Abstract
This mini-review describes recent discoveries demonstrating that experience can drive the production of epigenetic marks in the adult nervous system, and that the experience-dependent regulation of epigenetic molecular mechanisms in the mature CNS participates in the control of gene transcription underlying the formation of long-term memories. In the mammalian experimental systems investigated thus far, epigenetic mechanisms have been linked to associative fear conditioning, extinction of learned fear, and hippocampus-dependent spatial memory formation. Intriguingly, in one experimental system epigenetic marks at the level of chromatin structure (histone acetylation) have been linked to the recovery of memories that had appeared to be “lost”, i.e. not available for recollection. Environmental enrichment has long been known to have positive effects on memory capacity, and recent studies have suggested that these effects are at least partly due to the recruitment of epigenetic mechanisms by environmental enrichment. Finally, a an uncoupling of signal transduction pathways from the regulation of epigenetic mechanisms in the nucleus has been implicated in the closure of developmental critical periods. Taken together, these eclectic findings suggest a new perspective on experience-dependent dynamic regulation of epigenetic mechanisms in the adult nervous system, and their relevance to biological psychiatry.
Keywords: learning, memory, amygdala, hippocampus, LTP, histone, HDAC, DNA methylation, epigenetic, gene transcription, fear conditioning
Introduction
A central tenet in the epigenetics subfield has been that epigenetic marks, once laid down as part of development, are subsequently immutable. This concept has served developmental biologists well, and explains the permanence of, for example, cellular phenotype over the lifespan of an animal. However, recent studies of the CNS have indicated that while the permanence of epigenetic marks is a good general rule, there are some exceptions to that generalization in play in the nervous system. Thus, in some instances epigenetic molecular mechanisms appear to be recruited to help drive experience-dependent modifications in cognition and behavior.
This mini-review will focus on the emerging appreciation of roles for epigenetic molecular mechanisms in learning- and memory-related phenomena. I will first present an overview of the basic epigenetic molecular mechanisms operating in the CNS. I will then present a brief description of recent studies specifically indicating a role for epigenetic control of gene transcription in the context of associative fear conditioning, extinction of conditioned fear, and mammalian spatial learning and memory. I will then highlight a seminal series of recent experiments implicating epigenetic mechanisms in the positive effects of environmental enrichment on memory capacity, and in triggering a capacity to recollect memories that appeared to have been lost. Finally, I will briefly comment on a recent study suggesting that uncoupling of epigenetic mechanisms from cellular signal transduction cascades is a component of the processes that lead to the closure of developmental critical periods. Altogether, consideration of these studies provokes a new viewpoint concerning the role of epigenetic processes in the CNS, and suggests that dynamic regulation of epigenetic mechanisms is part of the normal gene-environment interface.
Please note that I will not deal with the various interesting studies concerning the role of epigenetic mechanisms in drug addiction, psychiatric and neurological illnesses, and the lasting effects of neonatal experience, because these are topics addressed in other recent and upcoming mini-reviews in this journal.
Whence epigenetics?
The term epigenetics is derived from Waddington (1). Waddington coined the term to describe a conceptual solution to a conundrum, a puzzle that arises as a fundamental consideration in developmental biology. All the different cells in your body have exactly the same genome, that is, exactly the same DNA nucleotide sequence, with only a few exceptions in your reproductive and immune systems. Thus, your liver cells have exactly the same DNA as your neurons. However, those two types of cells clearly are vastly different in terms of the gene products that they produce. How can two cells have exactly the same DNA but be so different? Especially when what makes them different is that they produce different gene transcripts that are read directly from the identical DNA? Waddington coined the term epigenesis to describe the conceptual solution to this problem. Some level of mechanism must exist, he reasoned, that was “above” the level of the genes encoded by the DNA sequence, that controlled the DNA readout. These are what we now refer to as epigenetic mechanisms. These epigenetic mechanisms specify in a neuron that genes A, C, D, L…‥ are turned into functional products, and in a liver cell that genes A, B, C, E …‥ are turned into functional products. Epigenetic marks are put in place during cell fate determination and serve as a cellular information storage system perpetuating cellular phenotype over the lifespan.
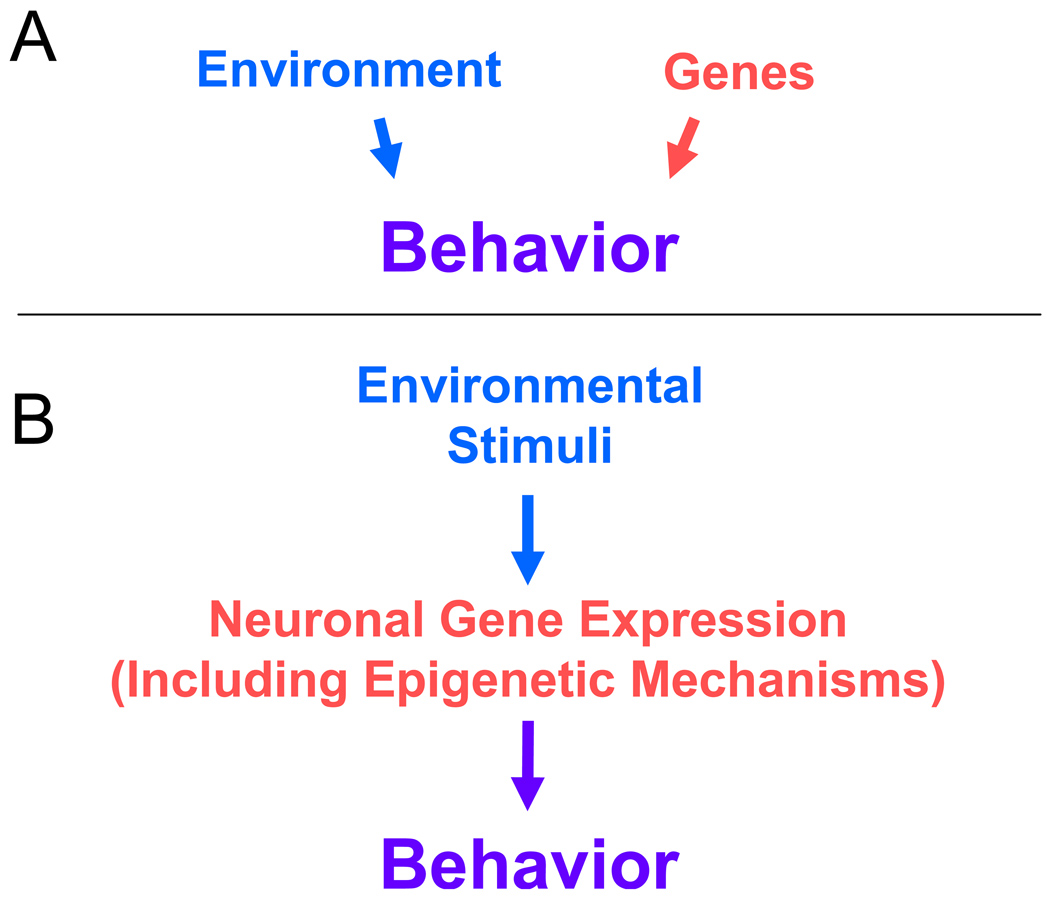
However, an additional aspect of epigenetic control of gene expression is now emerging from recent studies of epigenetic molecular mechanisms in the nervous system. Thus, convincing evidence is accumulating that epigenetic mechanisms do not just contribute to phenotypic hard-wiring at the cellular level. Rather, in the nervous system with its abundance of terminally differentiated, non-dividing cells, epigenetic mechanisms also play a role in acute regulation of gene expression in response to environmental signals, drugs of abuse, and experience. In addition, epigenetic mechanisms appear to contribute to both psychiatric and neurological disorders. In retrospect, these roles for epigenetic molecular mechanisms are perhaps not surprising. Epigenetic mechanisms, even in their role in development, sit at the interface of the environment and the genome. In the broadest terms of neuroscience and psychiatry, there is an emerging fascination with epigenetics because it has been a hidden mechanistic layer operating at the environment-genome interface. (Figure 1).
Figure 1.
The historical model with separate and distinct influences of genes and environment on behavior (Panel A) is now known to be over-simplified. Instead, contemporary studies have illustrated that environment and experience act in part through altering gene readout in the CNS in order to achieve their effects on behavior (Panel B). One component of the processes by which the environment and experience alter individual behavior includes epigenetic molecular mechanisms such as regulation of chromatin structure and DNA methylation. The historical dichotomy between “nature” (genes) and “nurture” (environment and experience) is too simplistic – genes and experience are mechanistically intertwined. Epigenetic molecular mechanisms contribute to this intertwining.
What are epigenetic marks and what do they do?
There are two basic molecular epigenetic mechanisms that are widely studied at present – regulation of chromatin structure through histone post-translational modifications, and DNA methylation. Other epigenetic molecular mechanisms such as regulation of gene expression through non-coding RNAs, and prion protein-based mechanisms, are also known to exist but I will not discuss them here.
The first major mechanism whereby the genome can be epigenetically marked is DNA methylation. Methylation of DNA is a direct chemical modification of a cytosine side-chain that adds a -CH3 group through a covalent bond. Methylation of DNA is catalyzed by a class of enzymes known as DNA methyltransferases (DNMTs) (2). DNMTs transfer methyl groups to cytosine residues within a continuous stretch of DNA, specifically at the 5-position of the pyrimidine ring (3,4). Not all cytosines can be methylated; usually cytosines must be immediately followed by a guanine in order to be methylated (5,6). These “CpG” dinucleotide sequences are highly underrepresented in the genome relative to what would be predicted by random chance; however about 70% of the CpG dinucleotides that are present are methylated (7). The rest of the normally unmethylated CpG dinucleotides occur in small clusters, known as “CpG islands” (8,9).
There are two variants of DNMTs: maintenance DNMTs and de novo DNMTs. The DNMT1 enzymatic isoform is the maintenance DNMT, DNMTs 3a and 3b are the de novo DNMT isoforms. Both maintenance and de novo DNMTs are expressed in most cells in the body. The two variants of DNMTs differ in one important respect, related to the conditions under which they will methylate DNA. De novo DNMTs methylate previously unmethylated CpG sites in DNA – sites which have no methyl-cytosine on either DNA strand. The maintenance DNMT isoform methylates hemi-methylated DNA – DNA which has a methylated CpG already present on one strand but no methyl-cytosine on the complementary strand. These two different isoforms thereby serve two distinct roles in the cell. De novo DNMTs place new methylation marks on DNA, for example when specific genes are first silenced as part of cell fate determination. Maintenance DNMTs perpetuate methylation marks after cell division. They regenerate the methyl-cytosine marks on the newly synthesized complementary DNA strand that arises from DNA replication.
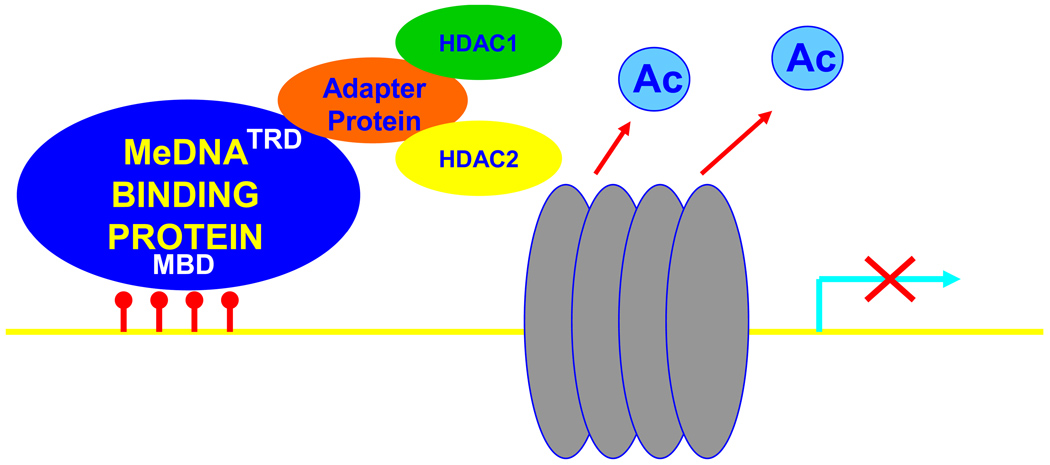
What are the functional consequences of DNA methylation? In most cases that have been studied so far, methylation of DNA is associated with suppression of gene transcription, and in many cases extensive DNA methylation triggers complete silencing of the associated gene. The precise molecular processes through which this occurs are complex and an area of intense investigation at present. However, one simplified model is shown in Figure 2. In essence, methylation of cytosines at CpG dinucleotides recruits methyl-DNA binding proteins, at specific sites in the genome. Proteins that bind to methylated DNA have both a methyl-DNA binding domain (MBD) and a transcription-regulatory domain (TRD). The TRD recruits adapter/scaffolding proteins, which in turn recruit histone de-acetylases (HDACs) to the site. The HDACs alter chromatin structure locally – “chromatin” is the term describing nuclear DNA/protein complexes. HDACs alter chromatin structure through removing acetyl groups from histone core proteins, leading to compaction of chromatin and transcriptional suppression. Thus, through this complex and highly regulated biochemical machinery, methylation of DNA triggers localized regulation of the three-dimensional structure of DNA and its associated histone proteins, resulting in a higher-affinity interaction between DNA and the histone core, and transcriptional repression by allosteric means. It is important to note that while DNA methylation is usually (and historically) associated with transcriptional suppression, recent studies have indicated that DNA methylation can also be associated with transcriptional activation, by mechanisms that have not yet been precisely determined (10,11).
Figure 2.
A simplified scheme for DNA methylation-dependent gene silencing. Methylation of cytosines at CpG dinucleotides (red lollipops) recruits methyl-DNA binding proteins locally to specific sites in the genome. All proteins that bind to methylated DNA have both a methyl-DNA binding domain (MBD) and a transcription-regulatory domain (TRD). The TRD recruits adapter proteins which in turn recruit histone de-acetylases (HDACs). The HDACs alter chromatin structure locally through removing acetyl groups (Ac) from histone core proteins (grey spheres), leading to compaction of chromatin and transcriptional suppression. It is important to note that while this is the traditional and well-established role of methyl-DNA binding proteins in transcriptional regulation, recent findings also support the idea that DNA methylation can also be associated with transcriptional activation.
Consideration of the mechanism of transcriptional silencing by DNA methylation thus leads us to the second major category of epigenetic marks, histone post-translational modifications.
Epigenetic tagging of histones
Histones are highly basic proteins whose function is to organize DNA within the nucleus. As mentioned above, in the nucleus, DNA is tightly packaged into chromatin, a DNA-protein complex that consists of DNA in a double helix, histone proteins, and various associated regulatory proteins. The interaction between histones, which form the core of the chromatin particle, and DNA is mediated in part by the N-terminal tail of histone proteins. One can imagine chromatin as a core of eight histone proteins (histones 2A, 2B, 3, and 4, with two copies of each molecule) with DNA wrapped around it like rope on a windlass. Structural studies indicate that the N-terminal tails of histones protrude beyond the DNA and are available for post-translational modifications (12).
Several specific sites of post-translational modification exist within the N-terminal tails of histone proteins, and modification of these sites modulates the overall structure of chromatin. Currently, four distinct post-translational modifications of histone tails have been well-characterized: acetylation, methylation, ubiquitination and phosphorylation. All of these modifications serve as epigenetic tags (13). However, for this mini-review I will only discuss histone acetylation because it has been the most extensively studied in the CNS.
Acetylation of histones occurs at lysine residues, specifically on their side-chain amino group, which effectively neutralizes their positive charge. Histone acetyltransferases (HATs) catalyze the direct transfer of an acetyl group from acetyl-CoA to the ε-NH+ group of the lysine residues within a histone (14–17). Histone acetylation is a reversible process, and the enzymes that catalyze the reversal of histone acetylation are known as histone deacetylases (HDACs).
Classical isoforms of HDACs catalyze the removal of acetyl groups from lysine residues through a Zn2+-dependent charge-relay system (18,19). The newly characterized SIR2 family of HDACs (the “Sirtuins”) operate through an NAD+-dependent mechanism, but I will not discuss them here (20). By way of background, there are a total of eleven different classical HDAC isoforms broadly divided into two classes. HDACs 1,2,3, and 8 are class I HDACs, while Class II encompasses HDAC isoforms 4,5,6,7, 9, 10, and 11.
HDAC inhibitors are undergoing a period of rapid development in the pharmaceutical industry because of their potential applicability in cancer treatment and the emerging possibility of their utility in neurological and psychiatric disorders. HDAC inhibitors are the principal way to manipulate the epigenome pharmacologically at present. In terms of some commonly available HDAC inhibitors, Trichostatin A (TsA) inhibits HDACs broadly across both Class I and Class II, while the inhibitors Sodium butyrate and suberoylanylide hydroxamic acid (SAHA, aka Vorinostat or Zolinza) select for class I HDACs. Valproate is also an HDAC inhibitor, but this drug also has additional targets and the role of HDAC inhibition in valproate’s clinical efficacy is unclear at this time.
The principal caveat to interpreting all studies utilizing HDAC inhibitors is the fact that “histone de-acetylase” is actually a misnomer. Histone de-acetylase enzymes should be more accurately described as “lysine de-acetylases”. Lysine amino-acid side chains are acetylated in a wide variety of different cellular proteins besides just histones. The list of known lysine-acetylated proteins is quite long, including transcription factors, cytoskeletal proteins, and a wide variety of metabolic enzymes. HDACs operate on all these proteins, not just their prototype substrate, histones. Therefore any behavioral effect of HDAC inhibitors might be due to alterations in acetylation of a wide variety of intracellular targets.
The signal transduction processes controlling histone acetylation in the mature CNS are just beginning to be investigated. However, two signaling cascades have been implicated in controlling histone acetylation and chromatin structure in the mature CNS thus far. One pathway is in the Mitogen-Acitvated Protein Kinase (MAPK) superfamily – exemplified by the ERK/MSK/CREB pathway. In this pathway the Extracellular-signal Regulated Kinase (ERK) activates its downstream target Mitogen- and Stress-activated Kinase (MSK), which in turn phosphorylates the Cyclic-AMP Regulatory Element Binding Protein (CREB, refs 21–24). This phosphorylation and activation of CREB recruits CREB Binding Protein (CBP), which is a histone acetyltransferase that regulates local chromatin structure as part of CREB-dependent activation of nuclear gene transcription.
The second known category of signaling pathway regulating chromatin structure in the mature CNS is the Nuclear Factor Kappa B (NFκB) signaling pathway. NFκB is a DNA-binding transcription factor that controls histone acetylation and chromatin structure in the CNS by mechanisms that are still being worked out (25,26). NFκB is controlled by its upstream regulator Inhibitor of KappaB Kinase (IKK), which itself is a target of multiple upstream regulatory signaling cascades. Thus, overall it is known at this point that both the ERK/MSK/CREB pathway and the IKK/NFκB pathway actively regulate chromatin structure in the mature CNS. One suspects that many important additional mechanisms await discovery (27–29).
Epigenetic mechanisms in learning and memory
In psychological terms memory describes the processes utilized by the brain for long-term storage of information. Early studies implicated both protein translation and gene transcription as vital to the formation of long-term memories (30,31). Subsequent studies have shown that formation of long-term memory is a complex process that requires the engagement of many distinct signaling pathways and the regulation of numerous genes (32–34).
A recent study by Levenson et al. (35) has suggested that the same processes that lead to formation of long-term memory also lead to epigenetic marking of the genome. Contextual fear conditioning is a hippocampus-dependent learning paradigm whereby an animal learns to associate a novel context with an aversive stimulus (36–38). Acetylation of hippocampal histone H3, but not H4, is significantly increased after an animal is trained using a contextual fear conditioning paradigm (35). Formation of long-term contextual fear memory requires NMDA-receptor-dependent synaptic transmission and the ERK MAPK signaling cascade in the hippocampus (39–42), and inhibition of either of these critical cellular processes blocks the memory-associated increase in acetylation of H3 (35). These observations were the first to demonstrate that epigenetic tagging of the genome occurred during consolidation of hippocampus-dependent long-term memory.
Interestingly, in these same studies Levenson et al found that a different form of long-term memory, latent inhibition, was associated with altered acetylation of histone H4, while H3 acetylation was unaltered by this paradigm. This finding along with many others suggests the intriguing possibility that a type of epigenetic code might exist for memory formation, whereby specific types of memories are associated with specific patterns of histone modifications (11, 43,44).
Addition of acetyl groups to lysine residues within histone proteins is accomplished via the action of histone acetyltransferases (HATs). If acetylation of histones is functionally significant for consolidation of long-term memory, then disruption of HAT activity would be predicted to interfere with long-term memory formation. CREB Binding Protein (CBP) is a transcriptional coactivator; an enzyme that contains endogenous HAT activity (45). Several studies have investigated long-term memory formation in genetically manipulated mice with impaired CBP function. One class of mice examined contain an allele of CBP that codes for a truncated form of the protein, which acts in a dominant negative manner (CBPDN+/−) (46). CBPDN+/− mice exhibit significant deficits in various forms of long-term memory including step-through passive avoidance, novel object recognition and cued fear conditioning (46,47). While these studies provided the first evidence that CBP might play a role in long-term memory formation, the widespread developmental derangements exhibited by CBPDN+/− animals prevented straightforward interpretation of the performance of these animals in various memory tasks(46).
To further elucidate the role of CBP in long-term memory formation, three very sophisticated recent studies have made CBP-deficient animals that lack the severe developmental problems present in the CBPDN+/− animals. The first study improved upon the CBPDN+/− mice by linking the dominant negative allele of CBP to an inducible promoter (CBPI-DN+/−)(48). Activation of the dominant negative allele after animals had developed normally, led to impaired acquisition of the spatial watermaze task and novel object recognition, two forms of hippocampus-dependent memory (48). In another series of experiments, animals that lacked one allele of CBP (CBP+/−) exhibited impairments in contextual and cued fear memory, and novel object recognition (49). In both studies, administration of an HDAC inhibitor restored normal long-term memory formation, suggesting that the balance of HAT/HDAC activity was altered in these mice (48,49) and causative of the memory deficits. Finally, mice that carry inactivating mutations in the CREB-binding (KIX) domain of CBP exhibit deficits in long-term memory for contextual fear conditioning and novel object recognition (50). These results support a role for CBP-mediated histone acetylation in memory formation and suggest that KIX-interacting transcription factors like CREB recruit CBP histone acetyltransferase activity during long-term memory storage.
The above studies demonstrate that histone acetylation is regulated by, and disruption of HAT activity impairs, long-term memory formation. Together, these observations suggest that any perturbations in the processes that regulate chromatin structure influence long-term memory formation in the behaving animal in vivo. However, can augmentation of histone acetylation enhance memory formation? To directly test this, several studies have investigated the effect of HDAC inhibitors on long-term memory formation. In a study by Yeh et al.(24), direct infusion of the HDAC inhibitor trichostatin A into the amygdala significantly enhanced formation of fear potentiated startle memory. Additionally, Levenson et al. (35) demonstrated that systemic administration of the HDAC inhibitor sodium butyrate enhanced formation of contextual fear memory, and Wood et al demonstrated that direct intrahippocampal infusion of an HDAC inhibitor enhanced hippocampus-dependent contextual fear conditioning (50). Finally, a fascinating recent study by Abel, Wood, and coworkers demonstrated that these memory-enhancing effects of HDAC inhibitors depend on the CREB/CBP pathway (51), nicely unifying the HDAC inhibitor studies with the CBP-HAT oriented studies described in the preceding paragraphs.
In the clinical setting, extinction of aversive and maladaptive memories is often a significant challenge. Might regulation of chromatin structure also be involved in extinction of memories? Bredy et al (52) have shown that extinction of fear conditioning in laboratory animals is associated with alterations in chromatin structure, implicating regulation of these processes in fear extinction. Moreover, using a conditioned fear paradigm, two different groups have shown that extinction of conditioned fear is accelerated when animals are administered HDAC inhibitors in vivo (53,54). Thus, extinction of learned fear, like fear learning itself, appears to involve chromatin modifications and be subject to enhancement with HDAC inhibitors. These studies both provide a new insight into molecular mechanisms of memory extinction, they suggest a potential new route for pharmacotherapy of maladaptive fear behaviors.
In a recent series of studies my laboratory has investigated the capacity of DNA methylation, the other major epigenetic molecular mechanism besides histone modification, to regulate synaptic plasticity and memory in adult animals (55,56). In our first series of studies in this area we found that inhibitors of DNMTs that likely block the net effects of both maintenance and de novo DNMTs could alter DNA methylation in adult CNS tissue and block hippocampal Long-term Potentiation (LTP) in physiologic studies vitro (55). In additional more recent studies we found that de novo DNMT gene expression (DNMT3a and DNMT3b) is upregulated in the adult rat hippocampus following contextual fear conditioning, and that generalized DNMT inhibition blocks memory formation in this same paradigm (56). In addition, fear conditioning was associated with rapid methylation and transcriptional silencing of the memory suppressor gene Protein Phosphatase 1 (PP1) and demethylation and transcriptional activation of the synaptic plasticity gene reelin. These findings have the surprising implication that both DNA methylation and demethylation might be involved in long-term memory consolidation. Overall these results suggest that DNA methylation is dynamically regulated in the adult nervous system and that this cellular mechanism is a crucial step in memory formation.
These studies of DNA methylation are at a very early stage and do not address several open questions. First, the biochemical mechanism for active DNA demethylation is unknown and indeed the general idea of active DNA demethylation has a contentious history (reviewed in 57). In addition, the mechanistic interplay between histone acetylation and DNA methylation/demethylation has not been worked out, and the possibility exists of reciprocal regulation between the two mechanisms (56). Finally, the signal transduction processes that might be controlling DNA methylation and demthylation in the adult CNS are completely unexplored at present.
Despite these numerous unanswered questions, taken together all these studies of histone acetylation and DNA methylation indicate that long-term behavioral memory processes regulate, and are regulated by, the epigenome.
Epigenetics in Human Cognition - Mental Retardation Syndromes and Cognitive Disorders
In the preceding section I discussed results from animal models implicating epigenetic mechanisms in learning and memory. However, there is a considerable body of evidence, albeit indirect, implicating disruption of epigenetic mechanisms as a causal basis for human cognitive dysfunction as well. In the following few paragraphs I will briefly review two cognitive disorders that are associated with epigenetic dysfunction: Rubinstein-Taybi Syndrome and Rett Syndrome.
In interpreting these findings in the present context, an important caveat applies. When considering these cases it is important to distinguish between a developmental need for epigenetic mechanisms, to allow formation of a normal nervous system, versus an ongoing need for these mechanisms as part of cognitive processing per se in the adult. The majority of the attention to date has justifiably focused on developmental roles for epigenetics in establishing the capacity for cognitive function in the adult. However, the experimental results outlined above also suggest the possibility of an ongoing and active role for epigenetic mechanisms in adult cognition.
Rubinstein-Taybi syndrome (RTS)was first described by Rubinstein and Taybi in 1963 (58). RTS occurs 1 in every 125,000 live births, and accounts for 1 in 300 patients with mental retardation. RTS is an inherited, autosomal dominant disease (59). Several studies have shown that RTS patients have a variety of mutations in CBP, including point mutations and 5’- or 3’-deletions (59,60). As mentioned above, CBP facilitates gene transcription coupled to activation of the transcription factor CREB and CBP contains endogenous HAT activity, although CBP has other molecular functions as well. The important implication of this work is its suggestion that altered HAT activity is causative of at least part of the cognitive deficits associated with RTS in humans. In other words, these findings are consistent with the idea of a role for epigenetic molecular mechanisms in human memory formation
Rett syndrome (RS) was first described in 1966 by Austrian pediatrician Andreas Rett. RS is an inherited, X-linked disease that afflicts about 1 in 15,000 females by ages 2 – 18 years of age, and is estimated to be the second leading cause of mental retardation in women (61). Development during approximately the first 6 months of life is normal in RS patients, with symptoms first appearing between 3 months to 3 years of age. The trademark of RS is a display of continuous, stereotypical hand movements, such as wringing, washing, clapping and/or patting. Other signs of RS include decreased growth (including microcephaly), abnormal respiration, gait ataxia, autism, seizures and other neurologic dysfunctions, along with learning disabilities and cognitive deficits. Recent studies indicate that mutation of the methyl CpG binding protein 2 (MeCP2) located in chromosomal region Xq28 causes Rett syndrome (62,63).
MeCP2 functionally connects DNA methylation to gene transcription. The role that disruption of this mechanism might play in the memory deficits observed in RS is still unclear, and MeCP2 likely plays a prominent role during development. However, as with RTS, recent discoveries that epigenetic mechanisms play a role in learning and memory suggest the possibility that all of the effects of MeCP2 deficiency on learning and cognition might not be purely developmental. Indeed, striking recent findings from Adrian Bird’s laboratory have supported the idea that an active, non-developmental role for MeCP2 is also involved in RS-related memory dysfunction (64).
Environmental Enrichment and Recovery of Lost Memories
As described above, HDAC inhibitors have been identified as being capable of improving memory formation in studies of normal rats and mice. In addition, a wide variety of prior laboratory animal studies pioneered by Bill Greenough’s laboratory (65) had demonstrated that environmental enrichment, i.e. making available a wide variety of toys, exercise apparati, and socially complex housing, boosts memory capacity as well.
Thus, two very different types of treatments, environmental enrichment and inhibition of histone deacetylases (HDACs), boost memory function in rodent experiments. Might these two observations be mechanistically related? Recent studies provide evidence that environmental enrichment achieves its effects through elevating histone acetylation in the hippocampus. Tsai and colleagues found that environmental enrichment is associated with increased histone acetylation in the hippocampus, an area of the CNS involved in long-term spatial memory formation (66). Tsai and co-workers also confirmed that environmental enrichment improves spatial memory capacity in mice, and found that this improvement in spatial memory was mimicked by HDAC inhibitors. These findings provided powerful evidence that regulation of chromatin structure is involved in spatial memory, and that environmental enrichment acts via increasing histone acetylation in the CNS.
In their studies Tsai and colleagues also found that HDAC inhibitors not only improve the capacity to form new memories, they restore the capacity to form memories in a mouse model of neurodegenerative disorders (66). They generated these findings by using a genetically engineered mouse model that they produced, that exhibits inducible neurodegeneration in its CNS. These engineered mice have neuronal loss in their hippocampus, and indeed Tsai and her colleagues have previously demonstrated that these mice have pronounced deficits in long-term spatial memory as assessed using a variety of behavioral assays. In their studies, they demonstrated that both HDAC inhibitors and environmental enrichment restored spatial memory capacity in these mice with neurodegeneration. This new study by Fisher et al thereby implicates HDAC inhibitors as a potential new therapeutic approach to human cognitive disorders arising from neurodegeneration. Indeed, their work and others’ suggests that HDAC inhibitors might be a useful general therapy for aging-related memory dysfunction as a broadly defined category (67).
In their studies Fisher et al also asked an intriguing question: are HDAC inhibitors capable of allowing an animal undergoing memory loss through neurodegeneration to recover memories that had apparently already been lost? This would seem almost beyond the realm of possibility, but that is exactly what Fisher et al observed to be an effect of HDAC inhibition in their mouse model of neurodegeneration. In a particularly fascinating set of experiments they trained a group of animals, let their memory for that training event decay over time (due directly or indirectly to neurodegeneration in their mouse model), and confirmed that the animals had lost the capacity to recall that memory. Amazingly, administration of an HDAC inhibitor then restored the capability of the animals to recall that memory, restoring access to a memory that had apparently already been lost. This is an extremely surprising finding and its cellular and neuronal circuit basis is quite mysterious. Overall, besides identifying HDAC inhibition as a potential new therapeutic target in neurodegenerative disorders, these findings complement an emerging literature suggesting an important role for epigenetic molecular mechanisms in memory function.
Epigenetics Mechanisms and Closure of Developmental Critical Periods
Recent findings have suggested a novel and exciting idea; that the capacity for chromatin modification specifically, and epigenetic molecular mechanisms in general, might be involved in switching off CNS cortical plasticity as a mechanism for closure of a developmental critical period (68,69).
In these laboratory experiments a developmental critical period is defined as a time-span during CNS development when young animals have a robustness for plastic change, both anatomical and functional. “Closure” of the critical period entails shutting off, at least to a large degree, this capacity for experience-driven anatomical and functional change. One very popular experimental system for studying critical periods involves ocular dominance column plasticity in the neocortex. In brief, ocular dominance plasticity is the process by which visual activity drives the apportioning of visual cortex neurons and synaptic connections to one eye or the other. The left eye is allocated a certain section of cortical territory, so to speak, versus that allocated to the right eye. This apportioning of territory is experience-driven based on relative visual stimulation of each eye and occurs selectively during a relatively brief neonatal period (a few weeks in rodents). After that, plasticity mechanisms are down-regulated and the visual cortex wiring remains largely static thereafter. This shutting down of the capacity for cortical plasticity is referred to as “closure” of the critical developmental period.
A recent paper by Pizzorusso and colleagues presents an interesting linkage between chromatin remodeling and experience-dependent plasticity in the visual cortex. In their work the authors identified a new candidate signaling and transcriptional regulation mechanism in OD column developmental plasticity; ERK/MAPK-dependent regulation of histone modifications. Their work specifically suggested that one mechanism for closure of the critical period is uncoupling activity-dependent MAPK regulation from one of its targets, histone acetylation. They also presented studies suggesting that administration of HDAC inhibitors could re-open the critical period, restoring the capacity for ocular dominance plasticity in the adult CNS. These fascinating studies both implicated epigenetic molecular mechanisms as a component of the machinery underlying developmental critical periods, and implicated uncoupling of these mechanisms as a component for closing the critical period.
Summary
In this brief overview I have presented an emerging new view of the epigenome and its role in the adult CNS. New studies are being published at a rapid pace demonstrating that epigenetic mechanisms are involved in mediating diverse experience-driven changes in the CNS. These experience-driven changes in the adult CNS are manifest at the molecular, cellular, circuit, and behavioral levels. Overall, these diverse observations support the view that the epigenome resides at the interface of the environment and the genome. Furthermore, it is now becoming clear that epigenetic mechanisms can influence behavior. Understanding the role of the epigenome in experience-dependent behavioral modification will clearly be important for, and relevant to the field of biological psychiatry.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement
Dr. Sweatt receives grant funding from the NIH, Evelyn F. McKnight Brain Research Foundation, the Rotary Clubs CART fund, and NARSAD. Dr. Sweatt reports no biomedical financial interests or potential conflicts of interest. The author also wishes to thank Felecia Hester for her assistance in preparing this review.
References
- 1.Waddington CH. The Strategy of the Genes. New York: MacMillan; 1957. [Google Scholar]
- 2.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 3.Chen L, et al. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991;30:11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- 4.Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- 5.Bird AP. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978;118:49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- 6.Cedar H, Solage A, Glaser G, Razin A. Direct detection of methylated cytosine in DNA by use of the restriction enzyme MspI. Nucleic Acids Res. 1979;6:2125–2132. doi: 10.1093/nar/6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet. 1989;83:181–188. doi: 10.1007/BF00286715. [DOI] [PubMed] [Google Scholar]
- 8.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 9.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 10.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen S, Zhou Z, Greenberg ME. Activating a repressor. Science. 2008;320:1172–1173. doi: 10.1126/science.1159146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 13.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 14.Tanner KG, et al. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J Biol Chem. 1999;274:18157–18160. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 15.Tanner KG, Langer MR, Kim Y, Denu JM. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J Biol Chem. 2000;275:22048–22055. doi: 10.1074/jbc.M002893200. [DOI] [PubMed] [Google Scholar]
- 16.Lau OD, et al. p300/CBP-associated factor histone acetyltransferase processing of a peptide substrate. Kinetic analysis of the catalytic mechanism. J Biol Chem. 2000;275:21953–21959. doi: 10.1074/jbc.M003219200. [DOI] [PubMed] [Google Scholar]
- 17.Tanner KG, Langer MR, Denu JM. Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry. 2000;39:11961–11969. doi: 10.1021/bi001272h. [DOI] [PubMed] [Google Scholar]
- 18.Buggy JJ, et al. Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem J. 1999;350(Pt 1):199–205. [PMC free article] [PubMed] [Google Scholar]
- 19.Finnin MS, et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 20.Buck SW, Gallo CM, Smith JS. Diversity in the Sir2 family of protein deacetylases. J Leukoc Biol. 2004;75:939–950. doi: 10.1189/jlb.0903424. [DOI] [PubMed] [Google Scholar]
- 21.Swank MW, Sweatt JD. Increased histone acetyltransferase and lysine acetyltransferase activity and biphasic activation of the ERK/RSK cascade in insular cortex during novel taste learning. J Neurosci. 2001;21:3383–3391. doi: 10.1523/JNEUROSCI.21-10-03383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chwang WB, O'Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13:322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brami-Cherrier K, Valjent E, Hervé D, Darragh J, Corvol JC, Pages C, Arthur SJ, Girault JA, Caboche J. Parsing molecular and behavioral effects of cocaine in mitogen- and stress-activated protein kinase-1-deficient mice. J Neurosci. 2005;25:11444–11454. doi: 10.1523/JNEUROSCI.1711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chwang WB, Arthur JS, Schumacher A, Sweatt JD. The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J Neurosci. 2007;27:12732–12742. doi: 10.1523/JNEUROSCI.2522-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lubin FD, Sweatt JD. The IkappaB kinase regulates chromatin structure during reconsolidation of conditioned fear memories. Neuron. 2007;55:942–957. doi: 10.1016/j.neuron.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh SH, Lin CH, Gean PW. Acetylation of nuclear factor-kappaB in rat amygdala improves long-term but not short-term retention of fear memory. Mol Pharmacol. 2004;65:1286–1292. doi: 10.1124/mol.65.5.1286. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira AM, Wood MA, McDonough CB, Abel T. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learn Mem. 2007;14:564–572. doi: 10.1101/lm.656907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SY, Levenson JM, Korsmeyer S, Sweatt JD, Schumacher A. Developmental regulation of Eed complex composition governs a switch in global histone modification in brain. J Biol Chem. 2007;282:9962–9972. doi: 10.1074/jbc.M608722200. [DOI] [PubMed] [Google Scholar]
- 29.Chandramohan Y, Droste SK, Reul JM. Novelty stress induces phospho-acetylation of histone H3 in rat dentate gyrus granule neurons through coincident signalling via the N-methyl-D-aspartate receptor and the glucocorticoid receptor: relevance for c-fos induction. J Neurochem. 2007;101:815–828. doi: 10.1111/j.1471-4159.2006.04396.x. [DOI] [PubMed] [Google Scholar]
- 30.Barondes SH, Jarvik ME. The Influence of Actinomycin-D on Brain Rna Synthesis and on Memory. J Neurochem. 1964;11:187–195. doi: 10.1111/j.1471-4159.1964.tb06128.x. [DOI] [PubMed] [Google Scholar]
- 31.Cohen HD, Barondes SH. Further studies of learning and memory after intracerebral actinomycin-D. J Neurochem. 1966;13:207–211. doi: 10.1111/j.1471-4159.1966.tb06793.x. [DOI] [PubMed] [Google Scholar]
- 32.Roberson ED, Sweatt JD. A biochemical blueprint for long-term memory. Learn Mem. 1999;6:381–388. [PMC free article] [PubMed] [Google Scholar]
- 33.Selcher JC, Weeber EJ, Varga AW, Sweatt JD, Swank M. Protein kinase signal transduction cascades in mammalian associative conditioning. Neuroscientist. 2002;8:122–131. doi: 10.1177/107385840200800208. [DOI] [PubMed] [Google Scholar]
- 34.Levenson JM, et al. A bioinformatics analysis of memory consolidation reveals involvement of the transcription factor c-rel. J Neurosci. 2004;24:3933–3943. doi: 10.1523/JNEUROSCI.5646-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levenson JM, et al. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 36.Selden NR, Everitt BJ, Jarrard LE, Robbins TW. Complementary roles for the amygdala and hippocampus in aversive conditioning to explicit and contextual cues. Neuroscience. 1991;42:335–350. doi: 10.1016/0306-4522(91)90379-3. [DOI] [PubMed] [Google Scholar]
- 37.Phillips RG, LeDoux JE. Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behav Neurosci. 1992;106:274–285. doi: 10.1037//0735-7044.106.2.274. [DOI] [PubMed] [Google Scholar]
- 38.Kim JJ, Rison RA, Fanselow MS. Effects of amygdala, hippocampus, and periaqueductal gray lesions on short- and long-term contextual fear. Behav Neurosci. 1993;107:1093–1098. doi: 10.1037//0735-7044.107.6.1093. [DOI] [PubMed] [Google Scholar]
- 39.Fanselow MS, Kim JJ, Yipp J, De Oca B. Differential effects of the N-methyl-D-aspartate antagonist DL-2-amino-5-phosphonovalerate on acquisition of fear of auditory and contextual cues. Behav Neurosci. 1994;108:235–240. doi: 10.1037//0735-7044.108.2.235. [DOI] [PubMed] [Google Scholar]
- 40.Atkins CM, Selcher JC, Petraitis JJ, Trzaskos JM, Sweatt JD. The MAPK cascade is required for mammalian associative learning. Nat Neurosci. 1998;1:602–609. doi: 10.1038/2836. [DOI] [PubMed] [Google Scholar]
- 41.Selcher JC, Atkins CM, Trzaskos JM, Paylor R, Sweatt JD. A necessity for MAP kinase activation in mammalian spatial learning. Learn Mem. 1999;6:478–490. doi: 10.1101/lm.6.5.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rampon C, et al. Enrichment induces structural changes and recovery from nonspatial memory deficits in CA1 NMDAR1-knockout mice. Nat Neurosci. 2000;3:238–244. doi: 10.1038/72945. [DOI] [PubMed] [Google Scholar]
- 43.Gräff J, Mansuy IM. Epigenetic codes in cognition and behaviour. Behav Brain Res. 2008 doi: 10.1016/j.bbr.2008.01.021. in press. [DOI] [PubMed] [Google Scholar]
- 44.Wood MA, Hawk JD, Abel T. Combinatorial chromatin modifications and memory storage: a code for memory. Learn Mem. 2006;13:241–244. doi: 10.1101/lm.278206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 46.Oike Y, et al. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: implications for a dominant-negative mechanism. Hum Mol Genet. 1999;8:387–396. doi: 10.1093/hmg/8.3.387. [DOI] [PubMed] [Google Scholar]
- 47.Bourtchouladze R, et al. A mouse model of Rubinstein-Taybi syndrome: defective long-term memory is ameliorated by inhibitors of phosphodiesterase 4. Proc Natl Acad Sci U S A. 2003;100:10518–10522. doi: 10.1073/pnas.1834280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alarcon JM, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 50.Wood MA, Attner MA, Oliveira AM, Brindle PK, Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn Mem. 2006;13:609–617. doi: 10.1101/lm.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14:268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bredy TW, Barad M. The histone deacetylase inhibitor valproic acid enhances acquisition, extinction, and reconsolidation of conditioned fear. Learn Mem. 2008;15:39–45. doi: 10.1101/lm.801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lattal KM, Barrett RM, Wood MA. Systemic or intrahippocampal delivery of histone deacetylase inhibitors facilitates fear extinction. Behav Neurosci. 2007;121:1125–1131. doi: 10.1037/0735-7044.121.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 56.Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 58.Rubinstein JH, Taybi H. Broad thumbs and toes and facial abnormalities. A possible mental retardation syndrome. Am J Dis Child. 1963;105:588–608. doi: 10.1001/archpedi.1963.02080040590010. [DOI] [PubMed] [Google Scholar]
- 59.Petrij F, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional coactivator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 60.Blough RI, et al. Variation in microdeletions of the cyclic AMP-responsive element-binding protein gene at chromosome band 16p13.3 in the Rubinstein-Taybi syndrome. Am J Med Genet. 2000;90:29–34. doi: 10.1002/(sici)1096-8628(20000103)90:1<29::aid-ajmg6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 61.Ellaway C, Christodoulou J. Rett syndrome: clinical characteristics and recent genetic advances. Disabil Rehabil. 2001;23:98–106. doi: 10.1080/09638280150504171. [DOI] [PubMed] [Google Scholar]
- 62.Sirianni N, Naidu S, Pereira J, Pillotto RF, Hoffman EP. Rett syndrome: confirmation of X-linked dominant inheritance, and localization of the gene to Xq28. Am J Hum Genet. 1998;63:1552–1558. doi: 10.1086/302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 64.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greenough WT, Wood WE, Madden TC. Possible memory storage differences among mice reared in environments varying in complexity. Behav Biol. 1972;7:717–722. doi: 10.1016/s0091-6773(72)80078-6. [DOI] [PubMed] [Google Scholar]
- 66.Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodeling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- 67.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Medini P, Pizzorusso T. Visual experience and plasticity of the visual cortex: a role for epigenetic mechanisms. Front Biosci. 2008;13:3000–3007. doi: 10.2741/2905. [DOI] [PubMed] [Google Scholar]
- 69.Putignano E, Lonetti G, Cancedda L, Ratto G, Costa M, Maffei L, Pizzorusso T. Developmental downregulation of histone posttranslational modifications regulates visual cortical plasticity. Neuron. 2007;53:747–759. doi: 10.1016/j.neuron.2007.02.007. [DOI] [PubMed] [Google Scholar]