annulus fibrosus cells, with 73 to 81% efficiency in mice of different ages (Fig. 1B and Table 1). In contrast, the targeting efficiency for outer annulus fibrosus cells was very poor. These results are expected since it is well documented that inner annulus fibrosus cells are enriched in type II collagen while outer annulus fibrosus cells have a high type I collagen content [16]. We did not observe X-Gal positive cells in the nucleus pulposus region (Fig. 1A). This is probably because of low type II collagen expression in nucleus pulposus cells. Immunohistochemistry (IHC) showed that type II collagen expression was undetectable in nucleus pulposus cells (data not shown). Another possible reason is that tamoxifen may not be able to reach the nucleus pulposus due to the lack of a blood supply to the nucleus pulposus region. However, this may not be the case because tamoxifen can induce Cre-recombination in nucleus pulposus cells in aggrecan-CreERT2 transgenic mice [17]. In this study, we provide evidence that Col2a1-CreERT2 transgenic mice can efficiently target growth plate chondrocytes and inner annulus fibrosus cells in the IVD, suggesting that these mice are an important tool to selectively target floxed genes specifically expressed in inner annulus fibrosus cells in the disc tissue and surrounding growth plate chondrocytes.
Fig. 1.


X-Gal staining of Col2a1-CreERT2;R26R mice. (A) Two-week-old Col2a1-CreERT2;R26R double transgenic mice were injected with tamoxifen (1 mg/10g body weight, x5 days, i.p. injection). Mice were sacrificed 1, 2, 3, and 4 months (a–d) after injections were completed and disc samples were fixed, decalcified and processed for frozen section preparation. X-Gal staining was performed and sections (L3/L4) were then counterstained with nuclear fast red. Col2a1-CreERT2 targeted cells showed lacZ-positivity and stained blue. Recombination efficiency was evaluated by counting lacZ positive cells divided by the total cell number in the growth plate, cartilage endplate, inner and outer annulus fibrosus, and nucleus pulposus regions. Three non-consecutive histological sections from four different transgenic mice (n=4) were analyzed. High Cre-recombination efficiency in growth plate and inner annulus fibrosus cells was observed in Col2a1-CreERT2 transgenic mice 1, 2, 3, and 4 months after tamoxifen induction. Relatively low Cre-recombination efficiency was found in cartilage endplate and outer annulus fibrosus cells. No lacZ-positive cells were found in the nucleus pulposus region. (B) Evaluation of Cre recombination efficiency. Tamoxifen was administered to 2-week-old Col2a1-CreERT2;R26R mice and mice were sacrificed 1 month after tamoxifen induction. X-Gal staining was performed and sections (L3/L4) were then counterstained with nuclear fast red. Total and X-Gal-positive cell numbers were counted in the growth plate, cartilage endplate, inner and outer annulus fibrosus, and nucleus pulposus regions.
Table 2.
Col2a1-CreERT2-mediated Cre-recombination efficiency in disc cells
| 1 | 2 | 3 | 4 (month) | |
|---|---|---|---|---|
| Growth plate | 79.9 | 74.7 | 73.5 | 70.6 |
| Cartilage endplate | 10.7 | 20.4 | 27.5 | 37.3 |
| Inner annulus fibrosus | 76.8 | 81.1 | 79.1 | 73.3 |
| Outer annulus fibrosus | 11.4 | 4.7 | 8.7 | 4.8 |
| Nucleus Pulposus | 0 | 0 | 0 | 0 |
Induction: 2-week-old; Sac: 1-, 2-, 3-, and 4-month-old after tamoxifen induction (n=4)
To determine the role of TGF-β signaling in the growth and maintenance of disc tissue at postnatal stage, we generated Tgfbr2Col2ER mice using the Col2a1-CreERT2 transgenic mice to specifically inactivate TGF-β signaling in inner annulus fibrosus cells in the IVD and surrounding growth plate chondrocytes. In P14 early postnatal mice, the cartilage endplate tissue was well developed in wild-type mice. In contrast, significant reduction in the length and area of cartilage endplate tissue was observed in Tgfbr2Col2ER mice. A significant loss of matrix tissue and endplate cartilage cells was observed in Tgfbr2Col2ER mice (Fig. 2A). In 3-month-old postnatal mice, significant loss of growth plate chondrocytes in disc tissue was found in Tgfbr2Col2ER mice. In addition, growth plate chondrocytes lost their columnar structure compared to their Cre-negative control mice. Disorganized chondrocytes formed clusters in the growth plate region of Tgfbr2Col2ER mice (Fig. 2B). These results demonstrated that TGF-β signaling is required for normal growth of disc and surrounding tissues at the postnatal stage. To determine changes in gene expression of disc cells, we isolated primary disc cells from these mice. We found that mRNA expression of Col10a1 (11-fold increase), Mmp13 (14-fold increase), Adamts4 (4-fold increase), and Adamts5 (15-fold increase) was significantly up-regulated in Tgfbr2Col2ER mice (Fig. 3A), indicating that disc cell differentiation process was accelerated and several genes (and probably their coding proteins) involved in matrix degradation were activated. To confirm the effect of TGF-β on Mmp13 expression in vitro, we treated RCS chondrogenic cells with 5ng/ml of TGF-β up to 48 hours and found that TGF-β significantly inhibited Mmp13 expression at 24 and 48 time points (Fig. 3B). This finding is consistent with the in vivo observation. To explore the possible interaction of TGF-β with PTHrP and Ihh signaling, we examined the expression of Pthr1 and Ptc1 in RCS cells in which Tgfbr2 siRNA was transfected. We found that silencing of Tgfbr2 expression significantly up-regulated Ptc1 expression but had no effect on Pthr1 expression (Fig. 3C), suggesting that TGF-β may have inhibitory effect on Ihh signaling in chondrocytes.
Fig. 2.


Inactivation of TGF-β signaling leads to defects in the development and the growth of intervertebral disc tissue. (A) P14 Tgfbr2Col2ER mice were analyzed by histology. Histologic sections of disc samples (L3/L4) of P14 mice were stained with AB/H&E. Four Cre-negative (n=4) and three Tgfbr2Col2ER mice (n=3) were analyzed. Significant reduction in the length and area of cartilage endplate tissue (green bar) was observed in Tgfbr2Col2ER mice. GP: growth plate; CE: cartilage endplate; and VB: vertebral body. (B) AB/H&E staining were performed using histologic sections (L3/L4) of Cre-negative (n=5) and Tgfbr2Col2ER mice (n=6) mice. Growth plate chondrocytes lost their columnar structure and disorganized chondrocytes formedclusters (red arrows) in the growth plate region of Tgfbr2Col2ER mice.
Fig. 3.

Gene expression analysis. (A) Total RNA was extracted from primary disc cells derived from P14 Tgfbr2Col2ER mice and Cre-negative littermates. The mRNA expression of Col2a1, Col10a1, Mmp13, Adamts4 and Adamts5 was analyzed by real-time PCR. The expression of Col10a1, Mmp13, Adamts4, and Adamts5 was significantly up-regulated in Tgfbr2Col2ER mice. (B) Chondrogenic RCS cells were treated with 5ng/ml of TGF-β for 4, 24 and 48 hours. The expression levels of Mmp13 mRNA (normalized to the levels of β-actin mRNA) were analyzed by real time PCR. TGF-β significantly inhibited Mmp13 mRNA expression at 24 and 48 hour time points. (C) RCS cells were trasfected with 20nM of Tgfbr2 siRNA or control siRNA for 48 hours and the total RNAs were extracted from cells and real time PCR was performed to assess changes in expression levels of Tgfbr2, Ptc1 and Pthr1 mRNA. Knocking-down of the Tgfbr2 expression significantly enhanced Ptc1 expression but had no effect on Pthr1 expression. Unpaired Student t-test, p<0.01, n=3.
MMP13 plays a critical role in cartilage function and the development of osteoarthritis [18, 19]. In order to determine if the phenotype observed in Tgfbr2Col2ER mice is due to the activation of Mmp13 expression, we have deleted the Mmp13 gene under the Tgfbr2 deletion background by generating Tgfbr2/Mmp13 double KO (dKO) mice. Histologic analysis of spine sections showed that proteoglycan levels and cell numbers in growth plate cartilage area were significantly decreased in Tgfbr2Col2ER mice. In contrast, increased proteoglycan levels and the preservation of the growth plate cartilage morphology were seen in Tgfbr2/Mmp13 dKO mice (Fig. 4). Deletion of the Mmp13 gene alone had no significant disc phenotype (Fig. 4). These results suggest that Mmp13 is a critical downstream target gene of TGF-β signaling in disc cells and inhibition of TGF-β signaling may lead to growth plate and endplate tissue destruction through activation of Mmp13 gene expression.
Fig. 4.
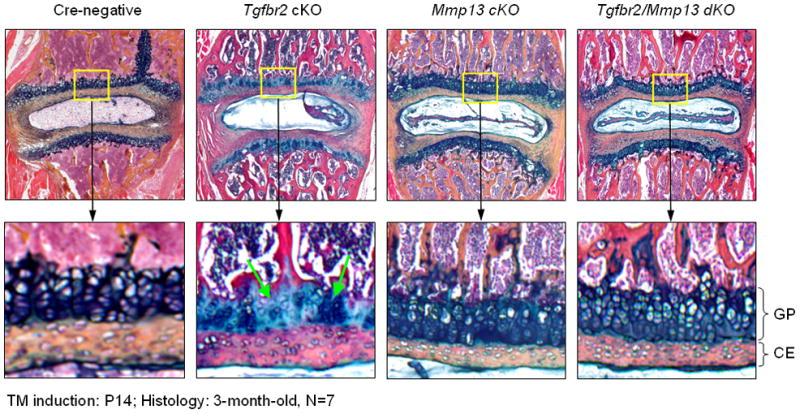
Deletion of the Mmp13 gene reverses the disc phenotype of 3-month-old Tgfbr2Col2ER mice. Two-week-old Tgfbr2Col2ER mice were injected with tamoxifen (1 mg/10g body weight, x5 days, i.p. injection) at 2-week-old. Mice were sacrificed at 3 months of age. The disc samples were fixed, decalcified and paraffin-embedded. The histological sections were stained with Alcian blue/H&E method.
Discussion
Two major cell types were identified in the nucleus pulposus region, notochord-like cells and chondrocyte-like cells. It has been reported that the immature disc nucleus pulposus consists of notochordal cells. With nucleus pulposus maturation, notochordal cells disappear and are replaced by chondrocyte-like mature nucleus pulposus cells in the human spine. It seems that the notochordal cells disappear at different stages in postnatal and adult animals in different species [20]. In humans, notochordal cells could be lost as early as 4 years of age [21]. In the rabbit, notochordal cells disappeared by 11 weeks of age [22]. In the mouse, notochordal cells are survived and greatly reduced at postnatal stage. The changes in disc phenotype coincide with early signs of disc degeneration [23]. Therefore, it is important to determine the origin of chondrocyte-like mature nucleus pulposus cells. It has been reported that postnatal transition from a notochordal to a cartilaginous nucleus pulposus is accomplished by the migration of chondrocyte-like cells from the hyaline cartilage endplate and inner annulus fibrosus region into the ectopic notochordal nucleus pulposus region at the postnatal stage [24–26]. A study using a rabbit model suggests that chondrocytes from the cartilage endplate can migrate toward the nuclear pulposus region as notochordal cells undergo apoptosis [21]. In vivo analysis of cell fate has been facilitated by the creation of Cre-dependent conditional reporter lines to track the fate of cell populations expressing Cre under the control of tissue-specific promoters [27]. Our findings from Col2a1-CreERT2;R26R mice indicate that no X-Gal positive cells migrated from the cartilage endplate and annulus fibrosus cells during the postnatal 4 month period, suggesting that chondrocyte-like cells in the nucleus pulposus region are not derived from the surrounding annulus fibrosus or cartilage endplate, at least at the early postnatal stage in mice.
Although TGF-β signaling has been shown to play a critical role in spine and disc tissue development at embryonic stage [8, 13], its role in disc tissue growth and maintenance at postnatal stage have not been reported. Since cellular environments and gene expression profiles could be different at embryonic and postnatal stages, functions of TGF-β signaling at these two developmental stages could be different. Our present study demonstrated that TGF-β signaling is required for normal growth plate and endplate cartilage function at the postnatal stage. Loss of TGF-β signaling in growth plate chondrocytes and inner annulus fibrosus cells led to the loss of matrix tissue and endplate cartilage cells and abnormal growth plate cartilage morphology. Since the expression of Mmp13 gene was significantly up-regulated in disc cells of Tgfbr2Col2ER mice, we also examined the role of Mmp13 in disc tissue phenotype of Tgfbr2Col2ER mice. We found that deletion of the Mmp13 gene under Tgfbr2 null background completely reversed disc phenotype observed in Tgfbr2Col2ER mice. These findings demonstrate that TGF-β-Mmp13 signaling plays an essential role in postnatal disc tissue growth and the maintenance.
Acknowledgments
This work was supported by Grants R01 AR055915, R01 AR054465 and Contract C024320 to D.C. from National Institute of Health and New York State Department of Health and Empire State Stem Cell Board. This work was also partially supported by Grants Z2111261 and Y2111184 to D.C. and H.J. from Zhejiang Natural Science Foundation, China.
Footnotes
Competing interest statement
The authors have no conflict of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Takahashi K, Aoki Y, Ohtori S. Resolving discogenic pain. Eur Spine J. 2008;17(Suppl 4):428–31. doi: 10.1007/s00586-008-0752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller JA, Schmatz C, Schultz AB. Lumbar disc degeneration: correlation with age, sex, and spine level in 600 autopsy specimens. Spine. 1988;13:173–178. [PubMed] [Google Scholar]
- 3.Dahia CL, Mahoney EJ, Durrani AA, Wylie C. Postnatal growth, differentiation, and aging of the mouse intervertebral disc. Spine. 2009;34:447–55. doi: 10.1097/BRS.0b013e3181990c64. [DOI] [PubMed] [Google Scholar]
- 4.Roughley PJ. Biology of intervertebral disc aging and degeneration involvement of the extracellular matrix. Spine. 2004;29:2691–2699. doi: 10.1097/01.brs.0000146101.53784.b1. [DOI] [PubMed] [Google Scholar]
- 5.Chen M, Lichtler AC, Sheu T, Xie C, Zhang X, O’Keefe RJ, Chen D. Generation of a transgenic mouse model with chondrocyte-specific and tamoxifen-inducible expression of Cre recombinase. Genesis. 2007;45:44–50. doi: 10.1002/dvg.20261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu M, Chen M, Lichtler AC, O’Keefe RJ, Chen D. Tamoxifen-inducible Cre-recombination in articular chondrocytes of adult Col2a1-CreERT2 transgenic mice. Osteoarthr Cartilage. 2008;16:129–130. doi: 10.1016/j.joca.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baffi MO, Slattery E, Sohn P, Moses HL, Chytil A, Serra R. Conditional deletion of the TGF-beta type II receptor in Col2a expressing cells results in defects in the axial skeleton without alterations in chondrocyte differentiation or embryonic development of long bones. Dev Biol. 2004;276:124–142. doi: 10.1016/j.ydbio.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 8.Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschmann T. TGF-b2 knockout mice have multiple developmental defects that are non-overlapping with other TGF-h knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, Annunziata N, Doetschman T. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulkarni AB, Huh CG, Becker D, Geiser A, Lyght M, Flanders KC, Roberts AB, Sporn MB, Ward JM, Karlsson S. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci USA. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diebold RJ, Eis MJ, Yin M, Ormsby I, Boivin GP, Darrow BJ, Saffitz JE, Doetschman T. Early-onset multifocal inflammation in the transforming growth factor beta 1-null mouse is lymphocyte mediated. Proc Natl Acad Sci USA. 1995;92:12215–12219. doi: 10.1073/pnas.92.26.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark JC, Wert SE, Brachurski CJ, Stahlman MT, Stripp BR, Weaver TE, Whitsett JA. Targeted disruption of the surfactant B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc Natl Acad Sci USA. 1995;92:7794–7798. doi: 10.1073/pnas.92.17.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baffi MO, Moran MA, Serra R. Tgfbr2 regulates the maintenance of boundaries in the axial skeleton. Dev Biol. 2006;296:363–374. doi: 10.1016/j.ydbio.2006.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soriano P. Generalized LacZ expression with the Rosa26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 15.Chytil A, Magnuson MA, Wright CV, Moses HL. Conditional inactivation of the TGF-beta type II receptor using Cre: Lox. Genesis. 2002;32:73–75. doi: 10.1002/gene.10046. [DOI] [PubMed] [Google Scholar]
- 16.Roughley Peter J. Biology of intervertebral disc aging and degeneration involvement of the extracellular matrix. Spine. 2004;29:2691–2699. doi: 10.1097/01.brs.0000146101.53784.b1. [DOI] [PubMed] [Google Scholar]
- 17.Henry SP, Jang CW, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis. 2009;47:805–14. doi: 10.1002/dvg.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stickens D, Behonick DJ, Ortega N, Heyer B, Hartenstein B, Yu Y, Fosang AJ, Schorpp-Kistner M, Angel P, Werb Z. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development. 2004;131:5883–95. doi: 10.1242/dev.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inada M, Wang Y, Byrne MH, Rahman MU, Miyaura C, López-Otín C, Krane SM. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc Natl Acad Sci USA. 2004;101:17192–17197. doi: 10.1073/pnas.0407788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter CJ, Matyas JR, Duncan NA. Cytomorphology of notochordal and chondrocytic cells from the nucleus pulposus: a species comparison. J Anat. 2004;205:357–62. doi: 10.1111/j.0021-8782.2004.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pazzaglia UE, Salisbury JR, Byers PD. Development and involution of the notochord in the human spine. J R Soc Med. 1989;82:413–5. doi: 10.1177/014107688908200714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott NA, Harris PF, Bagnall KM. A morphological and histological study of the postnatal development of intervertebral discs in the lumbar spine of the rabbit. J Anat. 1980;130:75–81. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao CQ, Wang LM, Jiang LS. The cell biology of intervertebral disc aging and degeneration. Ageing Research Reviews. 2007;6:247–261. doi: 10.1016/j.arr.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Kim KW, Lim TH, Kim JG, Jeong ST, Masuda K, An HS. The origin of chondrocytes in the nucleus pulposus and histologic findings associated with the transition of a notochordal nucleus pulposus to a fibrocartilaginous nucleus pulposus in intact rabbit intervertebral discs. Spine. 2003;28:982–90. doi: 10.1097/01.BRS.0000061986.03886.4F. [DOI] [PubMed] [Google Scholar]
- 25.Kim KW, Ha KY, Lee JS, Nam SW, Woo YK, Lim TH, An HS. Notochordal cells stimulate migration of cartilage end plate chondrocytes of the intervertebral disc in in vitro cell migration assays. Spine J. 2009;9:323–9. doi: 10.1016/j.spinee.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Thorsten G, Andreas N, Markus K. Sensitivity of notochordal disc cells to mechanical loading: an experimental animal study. Eur Spine J. 2010;19:113–121. doi: 10.1007/s00586-009-1217-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyle S, Misfeldt A, Chandlerk KJ, Deal KK, Southard-Smith EM, Mortlock DP, Baldwin HS, de Caestecker M. Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev Biol. 2008;313:234–45. doi: 10.1016/j.ydbio.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]


