Abstract
From January 2001 to December 2002, 587 strains of gram-negative bacterial isolates demonstrating resistance to ceftazidime and a combination of sulbactam and cefoperazone were subjected to a disk diffusion screening test using sodium mercaptoacetic acid; 431 strains (73.4%) appeared to produce metallo-β-lactamase (MBL). Of these 431 strains, 357 were found by PCR to carry genes for IMP-1 type MBL (blaIMP-1), while only 7 and 67 strains carried the IMP-2 gene (blaIMP-2) and the VIM-2 gene (blaVIM-2), respectively. Neither VIM-1 nor SPM-1 type MBL genes were found among the strains tested. Of 431 strains, 427 carried the intI1 gene, and 4 strains carrying both the intI1 and intI3 genes were reidentified as Pseudomonas putida harboring blaIMP-1. Of these four P. putida strains, three strains and one strain, respectively, were separately isolated from two hospitals located in the same prefecture, and the three strains showed very similar pulsed-field gel electrophoresis patterns. Of 357 blaIMP-1 carriers, 116, 53, 51, 47, and 30 strains were identified as Pseudomonas aeruginosa, Alcaligenes xylosoxidans, P. putida/fluorescens, Serratia marcescens, and Acinetobacter baumannii, respectively. Four strains carrying blaIMP-2 were reidentified as P. putida. Sixty-three P. aeruginosa strains and four P. putida strains carried blaVIM-2. Of 427 intI1-positive strains, 180, 53, 51, 47, and 35 were identified as P. aeruginosa, A. xylosoxidans, P. putida/fluorescens, S. marcescens, and A. baumannii, respectively. In the present study, it was confirmed that strains carrying blaIMP-1 with a class 1 integron are the most prevalent type in Japan, although several intI3 carriers have also been identified sporadically in this country.
Since metallο-β-lactamases (MBLs) can hydrolyze a very wide range of broad-spectrum β-lactams, MBL-producing gram-negative bacteria usually demonstrate consistent resistance to a variety of broad-spectrum β-lactams, including oxyiminocephalosporins, cephamycins, and carbapenems, which are the last resort for control of infections caused by gram-negative bacteria. Thus, MBL-producing gram-negative bacteria have been recognized to be among the most important nosocomial pathogens (3), and further proliferation of these strains in clinical settings will pose a serious global problem in the future (8). For this reason, aggressive surveillance of MBL producers with respect to the classification of the genetic determinant for MBLs as well as the integron will be extremely important.
At least three major groups of plasmid-mediated MBLs—the IMP, VIM, and SPM types—have been recognized worldwide, and their genetic determinants are often associated with integrons (5, 9). IMP-1-producing Serratia marcescens was initially identified in Japan in 1991 (14), and in 1997 VIM-1 and SPM-1 producers were also isolated, in Italy (10) and Brazil (23), respectively. After these reports, variants of these MBL types have been reported from almost every global region. For the IMP and VIM types of MBLs, at least 12 and 6 variants, respectively, have been published or reported to date (http://www.lahey.org/studies/other.stm#table 1). Moreover, genes for these MBLs are usually located in integrons that successfully accumulate many antibiotic-resistant gene cassettes as a gene cluster (22).
TABLE 1.
PCR primers for detection of MBL and integrase genes
| Gene | Primer name (sequence) | Expected size of amplicon (bp) | Reference or source |
|---|---|---|---|
| MBL genes | |||
| blaIMP-1 | F1 (5′-ACC GCA GCA GAG TCT TTG CC-3′) | 587 | This study |
| R1 (5′-ACA ACC AGT TTT GCC TTA CC-3′) | |||
| blaIMP-2 | F2 (5′-GTT TTA TGT GTA TGC TTC C-3′) | 678 | This study |
| R2 (5′-AGC CTG TTC CCA TGT AC-3′) | |||
| blaVIM-1 | F3 (5′-AGT GGT GAG TAT CCG ACA G-3′) | 261 | 24 |
| R3 (5′-ATG AAA GTG CGT GGA GAC-3′) | |||
| blaVIM-2 | F4 (5′-ATG TTC AAA CTT TTG AGT AAG-3′) | 801 | 17a |
| R4 (5′-CTA CTC AAC GAC TGA GCG-3′) | |||
| blaSPM-1 | F5 (5′-GCG TTT TGT TTG TTG CTC-3′) | 786 | This study |
| R5 (5′-TTG GGG ATG TGA GAC TAC-3′) | |||
| Integrase genes | |||
| intI1 | F6 (5′-GCA TCC TCG GTT TTC TGG-3′) | 457 | This study |
| R6 (5′-GGT GTG GCG GGC TTC GTG-3′) | |||
| intI2 | F7 (5′-CAC GGA TAT GCG ACA AAA AGG T-3′) | 789 | This study |
| R7 (5′-GTA GCA AAC GAG TGA CGA AAT G-3′) | |||
| intI3 | F8 (5′-ATC TGC CAA ACC TGA CTG-3′) | 922 | This study |
| R8 (5′-CGA ATG CCC CAA CAA CTC-3′) | |||
| Coamplification of the intI3-blaIMP-1 region | F9 (5′-GGT CTT GTA GGC TGT AAT TG-3′) | 609 | This study |
| R9 (5′-TTG TGG CTT GGA ACC TTT AC-3′) |
Since the nucleotide sequence appearing in reference 17 has a typographical error, the accurate sequence is shown in this table.
At least nine genetically different integrons have been identified in various bacterial species (5), and class 1 (9), class 2 (16), and class 3 (1) integrons are often found in pathogenic gram-negative bacilli including Pseudomonas aeruginosa, Pseudomonas putida, Acinetobacter spp., Escherichia coli, S. marcescens, Citrobacter freundii, and Salmonella spp. (15). Among these integrons, those in class 1 and class 3 have been reported to carry genetic determinants for MBLs. An IMP-1 type MBL associated with a class 1 integron was initially found in an S. marcescens clinical isolate in Japan (14), and integron-associated IMP-1 type MBLs were subsequently found in various gram-negative bacterial species such as Klebsiella pneumoniae, C. freundii, Enterobacter aerogenes, Enterobacter cloacae, E. coli, Proteus vulgaris, Acinetobacter spp., Alicaligenes spp., and P. putida (2, 20, 21).
In 1993, a class 3 integron-mediating IMP-1 was first identified by Arakawa et al. in an S. marcescens strain isolated in Japan (1). The organization of a class 1 integron that carries the gene for IMP-1 type MBL was characterized for P. aeruginosa (9). Although various class 1 integrons that convey MBL genes have been reported from a variety of gram-negative bacterial species isolated from different geographical regions, only a few isolates carrying class 3 integrons have been reported to date. Recently, intI3, a genetic determinant for class 3 integrons, was identified in a class A β-lactamase (GES-1)-producing K. pneumoniae strain isolated in Portugal (EMBL accession no. AY219651). This finding may suggest the potential for a future worldwide dissemination of bacteria carrying class 3 integrons in addition to the widely dispersal of class 1 integrons. In the present study, we characterized the types of MBLs and integrons found in various gram-negative bacteria isolated in Japanese clinical environments.
MATERIALS AND METHODS
Bacterial strains.
From January 2001 to December 2002, 978 strains belonging to gram-negative bacterial species were submitted to the reference laboratory for antibiotic resistance at the Department of Bacterial Pathogenesis and Infection Control, National Institute of Infectious Diseases, Tokyo, Japan, for typing and/or characterization of β-lactamases, including extended-spectrum β-lactamases (ESBLs), AmpC or CMY type class C cephamycinases, and class B MBLs. Of these strains, 587 demonstrating high-level resistance to both ceftazidime and sulbactam-cefoperazone (MICs, ≥128 μg/ml) were selected for screening of MBL production, because, from our experience, most MBL-producing gram-negative bacteria demonstrate very high levels of resistance to these agents. In the case of Acinetobacter baumannii, however, strains for which MICs of ceftazidime and sulbactam-cefoperazone were ≥16 μg/ml were also selected for screening in order to prevent overlooking of MBL producers. This is because several MBL-producing Acinetobacter strains were found to demonstrate low-level resistance to these agents in our preliminary study, and no data were available on the distribution of MICs of ceftazidime and sulbactam-cefoperazone for MBL-producing Acinetobacter spp. Stenotrophomonas maltophilia and Chryseobacterium spp., which produce chromosomal MBLs, were excluded from this study. Strains to be tested were collected from 108 Japanese hospitals, with only one strain selected from each patient.
Screening of MBL producers.
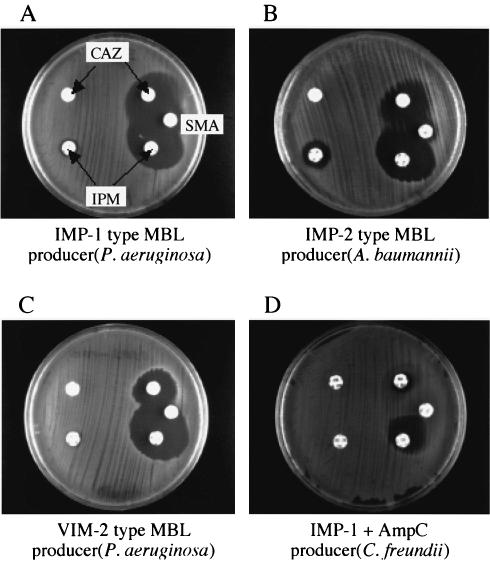
Strains selected by the criteria described above were subjected to a screening test for MBL production by using disks containing an MBL inhibitor (2). Two Kerby-Bauer disks containing 30 μg of ceftazidime and one disk containing 3 mg of sodium mercaptoacetic acid (SMA) (Eiken Chemical Co. Ltd., Tokyo, Japan) were used in the screening test. When the strains produce MBL together with a large amount of ESBL, AmpC cephalosporinase, or CMY type enzymes as well as bacterial membrane alterations, no evident expansion of the growth inhibition zone around the ceftazidime disk tends to appear. For such strains, a disk containing 10 μg of imipenem or meropenem was used instead of the ceftazidime disk. However, disks containing 10 μg of imipenem or meropenem are not suitable for screening all strains that produce MBL only, because the MICs of imipenem and meropenem for several MBL producers are lower than 8 μg/ml, as reported in previous studies (2, 20). Appearance of a large growth inhibition zone around the disks containing imipenem or meropenem sometimes leads to an incorrect judgment. Several samples of the SMA-test results are shown in Fig. 1.
FIG. 1.
Inhibition test for detection of MBL producers by use of SMA. Sample results of inhibition tests using SMA for detection of strains producing IMP-1, IMP-2, or VIM-2 type MBLs are shown. For MBL-producing strains, a clear expansion of the growth inhibition zone appears between the disks containing ceftazidime and SMA, respectively (A to C). In strains producing MBL as well as ESBLs or AmpC or CMY type cephalosporinases, the expansion of the growth inhibition zone is usually unclear. For such strains, the use of disks containing imipenem or meropenem instead of ceftazidime is recommended. By this substitution of disks, a clear growth inhibition zone appears between the disks containing imipenem and SMA, respectively (D). The MIC of imipenem or meropenem for MBL producers is sometimes lower than 32 μg/ml, but that of ceftazidime is usually higher than 64 μg/ml. Thus, disks containing 30 μg of ceftazidime are suitable for the first testing to detect MBL producers. IPM, imipenem; CAZ, ceftazidime.
PCR detection of genes for MBLs and integrases.
Although PCR analysis can predict only the approximate types of genes, it is suitable for testing a large number of samples at the same time. In the present study, therefore, we used this method for the rough classification of genetic determinants for MBLs and integrases. PCR analyses for the detection of MBL genes were carried out for all strains for which the screening test using SMA disks gave positive results. PCR amplification for the detection of genes for MBL and integrases was performed according to the method reported by Senda et al. (20), using each positive-control strain to avoid false-negative results. The PCR primers for blaVIM-1 and blaVIM-2 were used with the same PCR conditions as those for the other types of MBL genes. The five primer sets used in this study for amplifying MBL genes are shown in Table 1. PCR amplification of integrase genes was also performed as described above, by using three primer sets shown in Table 1.
Sequencing analyses of PCR amplicons.
Sequencing analyses on both strands were performed on five amplicons in each blaIMP-1-, blaIMP-2-, or blaVIM-2-positive strain and on five amplicons in the intI1-positive strain. All four amplicons from intI3-positive strains were sequenced from both sides. PCR amplicons were purified by use of the Qiaquick PCR purification kit (Qiagen K.K., Tokyo, Japan) prior to the labeling reaction. DNA sequences were determined by using BigDye Primer Cycle Sequencing Ready Reaction kits (Applied Biosystems, Foster City, Calif.) and an ABI model 377 DNA sequence analyzer (Perkin-Elmer Applied Biosystems, Foster City, Calif.). Nucleotide and amino acid sequences were analyzed by the GENETYX program (version 11.0, available at http://www.sdc.co.jp/genetyx/; SDC Co. Ltd., Tokyo, Japan) and were submitted to the DNA Data Bank of Japan (DDBJ) database to check the identity or similarity of each sequence by using FASTA (http://www.ddbj.nig.ac.jp/E-mail/homology-j.html).
PCR analysis of the relationship between the intI3 and blaIMP-1 genes.
A PCR primer set (Table 1) was made for coamplification of the intI3 and blaIMP-1 genes. By this PCR analysis, a fragment containing the 5′ regions of both the intI3 and blaIMP-1 genes is able to be amplified, so that the distance and relationship between the intI3 and blaIMP-1 genes can be measured.
Reidentification of blaVIM-2- or intI3-positive strains of P. putida/fluorescens.
It is sometimes difficult to distinguish P. putida from Pseudomonas fluorescens by the routine identification protocol that depends on biochemical profiles. Therefore, four strains of P. putida/fluorescens carrying blaVIM-2 were reidentified by a sequencing analysis of their 16S rRNA according to the method described previously (19). Four intI3-positive strains of P. putida/fluorescens were also reidentified in the same manner.
Pulsed-field gel electrophoretic (PFGE) analysis of intI3-positive strains.
Chromosomal DNAs prepared from four strains of P. putida were embedded in agarose gel plugs (InCert; Bio-Whittaker Molecular Applications, Rockland, Maine). Plugs were mounted in wells of agarose and electrophoresed after treatment with reaction mixtures according to the method described previously (20).
RESULTS
Screening of MBL production by using an SMA disk.
Of 587 strains tested, 431 (73.4%) appeared to produce MBL, as determined by the screening tests using disks containing a thiol compound (2), SMA. P. aeruginosa (n = 116), Alcaligenes xylosoxidans (n = 53), P. putida/fluorescens (n = 51), S. marcescens (n = 47), A. baumannii (n = 30), K. pneumoniae (n = 23), and E. coli (n = 17) were the main species of SMA test-positive strains (Table 2). One hundred fifty-six strains were SMA negative even when a combination of disks containing imipenem and SMA, respectively, were used together. Since many of these strains were susceptible to meropenem, they were speculated to be either producers of class A enzymes such as ESBLs, hyperproducers of chromosomal class C enzymes, or producers of plasmid-mediated AmpC and CMY type cephamycinases; some of them may also acquire altered membrane permeability to β-lactams. Further characterization of these strains will be performed in the next study.
TABLE 2.
Results of screening tests using SMA disks
| Bacterial species | No. of strains testing:
|
Total | |
|---|---|---|---|
| Positive | Negative | ||
| Pseudomonas aeruginosa | 180 | 48 | 228 |
| Pseudomonas putida/fluorescens | 55 | 1 | 56 |
| Alcaligenes xylosoxidans | 53 | 0 | 53 |
| Serratia marcescens | 47 | 9 | 56 |
| Acinetobacter baumannii | 35 | 13 | 48 |
| Klebsiella pneumoniae | 23 | 16 | 39 |
| Escherichia coli | 17 | 40 | 57 |
| Enterobacter cloacae | 5 | 8 | 13 |
| Burkholderia cepacia | 5 | 2 | 7 |
| Citrobacter freundii | 3 | 8 | 11 |
| Klebsiella oxytoca | 2 | 4 | 6 |
| Providencia rettgeri | 2 | 0 | 2 |
| Alcaligenes faecalis | 1 | 0 | 1 |
| Morganella morganii | 1 | 2 | 3 |
| Acinetobacter lwoffii | 1 | 1 | 2 |
| Enterobacter aerogenes | 1 | 3 | 4 |
| Proteus spp. | 0 | 1 | 1 |
| Total | 431 | 156 | 587 |
PCR typing of MBL genes.
Of 431 SMA-positive strains, 357 appeared to carry blaIMP-1 or one of its mutants such as blaIMP-3 or blaIMP-6, which are indistinguishable from blaIMP-1 with the PCR primers and conditions used in this study. Of these 357 strains, 116, 53, 51, 47, 30, and 23 were identified as P. aeruginosa, A. xylosoxidans, P. putida/fluorescens, S. marcescens, A. baumannii, and K. pneumoniae, respectively (Table 3). Sixty-seven and seven strains appeared to carry blaVIM-2 and blaIMP-2, respectively, and all these strains harbored the intI1 gene (Tables 3 and 4). No isolate carrying two or more different types of MBL genes together was found among the strains tested. Of 67 blaVIM-2-positive strains, 63 were P. aeruginosa and the remaining 4 were reidentified as P. putida (Table 3) by 16S rRNA sequencing. As for seven blaIMP-2-positive strains, one was P. aeruginosa and five were A. baumannii (Table 3). All SMA-positive strains were found to carry one of the MBL genes tested in this study, although no strain harboring blaVIM-1 or blaSPM-1 was detected among them.
TABLE 3.
Number of strains of each MBL type among SMA test-positive strains
| Bacterial species | No. of strains of type:
|
Total no. of strains | ||
|---|---|---|---|---|
| IMP-1 | IMP-2 | VIM-2 | ||
| Pseudomonas aeruginosa | 116 | 1 | 63 | 180 |
| Pseudomonas putida/fluorescens | 51 | 0 | 4a | 55 |
| Alcaligenes xylosoxidans | 53 | 0 | 0 | 53 |
| Serratia marcescens | 47 | 0 | 0 | 47 |
| Acinetobacter baumannii | 30 | 5 | 0 | 35 |
| Klebsiella pneumoniae | 23 | 0 | 0 | 23 |
| Escherichia coli | 17 | 0 | 0 | 17 |
| Enterobacter cloacae | 5 | 0 | 0 | 5 |
| Burkholderia cepacia | 5 | 0 | 0 | 5 |
| Citrobacter freundii | 3 | 0 | 0 | 3 |
| Klebsiella oxytoca | 2 | 0 | 0 | 2 |
| Providencia rettgeri | 2 | 0 | 0 | 2 |
| Alcaligenes faecalis | 1 | 0 | 0 | 1 |
| Morganella morganii | 1 | 0 | 0 | 1 |
| Acinetobacter lwoffii | 0 | 1 | 0 | 1 |
| Enterobacter aerogenes | 1 | 0 | 0 | 1 |
| Total | 357 | 7 | 67 | 431 |
These strains were reidentified as P. putida.
TABLE 4.
Combination of MBL and intI genes among all SMA test-positive strains
| Type of MBL | No. of strains with the following type of integrase gene:
|
Total no. of strains | |
|---|---|---|---|
| intI1 | intI1 + intI3 | ||
| IMP-1 | 353 | 4a | 357 |
| IMP-2 | 7 | 0 | 7 |
| VIM-2 | 67 | 0 | 67 |
| Total | 427 | 4 | 431 |
Reidentified as P. putida.
PCR typing of integrase genes.
Of 431 SMA-positive strains, 427 appeared to carry the intI1 gene. Among these 427 strains, 180, 53, 51, 47, 35, and 23 were identified, respectively, as P. aeruginosa, A. xylosoxidans, P. putida/fluorescens, S. marcescens, A. baumannii, and K. pneumoniae. Only four strains appeared to carry both the intI1 and intI3 genes, and these were reidentified as P. putida (Table 4) by sequencing of 16S rRNA as well as by the conventional identification protocol depending on the characteristic biochemical properties of each bacterial species. Of 51 P. putida/fluorescens strains harboring only the intI1 gene, 47 and 4 appeared to carry blaIMP-1 and blaVIM-2, respectively (Table 5). All four P. putida strains positive for both the intI1 and intI3 genes carried the IMP-1 type MBL gene. The four intI1-positive strains carrying the VIM-2 type MBL gene were also reidentified as P. putida (Table 5).
TABLE 5.
Combination of MBL and intI genes in P. putida/fluorescens
| Type of MBL | No. of strains with the following type of integrase gene:
|
Total no. of strains | |
|---|---|---|---|
| intI1 | intI1 + intI3 | ||
| IMP-1 | 47 | 4a | 51 |
| VIM-2 | 4a | 0 | 4 |
| Total | 51 | 4 | 55 |
Reidentified as P. putida.
Sequencing analyses of bla and intI genes.
The nucleotide sequences of 15 amplicons, consisting of 5 blaIMP-1, 5 blaIMP-2, and 5 blaVIM-2 amplicons, were consistent with the types predicted by the preceding PCR analyses, at least within the sequenced areas. Similarly, the nucleotide sequences of nine amplicons, consisting of five intI1 and four intI3 genes, also coincided with the results predicted by the PCR analyses.
Relationships of the intI3 and blaIMP-1 genes.
According to the result of a PCR analysis using a set of primers for amplification of a 609-bp fragment in the intI3-blaIMP-1 region (Table 1), the intI3 gene was suggested to be located adjacent to the blaIMP-1 gene in the configuration reported previously (1) in all four P. putida strains carrying both the intI3 and blaIMP-1 genes.
P. putida strains carrying both the intI1 and intI3 genes.
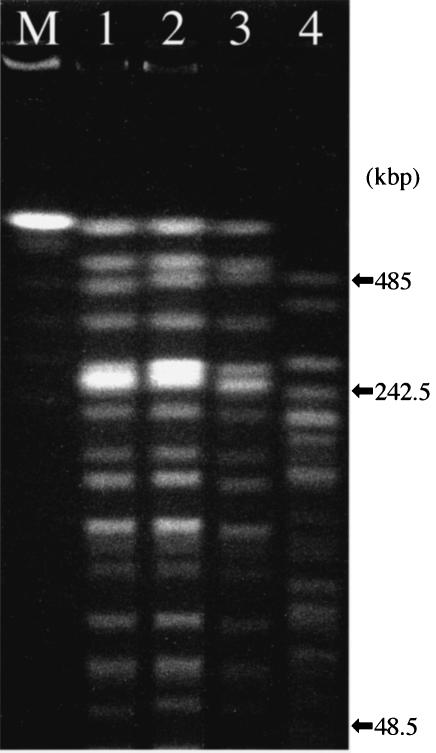
Of four P. putida strains carrying both the intI1 and intI3 genes, three were isolated at hospital A and one was isolated at hospital B; both these hospitals are located in Mie Prefecture. Table 6 shows the clinical associations of these four strains. The SpeI-digested genomic DNA patterns of the four clinical isolates were classified into two types (Fig. 2). The three strains isolated in hospital A, NCB 01-121, NCB 02-182, and NCB 02-204, demonstrated very similar PFGE patterns, suggesting a clonal lineage.
TABLE 6.
Clinical associations of P. putida isolates carrying both the intI1 and intI3 genesa
| Strain no. | Age of patient (yr) | Sex of patient | Disease | Specimen type | Hospital | Date of isolation (mo/yr) |
|---|---|---|---|---|---|---|
| NCB 01-121 | 77 | Male | Prostatic cancer | Urine | A | 6/2001 |
| NCB 02-182 | 79 | Male | Cerebral infarction | Urine | A | 5/2002 |
| NCB 02-190 | 66 | Female | Cerebral infarction | Sputum | B | 5/2002 |
| NCB 02-204 | 76 | Female | Gallbladder cancer | Biliary tract drainage tube | A | 6/2002 |
All these strains were isolated in Mie Prefecture.
FIG. 2.
PFGE of four P. putida strains carrying both the intI1 and intI3 genes. Lanes: M, size marker for PFGE; 1, P. putida strain NCB 01-121; 2, strain NCB 02-182; 3, strain NCB 02-204; 4, strain NCB 02-190. Clinical associations of the strains subjected to this PFGE analysis are listed in Table 6.
DISCUSSION
Since 1988, transferable carbapenem resistance has been found in several P. aeruginosa strains isolated in Toyama Prefecture, Japan (12, 25). In 1991, an IMP-1 type MBL, initially characterized in a strain of S. marcescens, gave high-level resistance to various broad-spectrum β-lactams including imipenem (6, 14). This strain was isolated in a hospital in Aichi Prefecture and had the intI1 gene just upstream of the blaIMP-1 gene cassette on the chromosome. In 1993, several S. marcescens strains harboring a plasmid-mediated blaIMP-1 gene were also identified in Aichi Prefecture (1, 6). Several of these strains were found to carry a novel integron-like element that was classified as a class 3 integron (5). The newly identified class 3 integrase, IntI3, has about 60.9% identity to the previously identified IntI1 at the amino acid sequence level (1). The genomic organization of a class 1 integron was characterized in a blaIMP-1-carrying strain (9) which was isolated in Japan. The emergence and development of carbapenem resistance through the acquisition of genes for MBLs has since become a matter of general concern, especially in gram-negative bacteria (13, 21).
At least three genetically different clusters of MBLs have been found to date: IMP-1 to IMP-12 (4), VIM-1 to VIM-6 (EMBL accession no. AY165025), and SPM-1 (23) have been published or registered with the EMBL/GenBank database. Almost all of their genetic determinants are associated with class 1 integrons, and only a few strains have been found to carry class 3 integrons to date. In the present study, almost all MBL genes were found to correlate with class 1 integrons. To our knowledge, only S. marcescens strain AK9373 and a small number of strains isolated in Aichi Prefecture carry the intI3 gene (N. Shibata, Y. Arakawa, H. Kurokawa, Y. Doi, and K. Shibayama, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. C524, 2001). Recently however, a class 3 integron mediating a gene for a GES-1 type class A β-lactamase, a kind of ESBL, was submitted from Portugal to EMBL/GenBank (accession no. AY219651). Because intI genes are usually accompanied by genes for aminoglycoside-modifying enzymes such as aacA4 or aadA1, these gene cassettes in integrons may be derived from some nonpathogenic bacteria such as aminoglycoside-producing actinomycetes, which are widely distributed in the natural environment. Therefore, the two strains possessing class 3 integrons found in Japan (1) and Portugal may have been generated independently of each other by the integration of separate β-lactamase gene cassettes in clinical environments, where the strong influence of antimicrobial agents has continued.
In the present study, we confirmed that class 1 is the most abundant type of integron in Japan, although four P. putida strains carrying both class 1 and class 3 integrons have been newly identified. Although the reason why these four strains carry two types of integron is not well understood at present, analyses of the organization and function of class 1 and class 3 integrons in these strains may elucidate their biological advantage in the future. In any case, since intI3 has already been identified in at least three gram-negative bacterial species (S. marcescens, K. pneumoniae, and P. putida) isolated from geographically different areas, the emergence and proliferation of class 3 integrons that carry various gene cassettes responsible for multiple antimicrobial resistance may become not so much a local problem as a global issue.
It was confirmed that the IMP-1 type of MBL is the most common MBL in Japan at present, although IMP-3 (7) and IMP-6 (27) have also been identified in Japan. Since very few amino acid substitutions exist among IMP-1, IMP-3, and IMP-6, the latter two MBLs are fundamentally variants of IMP-1. It is difficult to confirm whether all the blaIMP-1-positive strains found in this study carry genuine blaIMP-1, since the primer sets used in this study cannot distinguish blaIMP-3 and blaIMP-6 from blaIMP-1. Strains producing IMP-3 or IMP-6 usually demonstrate susceptibility or low-level resistance to imipenem (MICs, 1 to 16 μg/ml) (7, 27). However, almost all blaIMP-1-positive strains identified in this study, especially in P. aeruginosa and S. marcescens, demonstrated high-level resistance to imipenem (MICs, >32 μg/ml), as was reported previously (6, 18, 20). In any event, the blaIMP-1-positive strains detected in this study appeared to produce MBLs belonging to the IMP-1 complex, consisting mainly of IMP-1 and probably small amounts of its variants, IMP-3 and IMP-6. Detailed typing of the MBLs produced by the 357 blaIMP-1-positive strains will be continued in the next study.
IMP-2 was initially found in A. baumannii in Italy (18), and its variant IMP-8 was also identified in K. pneumoniae in Taiwan (26). In the present study, one strain of P. aeruginosa and five strains of A. baumannii were found to carry the blaIMP-2 gene in Japan. IMP-2 is not a variant of IMP-1, since many amino acid substitutions are found between them (18). One has to wonder why IMP-2 has been found in such widely separated geographical regions as Europe and the Far East. The increased frequency of international travel and transportation may well be involved in the transmission of IMP-2 producers, although they may have emerged independently in each area. Precise characterization of the genetic organization of each integron is necessary to determine their origins and relationships.
The present study suggests that VIM-2 producers are also increasing in Japan, especially in Pseudomonas spp. VIM-2 was initially identified in France in 1996 (17). VIM-2 producers have occasionally caused outbreaks in many countries recently, including Korea (11), where VIM-2 producers are reportedly increasing (28). It is not well understood why VIM-2 producers are becoming predominant in Korea, a country adjacent to Japan, where IMP-1 producers are very common. A similar observation was made in the isolation of ESBL producers. Strains producing TEM- or SHV-derived ESBLs have been increasing in Korea, while such ESBL producers still remain rare in Japan, though producers of CTX-M type enzymes, including Toho-1, are becoming common in Japan. These discrepancies may depend on differences in the administration of antimicrobial agents.
In the present study, 16 gram-negative bacterial species were found to carry genes for at least three types of MBLs. Indeed, although P. aeruginosa, A. xylosoxidans, P. putida/fluorescens, and S. marcescens are the most frequent carriers of MBL genes among these 16 bacterial species, blaIMP-1 genes have already been transmitted into various bacterial species belonging to the family Enterobacteriaceae, such as E. coli, E. cloacae, E. aerogenes, C. freundii, K. pneumoniae, Providencia rettgeri, and Morganella morganii, through the mediation of class 1 integrons, which are usually carried on transferable plasmids. Isolation of MBL producers belonging to these bacterial species is still very rare even in Japan, which may be due to the instability of plasmids carrying MBL and integrase genes in these strains. However, the frequency of isolation of MBL producers belonging to these species is also likely to increase in the future, as it already has in P. aeruginosa and S. marcescens, if the gene cassettes or gene clusters locating in integron structures are transposed to stable resident plasmids or chromosomes in each bacterial species.
In hospital A, three strains of P. putida carrying intI1, intI3, and blaIMP-1 have been isolated from different patients for more than 12 months. Since these strains demonstrated very similar PFGE patterns, it is suggested that they are a clonal lineage and have been existing in the hospital for nearly 1 year. Since P. putida can grow at temperatures below 10°C and tends to be isolated from damp materials and environments such as plant surfaces, mops, sponges, and sinks, these strains might have survived in such nosocomial environments. Indeed, P. putida sometimes causes opportunistic infections such as respiratory and urinary tract infections, but its virulence is not as strong as that of P. aeruginosa. However, it can fully work as a reservoir of integrons and MBL genes. Thus, we should pay continuous attention even to such low-virulence bacterial species when they carry special genetic determinants such as MBL genes.
The blaIMP-1 genes move together with class 1 or class 3 integrons in many nosocomial gram-negative bacilli. Furthermore, genes for blaIMP-2 and blaVIM-2 have also been dispersing into various non-glucose-fermenting bacteria such as P. putida and A. baumannii. Hence, we should take special precautions against the further global dissemination of integron-associated MBL genes among a variety of nosocomial gram-negative bacterial species.
Acknowledgments
We thank all the medical institutions that submitted bacterial strains to the national reference laboratory. We are also much indebted to Masumi Matsuda and Manabu Nakano for kindly providing P. putida strains carrying both the intI1 and intI3 genes.
This work was mainly supported by four grants (H12-Shinkou-19, H12-Shinkou-20, H15-Shinkou-9, and H15-Shinkou-10) from the Ministry of Health, Labor and Welfare of Japan. PCR typing of MBL genes was supported by grant 13770141 from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Arakawa, Y., M. Murakami, K. Suzuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa, Y., N. Shibata, K. Shibayama, H. Kurokawa, T. Yagi, H. Fujiwara, and M. Goto. 2000. Convenient test for screening metallo-β-lactamase-producing gram-negative bacteria by using thiol compounds. J. Clin. Microbiol. 38:40-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush, K. 1998. Metallo-β-lactamases: a class apart. Clin. Infect. Dis. 27(Suppl. 1):S48-S53. [DOI] [PubMed] [Google Scholar]
- 4.Docquier, J. D., M. L. Riccio, C. Mugnaioli, F. Luzzaro, A. Endimiani, A. Toniolo, G. Amicosante, and G. M. Rossolini. 2003. IMP-12, a new plasmid-encoded metallo-β-lactamase from a Pseudomonas putida clinical isolate. Antimicrob. Agents Chemother. 47:1522-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall, R. M., C. M. Collis, M. J. Kim, S. R. Partridge, G. D. Recchia, and H. W. Stokes. 1999. Mobile gene cassettes and integrons in evolution. Ann. N. Y. Acad. Sci. 870:68-80. [DOI] [PubMed] [Google Scholar]
- 6.Ito, H., Y. Arakawa, S. Ohsuka, R. Wacharotayankun, N. Kato, and M. Ohta. 1995. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob. Agents Chemother. 39:824-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyobe, S., H. Kusadokoro, J. Ozaki, N. Matsumura, S. Minami, S. Haruta, T. Sawai, and K. O'Hara. 2000. Amino acid substitutions in a variant of IMP-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 44:2023-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurokawa, H., T. Yagi, N. Shibata, K. Shibayama, and Y. Arakawa. 1999. Worldwide proliferation of carbapenem-resistant gram-negative bacteria. Lancet 354:955. [DOI] [PubMed] [Google Scholar]
- 9.Laraki, N., M. Galleni, I. Thamm, M. L. Riccio, G. Amicosante, J. M. Frere, and G. M. Rossolini. 1999. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob. Agents Chemother. 43:890-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, K., J. B. Lim, J. H. Yum, D. Yong, Y. Chong, J. M. Kim, and D. M. Livermore. 2002. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob. Agents Chemother. 46:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minami, S., M. Akama, H. Araki, Y. Watanabe, H. Narita, S. Iyobe, and S. Mitsuhashi. 1996. Imipenem and cephem resistant Pseudomonas aeruginosa carrying plasmids coding for class B β-lactamase. J. Antimicrob. Chemother. 37:433-444. [DOI] [PubMed] [Google Scholar]
- 13.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 14.Osano, E., Y. Arakawa, R. Wacharotayankun, M. Ohta, T. Horii, H. Ito, F. Yoshimura, and N. Kato. 1994. Molecular characterization of an enterobacterial metallo-β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ploy, M. C., D. Chainier, N. H. Tran Thi, I. Poilane, P. Cruaud, F. Denis, A. Collignon, and T. Lambert. 2003. Integron-associated antibiotic resistance in Salmonella enterica serovar Typhi from Asia. Antimicrob. Agents Chemother. 47:1427-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ploy, M. C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki, T., T. Nishiyama, M. Shintani, and T. Kenri. 1997. Evaluation of a new method for identification of bacteria based on sequence homology of 16S rRNA gene. PDA J. Pharm. Sci. Technol. 51:242-247. [PubMed] [Google Scholar]
- 20.Senda, K., Y. Arakawa, S. Ichiyama, K. Nakashima, H. Ito, S. Ohsuka, K. Shimokata, N. Kato, and M. Ohta. 1996. PCR detection of metallo-β-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactams. J. Clin. Microbiol. 34:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senda, K., Y. Arakawa, K. Nakashima, H. Ito, S. Ichiyama, K. Shimokata, N. Kato, and M. Ohta. 1996. Multifocal outbreaks of metallo-β-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum β-lactams, including carbapenems. Antimicrob. Agents Chemother. 40:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 23.Toleman, M. A., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, R. N. Jones, and T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed] [Google Scholar]
- 24.Tsakris, A., S. Pournaras, N. Woodford, M. F. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe, M., S. Iyobe, M. Inoue, and S. Mitsuhashi. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan, J. J., W. G. Ko, and J. J. Wu. 2001. Identification of a plasmid encoding SHV-12, TEM-1, and a variant of IMP-2 metallo-β-lactamase, IMP-8, from a clinical isolate of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:2368-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yano, H., A. Kuga, R. Okamoto, H. Kitasato, T. Kobayashi, and M. Inoue. 2001. Plasmid-encoded metallo-β-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob. Agents Chemother. 45:1343-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yum, J. H., K. Yi, H. Lee, D. Yong, K. Lee, J. M. Kim, G. M. Rossolini, and Y. Chong. 2002. Molecular characterization of metallo-β-lactamase-producing Acinetobacter baumannii and Acinetobacter genomospecies 3 from Korea: identification of two new integrons carrying the blaVIM-2 gene cassettes. J. Antimicrob. Chemother. 49:837-840. [DOI] [PubMed] [Google Scholar]