Abstract
Lipoarabinomannan (LAM) is a major glycolipid in the mycobacterial cell envelope. LAM consists of a mannosylphosphatidylinositol (MPI) anchor, a mannan core and a branched arabinan domain. The termini of the arabinan branches can become substituted with one to three α(1→2)-linked mannosyl residues, the mannose cap, producing ManLAM. ManLAM has been associated with a range of different immunomodulatory properties of Mycobacterium tuberculosis during infection of the host. In some of these effects, the presence of the mannose cap on ManLAM appears to be crucial for its activity. So far, in the biosynthesis of the mannose cap on ManLAM, two enzymes have been reported to be involved: a mannosyltransferase that adds the first mannosyl residue of the mannose caps to the arabinan domain of LAM, and another mannosyltransferase that elongates the mannose cap up to three mannosyl residues. Here, we report that a third gene is involved, MMAR_2380, which is the Mycobacterium marinum orthologue of Rv1565c. MMAR_2380 encodes a predicted transmembrane acyltransferase. In M. marinum ΔMMAR_2380, the LAM arabinan domain is still intact, but the mutant LAM lacks the mannose cap. Additional effects of mutation of MMAR_2380 on LAM were observed: a higher degree of branching of both the arabinan domain and the mannan core, and a decreased incorporation of [1,2-14C]acetate into the acyl chains in mutant LAM as compared with the wild-type form. This latter effect was also observed for related lipoglycans, i.e. lipomannan (LM) and phosphatidylinositol mannosides (PIMs). Furthermore, the mutant strain showed increased aggregation in liquid cultures as compared with the wild-type strain. All phenotypic traits of M. marinum ΔMMAR_2380, the deficiency in the mannose cap on LAM and changes at the cell surface, could be reversed by complementing the mutant strain with MMAR_2380. Strikingly, membrane preparations of the mutant strain still showed enzymic activity for the arabinan mannose-capping mannosyltransferase similar to that of the wild-type strain. Although the exact function of MMAR_2380 remains unknown, we show that the protein is essential for the presence of a mannose cap on LAM.
INTRODUCTION
The complex mycobacterial cell envelope contains an exceptionally high amount of lipids. One of the major glycolipids present is lipoarabinomannan (LAM) (Briken et al., 2004; Chatterjee & Khoo, 1998; Nigou et al., 2002, 2003). With its mannosylphosphatidylinositol (MPI) anchor, LAM is probably inserted in both the cytoplasmic membrane and the outer membrane (Hoffmann et al., 2008; Pitarque et al., 2008). The glycan portion of LAM comprises a mannan core, which consists of α(1→6)-linked mannosyl residues substituted at some positions with single α(1→2)-linked mannoses, and an arabinan domain consisting of α(1→5)-linked arabinose (see Supplementary Fig. S1). This latter domain is branched with side chains that always end in β(1→2)-linked arabinosyl units (AraLAM). In mannose-capped LAM (ManLAM), a part of these terminal arabinosyl residues is substituted with an α(1→5)-linked mannosyl residue which can be further elongated by the addition of one or two α(1→2)-linked mannosyl residues, forming the mannose cap (Supplementary Fig. S1). Whether mannose caps are present on LAM depends on the Mycobacterium species (Pitarque et al., 2005).
Several immunomodulating properties of LAM have been reported which could codetermine the host–pathogen interaction during infection with pathogenic mycobacteria, such as Mycobacterium tuberculosis. Some of these have been linked specifically to the presence of the mannose cap on LAM, e.g. the binding of ManLAM to pattern recognition receptors such as the dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN) (Geijtenbeek et al., 2003; Maeda et al., 2003) and the macrophage mannose receptor (MMR) (Schlesinger et al., 1994), and its role in phagosome maturation arrest (Kang et al., 2005; Welin et al., 2008).
So far, two mannosyltransferases have been identified as being involved in the biosynthesis of the mannose cap on ManLAM. The first, encoded by Rv1635c, adds the first mannosyl residue of the mannose cap to the non-reducing arabinan termini (Dinadayala et al., 2006). Rv2181 encodes the second, cap-elongating mannosyltransferase, which besides lengthening of the mannose cap, substitutes the mannan core of LAM with the single α(1→2)-linked mannose moieties (Kaur et al., 2006, 2008). Both enzymes use polyprenylphosphomannose (PPM) as a sugar donor (Appelmelk et al., 2008; Kaur et al., 2006). A potential glycosyltransferase function of these enzymes has been predicted using bioinformatics tools. Although very successful, this approach is biased towards finding glycosyltransferases directly involved in the biosynthesis of glycolipids or glycoproteins, and proteins that exert more unexpected functions may be missed.
In an alternative approach to identify novel genes with a role in the biosynthesis of ManLAM, we have used a colony blot screen of a transposon-insertion mutant library in Mycobacterium marinum in which single-transposon mutants were probed with mannose cap-recognizing mAb 55.92.1A1 (Appelmelk et al., 2008). As recently described, this screen yielded the mutant in MMAR_2439, the M. marinum orthologue of Rv1635c. Subsequent mannosyltransferase assays and chemical analysis confirmed that the function of MMAR_2439 is the addition of the first mannosyl residue of the mannose cap to LAM, identical to Rv1635c (Appelmelk et al., 2008).
We have extended our search using the screening approach with the mannose cap-recognizing mAb. In this paper, we reveal a third gene involved in the biosynthesis of the mannose cap on LAM. In M. marinum ΔMMAR_2380, the LAM molecule is still intact, apart from lacking its mannose cap. Additional effects of the mutation of MMAR_2380 were seen in a higher degree of branching of both the arabinan domain and the mannan core in mutant LAM, decreased [1,2-14C]acetate incorporation in the acyl chains of mutant LAM and related lipoglycans, and an altered cell surface of the mutant strain as compared with wild-type M. marinum. MMAR_2380 encodes a predicted transmembrane acyltransferase, and here we explored potential roles for MMAR_2380 in the biosynthesis of LAM in M. marinum.
METHODS
Bacterial strains and growth conditions.
M. marinum strain E11 (Puttinaowarat et al., 1999) and Mycobacterium smegmatis mc2155 were grown in Middlebrook 7H9 broth (Difco) with 10 % Middlebrook albumin dextrose catalase (ADC) enrichment (BBL) and 0.05 % (v/v) Tween 80, or on Middlebrook 7H10 agar (Difco) with 10 % Middlebrook oleic acid albumin dextrose catalase (OADC) enrichment (BBL) at 30 and 37 °C, respectively. Escherichia coli DH5α was grown on LB at 37 °C. Mycobacterial growth was measured by determining OD600 with a UV/visible spectrophotometer (Jenway). The concentrations of antibiotics used were 25 μg kanamycin ml−1 and 50 μg hygromycin ml−1 for mycobacteria, and 100 μg hygromycin ml−1 for E. coli.
Immunoblotting assay.
Bacteria were grown until the late-exponential phase and disrupted with a Beadbeater (BioSpec). Protein concentration was measured with a bicinchoninic acid (BCA) Protein Assay (Pierce). Supernatants were run on a 10 % SDS-PAGE gel and immunostained after electroblotting on a PVDF membrane (Millipore). Murine mAbs of the IgM class were used. mAb F30-5 recognizes the branches of the arabinan domain of LAM (Appelmelk et al., 2008; Kolk et al., 1984). mAb 55.92.1A1 binds the mannose cap on ManLAM (Appelmelk et al., 2008). First the PVDF membranes were incubated in 0.5 % (w/v) blocking reagent (Boehringer) for 1 h at 37 °C, followed by another incubation of 2 h with the mAb diluted in 1 : 1 blocking buffer and PBS+0.05 % (v/v) Tween 80 (PBST). After thorough washing in PBST, the membranes were incubated with horseradish peroxidase (HRP)-labelled goat anti-mouse IgM (American Qualex) in PBST with 0.5 % (v/v) normal goat serum. After washing, the membranes were stained with 4-chloro-1-naphthol (4-CN; Bio-Rad) and 3,3′-diaminobenzidine.4HCl (DAB; Sigma).
Identification of a novel M. marinum mutant deficient in the biosynthesis of the mannose cap on LAM.
A transposon insertion mutant library was created in M. marinum E11 with the Himar1-based mariner transposon, which has little insertion site specificity (dinucleotide TA) (Rubin et al., 1999). The transposon was introduced via phagemid ϕMycoMarT7, as described by Sassetti et al. (2001). Mutants selected on kanamycin were screened in a colony blot assay for the absence of the mannose cap on LAM using mAb 55.92.1A1, as described previously (Appelmelk et al., 2008). In brief, single colonies were transferred to PVDF membranes with sterile toothpicks, after which the membranes were baked for 1 h at 70 °C and subsequently treated as in the immunoblotting assay described above. Mutants that were repeatedly negative with 55.92.1A1 in colony blots were examined further by SDS-PAGE and immunostaining. The location of the transposon insertion was determined by ligation-mediated PCR, as described previously (Abdallah et al., 2006). The exact insertion position of the transposon in gene MMAR_2380 was confirmed by standard PCR with the pMyco1-primer and a second primer, cap-RV, which binds about 210 bp downstream from MMAR_2380. The PCR product was sequenced on an ABI Prism 300 DNA sequencer (Applied Biosystems). To determine the direction of the kanamycin-resistance cassette in the transposon in MMAR_2380, two primers were used, MMAR_2380-FW and MMAR_2380-RV, with a binding site upstream or downstream of the transposon in gene MMAR_2380, respectively, and two primers binding to sequences within the transposon (the kanamycin cassette), Kana-FW and Kana-RV. See Supplementary Table S1 for primer sequences.
Complementation of the M. marinum ΔMMAR_2380 mutant strain.
MMAR_2380 was amplified from genomic M. marinum E11 wild-type DNA with primers cap-FW and cap-RV (Supplementary Table S1) using the Expand High Fidelity PCR kit (Roche). The obtained PCR product (2973 bp) was cloned into pCRII-TOPO. This plasmid was digested with HindIII and EcoRV and the fragment was ligated into HindIII/EcoRV-digested pSMT3-eGFP (Abdallah et al., 2006). In the resulting plasmid (pSMT3-eGFP-MMAR_2380), the gene of interest was located behind the GFP gene under the control of heat-shock promoter 60 (HSP60). The complementation plasmid was isolated from E. coli DH5α cells using the QIAprep Miniprep kit (Qiagen) and electroporated into M. marinum ΔMMAR_2380. Transformants (M. marinum ΔMMAR_2380 comp.) were selected on hygromycin.
Chemical analysis of the mannose cap on LAM.
The presence of the mannose cap in M. marinum wild-type and M. marinum ΔMMAR_2380 strains was analysed by capillary electrophoresis (CE), as described previously (Appelmelk et al., 2008; Nigou et al., 2000). In brief, purified LAM was partially degraded by controlled acid hydrolysis (0.1 M HCl for 20 min at 110 °C), and the oligosaccharides liberated were tagged with the fluorescent label 8-aminopyrene-1,3,6-trisulfonate (APTS). During CE, the labelled oligosaccharides were separated and peaks detected by laser-induced fluorescence, and elution times were compared with the appropriate standards.
Mannosyltransferase assay.
Membranes of M. marinum E11 wild-type, and the ΔMMAR_2380 and ΔMMAR_2380 complemented strains were prepared as follows: mycobacterial cells (10 g wet weight) were washed and resuspended in 30 ml buffer A, containing 50 mM MOPS (adjusted to pH 8.0 with KOH), 5 mM β-mercaptoethanol and 10 mM MgCl2 at 4 °C and subjected to probe sonication (Soniprep 150, MSE Sanyo; 1 cm probe) for a total time of 10 min in 60 s pulses with 90 s cooling intervals between pulses. The sonicate was centrifuged at 27 000 g for 20 min at 4 °C. Membrane fractions were obtained by centrifugation of the clarified lysate at 100 000 g for 1 h at 4 °C. The supernatant was carefully removed and the membranes gently resuspended in buffer A at a protein concentration of 20 mg ml−1. Protein concentrations were determined using the BCA Protein Assay Reagent kit (Pierce) (Lee et al., 1998). Assay conditions consisted of buffer A, 1 mM ATP, membranes (500 μg), 0.1 mM decaprenyl phosphate (stored in 1 % CHAPS as a 1 mM stock solution), acceptor Ara6 (1 mM) and GDP-[14C]mannose [0.25 μCi (9.25 kBq), 11 GBq mmol−1, Amersham] to a final reaction volume of 120 μl. Ara6 is synthetic aminooctyl-linked hexaarabinofuranoside, a motif representing the terminal, non-reducing part of LAM (Gadikota et al., 2003). The reaction mixtures were then incubated at 37 °C for 1 h. The enzymically synthesized products were extracted using ‘E-soak’ (water : ethanol : diethyl-ether : pyridine : ammonium hydroxide; 15 : 15 : 5 : 1 : 0.017 by vol.), which was added to the incubation tubes. These were mixed for 30 min and then centrifuged at 18 000 g. The supernatant was recovered and dried under reduced pressure with a Genevac EZ-2 evaporator, resuspended in water (1 ml) and loaded onto a pre-equilibrated Alltech C18 Sep-Pak cartridge (600 mg). The cartridge was washed with 5 ml water, and the enzymically synthesized products were eluted with methanol (5 ml) and subsequently dried under vacuum as described above. The dried eluate was resuspended in water (1 ml) and loaded onto a pre-equilibrated Supelco strong anion exchange (SAX) cartridge, which was washed with 5 ml water. The water eluate was collected, dried under vacuum and resuspended in 200 μl water, and contained the 14C-mannosylated acceptor Ara6 product. The total c.p.m. of recovered radiolabelled material with this two-step separation protocol was measured by scintillation counting of 10 % of the labelled material in 10 ml EcoScintA (National Diagnostics). The incorporation of [14C]mannose was determined by subtracting counts present in control assays (reaction components incubated in the absence of acceptor Ara6) (Appelmelk et al., 2008).
NMR analysis of the mannose cap, arabinan domain and mannan core of LAM.
NMR spectra of LAM from M. marinum wild-type and M. marinum ΔMMAR_2380 were recorded on a Bruker DMX-500 NMR spectrometer equipped with a double resonance (1H/X)-BBi z-gradient probe head. All samples were exchanged in D2O (99.97 % D, Euriso-top), with intermediate lyophilization, and then dissolved in 0.5 ml D2O and analysed at 313 K. The 1H and 13C NMR chemical shifts were referenced relative to internal acetone at 2.225 and 34.00 p.p.m. respectively. All the details concerning NMR sequences used and experimental procedures have been described in previous studies (Gilleron et al., 2000).
Glycosidic linkage analysis of LAM.
Glycosyl linkage composition was performed according to the modified procedure of Ciucanu & Kerek (1984). The per-O-methylated LAM was hydrolysed using 500 μl 2 M trifluoroacetic acid (TFA) at 110 °C for 2 h, reduced using 350 μl 10 mg ml−1 sodium borodeuteride [NaBD4 (1 M NH4OH : C2H5OH, 1 : 1, v/v)] and per-O-acetylated using 300 μl acetic anhydride for 1 h at 110 °C. The resulting alditol acetates were solubilized in cyclohexane before analysis by GC and GC/MS.
[1,2-14C]Acetate incorporation in total lipids.
M. marinum strains were grown as described above and metabolically labelled using 5 μCi ml−1 [1,2-14C]acetate [50–62 mCi mmol−1(1850–2294 MBq mmol−1), GE Healthcare, Amersham Bioscience] at OD600 0.4, and cultures were grown for a further 4 h at 37 °C with gentle shaking. Cells were harvested by centrifugation and washed once with PBS, and a small-scale apolar and polar lipid extraction was performed as described by Dobson et al. (1985). The lipid extracts were dried and resuspended in CHCl3 : CH3OH (2 : 1), and equal amounts of crude lipid (25 000 c.p.m.) were applied to the corners of 6.6×6.6 cm pieces of Merck 5554 aluminium-backed TLC plates. The plates were developed using solvent system E for polar lipids: chloroform : methanol : water (60 : 30 : 6, by vol.) in the first direction, and chloroform : acetic acid : methanol : water (40 : 25 : 3 : 6, by vol.) in the second direction (Besra, 1998; Dobson et al., 1985). Plates were dried and autoradiograms were produced by overnight exposure of Kodak X-Omat AR film to the TLC plates to reveal [1,2-14C]acetate-labelled lipids.
[1,2-14C]Acetate incorporation in lipoglycans.
Lipoglycans were extracted from [1,2-14C]acetate-labelled delipidated cells as previously described (Ludwiczak et al., 2001). Briefly, cells were broken by sonication (MSE Soniprep 150, 12 μm amplitude, 60 s on, 90 s off for 10 cycles, on ice) and the cell debris was refluxed five times with 50 % C2H5OH at 68 °C, for 12 h intervals. The cell debris was removed by centrifugation and the supernatant containing lipoglycans and neutral glycans was dried. The dried extract was then treated with 80 % hot phenol at 85 °C. The aqueous phase was dialysed and dried, followed by extensive treatments with α-amylase, DNase, RNase, chymotrypsin and trypsin to degrade any contamination. This fraction was dialysed to remove the low-molecular-mass breakdown products formed after the enzyme treatment, thus yielding the crude lipoglycan fraction. The crude lipoglycan extract was dried and resuspended in buffer A (50 mM ammonium acetate and 15 % propan-1-ol) and subjected to Octyl Sepharose CL-4B chromatography. The column (2.5×50 cm) was washed initially with four column volumes of buffer A to ensure removal of neutral glycans, followed by buffer B (50 mM ammonium acetate and 50 % propan-1-ol). The eluent was collected and concentrated to approximately 1 ml and precipitated using 5 ml C2H5OH. The sample was dried using a Savant Speedvac concentrator and resuspended in water, and the incorporation of [1,2-14C]acetate into lipoglycans was determined. Furthermore, 25 μg lipoglycans was loaded on a 15 % SDS-PAGE gel and developed using autoradiograms, as described previously (Mishra et al., 2008).
RESULTS AND DISCUSSION
M. marinum MMAR_2380 is essential for the presence of the mannose cap on LAM
Two genes have been reported to be involved in the biosynthesis of the mannose cap on ManLAM, Rv1635c and Rv2181 (Dinadayala et al., 2006; Kaur et al., 2008). An M. marinum mutant strain in which the homologue of Rv1635c (MMAR_2439) is mutated has also been isolated in a colony blot screen with mannose cap-recognizing mAb 55.92.1A1 (anti-ManLAM; α-ManLAM) (Appelmelk et al., 2008).
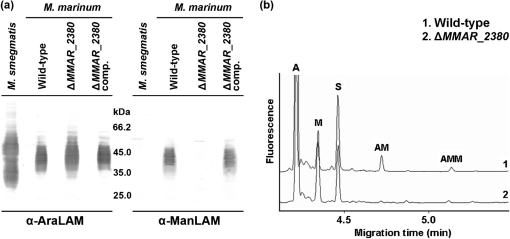
By continuing with this screen, we identified a novel gene involved in mannose-capping. LAM from an M. marinum mutant strain with a transposon inserted in MMAR_2380 at 91 bp upstream of its 3′ end (Supplementary Fig. S2) reacted well with an anti-AraLAM (α-AraLAM) mAb (mAb F30-5) which recognizes the arabinan branches of LAM, but not with α-ManLAM (Fig. 1a). The absence of the mannose cap on LAM in M. marinum ΔMMAR_2380 was further verified by chemical analysis. Both the mono- and di-mannoside caps, which are the dominant cap-lengths in the wild-type strain of M. marinum (Pitarque et al., 2005), were not detectable in the mutant strain (Fig. 1b). By complementing M. marinum ΔMMAR_2380 with a wild-type copy of MMAR_2380 (region indicated in Supplementary Fig. S2), the biosynthesis of the mannose cap on LAM was restored (Fig. 1a). This also excluded the possibility that the transposon affected the treXYZ-operon located downstream of MMAR_2380 rather than MMAR_2380 (Supplementary Fig. S2).
Fig. 1.
Absence of the mannose cap on mutant LAM. (a) Cell lysates from M. marinum wild-type, ΔMMAR_2380 and complemented ΔMMAR_2380 (ΔMMAR_2380 comp.) strains and M. smegmatis wild-type (control for ‘capless’ LAM) were immunoblotted with α-AraLAM antibody F30-5, which recognizes LAM, and α-ManLAM antibody 55.92.1A1, which recognizes the mannose cap. (b) Mannooligosaccharide cap analysis of LAM. Purified and partially degraded LAM was analysed for the presence of the mannose caps by CE. Shown are the profiles of LAM purified from M. marinum E11 (trace 1) and M. marinum ΔMMAR_2380 (trace 2). A, Ara-APTS; M, Man-APTS; S, internal standard, mannoheptose-APTS; AM, Manp-α(1→5)-Ara-APTS (monomannoside cap); AMM, Manp-α(1→2)-Manp-α(1→5)-Ara-APTS (dimannoside cap).
Mannosyltransferase capping enzymic activity is still present in the M. marinum ΔMMAR_2380 mutant strain
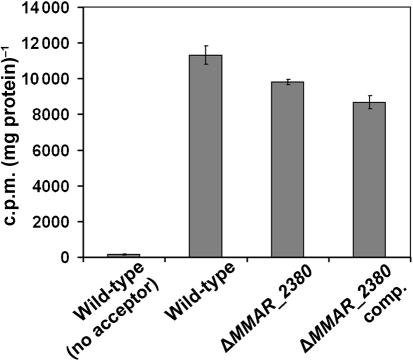
Since the full-length mannose cap is missing in the mutant strain, we checked the potential capability of the latter to add the first mannosyl unit of the cap. We used a mannosyltransferase assay previously set up to characterize the enzymic activity of the MMAR_2439 (Rv1635c) gene product (Appelmelk et al., 2008). A membrane preparation from M. marinum wild-type cells is able to transfer mannose from an in situ-produced decaprenylphosphomannose donor to a synthetic Ara6 acceptor, but a membrane preparation of M. marinum ΔMMAR_2439 does not show this enzyme activity (Appelmelk et al., 2008). In contrast to this previous mutant, a membrane preparation from M. marinum ΔMMAR_2380 was able to produce a mannosylated Ara6 acceptor similar to that of the wild-type strain (Fig. 2). This means that the arabinan mannose-capping enzymic activity is still present in the membrane of M. marinum ΔMMAR_2380; in other words, the gene product of MMAR_2380 is not involved in the direct functioning of the LAM-capping mannosyltransferase.
Fig. 2.
Enzymic analysis. Mannosyltransferase capping assay with synthetic Ara6 as acceptor and membranes from M. marinum wild-type, ΔMMAR_2380 and complemented ΔMMAR_2380 (ΔMMAR_2380 comp.), and as control, membranes from M. marinum wild-type without acceptor. Shown are means of triplicates; error bars, sd.
Structural differences between the glycan parts of wild-type and mutant LAM
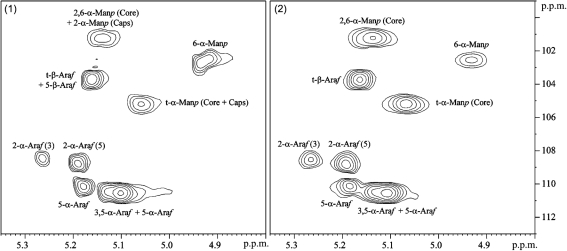
To investigate whether an alteration in the structure of the arabinan domain of mutant LAM might have caused the inability of the mutant strain to add the mannose cap, the glycan part of wild-type and mutant LAM was analysed by NMR. The NMR profile showed that the mutant LAM contained all mannosyl (Manp) and arabinosyl (Araf) residues that are present in the wild-type LAM, even the single α(1→2)-linked mannoses added to the mannan core (Fig. 3). Most importantly, the β-arabinosyl residues which form the non-reducing termini of the branches of the arabinan domain and which are the acceptors for the mannose-capping enzyme MMAR_2439 (Supplementary Fig. S1), were still part of M. marinum ΔMMAR_2380 LAM, and hence could, at least in theory, become substituted with a mannose cap.
Fig. 3.
NMR analysis of the glyco part of LAM from M. marinum wild-type (pattern 1) and ΔMMAR_2380 (pattern 2). 2D 1H-13C heteronuclear multiple quantum coherence spectroscopy (1H-13C HMQC) spectra of LAM in D2O at 313 K are shown with expanded regions (δ 1H, 4.80–5.35; δ 13C, 99–113). The different resonances are labelled with the abbreviated name of the corresponding glycosyl units. See Supplementary Fig. S1 for structure of ManLAM.
Further analysis, however, did indicate an increase in the degree of branching of the mannan core in the mutant LAM, as revealed by the 2,6-α-Manp : (2,6-α-Manp+6-α-Manp) ratio, which was 78 % in the mutant and 28 % in the wild-type LAM. In a similar way, the arabinan domain of the mutant LAM seemed to harbour more lateral branches, with a 2-α-Araf : 5-α-Araf ratio of 0.35 in the mutant as compared with 0.29 in the wild-type LAM. Methylation analysis corroborated these conclusions, although the ratio values were somewhat different: the 2,6-α-Manp : (2,6-α-Manp+6-α-Manp) ratio was 50 % for mutant LAM and 36 % for the wild-type LAM, and the 2-α-Araf : 5-α-Araf ratio was 0.23 and 0.15 for the mutant and wild-type LAM, respectively.
Accordingly, in the biosynthesis of ManLAM, the function of the MMAR_2380 protein appears essential for the presence of the mannose cap, but its mutation also results in increased branching of both the mannan core and the arabinan domain of LAM.
Acylation patterns of lipoglycans from wild-type and mutant strains
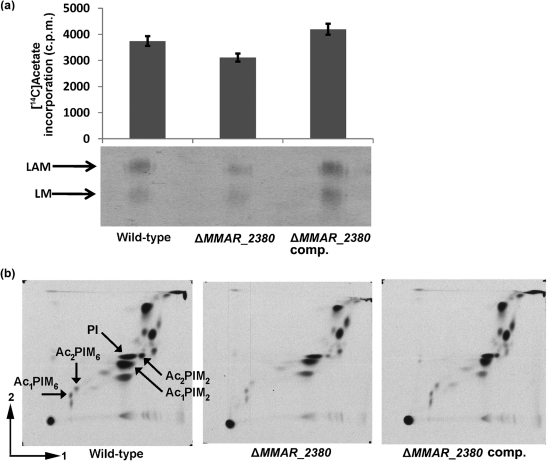
As MMAR_2380 is predicted to contain an acyltransferase domain, the acylation of mutant LAM was examined. To investigate this, M. marinum was cultured in the presence of [1,2-14C]acetate, after which lipoglycans were extracted and counted for the incorporation of [1,2-14C]acetate in the acyl chains (Fig. 4a). The data show that as compared with lipoglycans from wild-type M. marinum, lipoglycans from M. marinum ΔMMAR_2380 have a reduced [1,2-14C]acetate incorporation. The complementation of M. marinum ΔMMAR_2380 with wild-type MMAR_2380 resulted in an increase of [1,2-14C]acetate incorporation. LAM and the related lipoglycan lipomannan (LM) were further examined by SDS-PAGE/autoradiography (Fig. 4a), which also confirmed the reduction of [1,2-14C] acetate incorporation in the mutant LAM and showed a similar effect for mutant LM.
Fig. 4.
Reduced [1,2-14C]acetate incorporation in the polar lipids PIM6, LM and LAM from M. marinum ΔMMAR_2380. (a) Extracted crude lipoglycans from acetate-labelled delipidated cells (25 μg) from M. marinum wild-type, ΔMMAR_2380 and complemented ΔMMAR_2380 (ΔMMAR_2380 comp.) were counted for the incorporation of [1,2-14C]acetate and analysed by SDS-PAGE/autoradiography. Shown is the average of two independent experiments. (b) [1,2-14C]Acetate-labelled M. marinum cultures were processed and polar lipids were applied (25 000 c.p.m.) to the corners of 6.6×6.6 cm pieces of aluminium-backed TLC plates and analysed using the 2D solvent system E as described in Methods. Plates were dried and autoradiograms were produced by overnight exposure of Kodak X-Omat AR film to the TLC plates to reveal [1,2-14C]acetate-labelled lipids. PI, non-mannosylated, diacylated phosphatidylinositol anchor; PIM2, phosphatidylinositol dimannoside; PIM6, phosphatidylinositol hexamannoside; Ac1PIM, tri-acylated PIM; Ac2PIM, tetra-acylated PIM. Unassigned spots in the figure are lipooligosaccharides, and in the upper-right corner are diphosphatidylglycerol, phosphatidylethanolamine and unknown phospholipids (Burguière et al., 2005). Two independently prepared lipid extracts per strain were tested.
As similar effects of mutation of MMAR_2380 on [1,2-14C]acetate incorporation were seen for LM and LAM, we also analysed acylation of the structurally related phosphatidylinositol mannosides (PIMs). [1,2-14C]acetate-labelled lipid extracts were examined by 2D TLC using solvent system E for polar lipids (Besra, 1998; Dobson et al., 1985), which indeed showed differences in the polar lipid profiles of the wild-type and mutant strains. The lipid extract from wild-type M. marinum showed [1,2-14C]acetate incorporation into phosphatidylinositol (PI), tri- and tetra-acylated phosphatidylinositol dimannosides (Ac1/Ac2PIM2) and phosphatidylinositol hexamannosides (Ac1/Ac2PIM6) (Fig. 4b). Interestingly, in the lipid profile of M. marinum ΔMMAR_2380, a subtle but reproducible reduction in [1,2-14C]acetate incorporation in both Ac1PIM6 and Ac2PIM6 could be observed (Fig. 4b). The incorporation of [1,2-14C]acetate in Ac1/Ac2PIM2, however, showed some variations between the experiments, and therefore was less clear.
Phosphatidylinositol dimannosides (PIM2) have been hypothesized to be precursors for phosphatidylinositol hexamannosides (PIM6) as well as LAM and LM, with phosphatidylinositol tetramannosides (PIM4) as a putative branching point (Besra et al., 1997; Kovacevic et al., 2006), and therefore alterations at or before this point of the biosynthesis pathway of LAM may carry forward into the final structure of LAM, as well as LM and PIM6. Because [1,2-14C]acetate incorporation analysis only provides information on potential quantitative differences between the wild-type and mutant lipoglycans, and to verify the results obtained from the TLC assays, we also subjected extracted PIMs to direct structural analysis by MALDI-TOF MS in an independent experiment. This technique can discriminate between the various forms of PIMs in terms of mannosylation and acylation (Gilleron et al., 2006). The crude lipid extract from the mutant strain M. marinum ΔMMAR_2380 contained all the different forms of PIMs present in the wild-type strain but with an apparent reduction of the most acylated forms (Supplementary Fig. S3), which might explain the decrease of [1,2-14C]acetate incorporation in the mutant.
Hence, while LAM, LM and all forms of PIMs are present in the mutant strain, small quantitative differences in [1,2-14C]acetate incorporation in PIMs, LM, and LAM between the wild-type and the mutant strain can be observed, which might arise from a reduction of the relative abundance of the most acylated forms.
M. marinum ΔMMAR_2380 has altered cell-surface properties
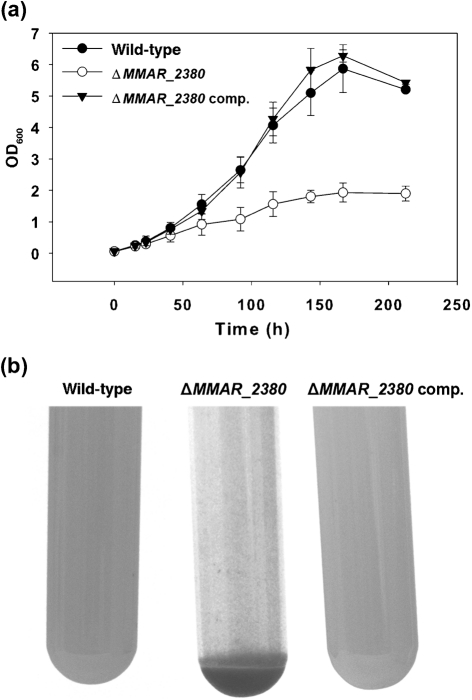
Along with the biosynthesis by M. marinum ΔMMAR_2380 of LAM that is devoid of the mannose cap, another phenotypic trait was observed for this mutant strain. As compared with the wild-type strain, M. marinum ΔMMAR_2380 showed severe aggregation. When grown as liquid cultures in 7H9 medium supplemented with 0.05 % (v/v) Tween 80, the mutant strain showed a reduced OD600 over time as compared with both its parent and the complemented strain (Fig. 5a). While the wild-type and complemented strain showed smooth yellow suspensions, the mutant strains showed aggregation and irregular pink clumps (Fig. 5b, and Supplementary Fig S4 for colour pictures). We cannot determine whether MMAR_2380 is essential for optimal growth in vitro or, alternatively, whether the reduced OD600 in liquid cultures is solely due to aggregation. On 7H10 agar plates, the mutant strain showed the same colony morphology and growth rate as the wild-type strain (not shown). The effect of mutation of MMAR_2380 on the cell-surface properties of M. marinum ΔMMAR_2380 in liquid cultures was consistent with earlier reported results for a Mycobacterium avium mutant strain in which the homologous gene was mutated (Yamazaki et al., 2006a, b). This M. avium mutant was impaired in its ability to produce large, wild-type amounts of biofilm (Yamazaki et al., 2006b), which also negatively influenced its invasiveness of human bronchial epithelial cells (Yamazaki et al., 2006a). On the other hand, in a designer arrays for defined mutant analysis (DeADMAn) screen of a transposon mutant library in M. tuberculosis CDC1551 for survival in mice, the mutant strain with a transposon in the homologous gene (ΔMT1616) was tested and did not appear to be attenuated in in vivo growth (Lamichhane et al., 2005).
Fig. 5.
Physical appearance of liquid-grown M. marinum wild type, ΔMMAR_2380 and complemented ΔMMAR_2380 (ΔMMAR_2380 comp.). (a) OD600 measurements during growth in 7H9+0.05 % Tween 80 with agitation. Shown are means of three independently grown cultures per strain; error bars sd. (b) M. marinum ΔMMAR_2380 shows increased aggregation as compared with the wild-type and the complemented strains (see Supplementary Fig. S4 for colour pictures).
The glycolipid profile and mannosylation pattern of proteins in M. marinum ΔMMAR_2380 appear identical to wild-type patterns
The above-mentioned effect of the transposon insertion in MMAR_2380, the aggregation of the mutant strain, suggested a more pleiotropic effect on the bacterial surface, and hence could have affected, for example, the biosynthesis of glycolipids other than LAM. Other alterations in phenotypic traits, such as morphology, smooth/rough transition of colonies and biofilm formation, have often been linked to changes in glycolipid constituents (Belisle et al., 1993; Belisle & Brennan, 1989; Daffé et al., 1991; Howard et al., 2006). Therefore, we performed an extensive lipid profile study in which polar and apolar extractable lipids were run in a range of four different TLC solvent systems, A to D, in addition to system E discussed above. Strikingly, no differences between the wild-type and the mutant strain could be observed (Supplementary Fig. S5).
Next, we checked for the mannosylation of proteins in M. marinum ΔMMAR_2380 as compared with the wild-type strain, because of the specific role of MMAR_2380 in the mannose-capping of LAM. Whole-cell lysates were examined by an SDS-PAGE/immunoblot with the lectin concanavalin A (ConA), which recognizes α-linked mannose. Overall, the mannosylation pattern of proteins from M. marinum ΔMMAR_2380 appeared similar to those of mannosylated proteins from the M. marinum wild-type strain, and the only difference observed in staining was at the position of (Man-)LAM on the blot (40–45 kDa) (Supplementary Fig. S6).
Prediction of gene function
MMAR_2380 encodes a 729 aa predicted transmembrane protein of unknown function (Supplementary Fig. S2). Therefore, a standard blastp search was performed against the nonredundant (nr) database at NCBI for the protein MMAR_2380 along with a search against the Conserved Domain Database (CDD) (Marchler-Bauer et al., 2009). In contrast to the mannose-capping enzyme Rv1635c, MMAR_2380 (Rv1565c) is also present with high similarity in Mycobacterium species known to produce LAM without a mannose cap, e.g. in M. smegmatis mc2155 (MSMEG_3187; identities 75 %, positives 85 %). Its homologue in M. tuberculosis strain CDC1551 (MT1616) is annotated as putative lipopolysaccharide biosynthesis protein WbpC. Unfortunately, the exact function of the WbpC protein has not yet been determined. At present, WbpC is annotated as a possible O-acetyltransferase in the O-antigen biosynthesis cluster of Pseudomonas aeruginosa (King et al., 2009). The N-terminal half of the protein contains a putative conserved domain of the acyltransferase-3 superfamily (Supplementary Fig. S2). As expected, many hits were found with predicted lipopolysaccharide biosynthesis acyltransferases from other non-mycobacterial species that contain the same conserved domain. MMAR_2380 was further predicted to be a protein with 10 transmembrane helices and a large C-terminal tail of approximately 300 amino acids in the periplasm (TMHMM 2.0) (Sonnhammer et al., 1998).
Considering the effects of mutation of MMAR_2380 described above, we postulate three hypotheses for how mutation of MMAR_2380 may affect the biosynthesis of LAM and the related lipoglycans, LM and PIMs. The first possibility is that MMAR_2380 is a redundant acyltransferase involved in the acylation of LAM, LM and PIMs. The phosphatidylinositol synthase required for the MPI anchor is PgsA1 (encoded by Rv2612c; MMAR_2090 in M. marinum) (Briken et al., 2004; Jackson et al., 2000), and acyltransferase Rv2611c has been shown to be involved in further acylation of PIMs (position C6 of the Manp linked to the C2 of the inositol) (Korduláková et al., 2003). Importantly, the Rv2611c-mutant M. smegmatis strains still produce tri- and tetra-acylated PIMs (Korduláková et al., 2003), and hence other acyltransferases are yet to be identified. From the TLC pattern (Fig. 4b), it is not possible to determine the position of acylation at which MMAR_2380 may be involved (position C3 of the inositol or position C6 of the Manp linked to the C2 of the inositol). However, the differences of [1,2-14C]acetate incorporation in PIMs, LAM and LM were small, and other effects of mutation of MMAR_2380 were observed, suggesting a different function for MMAR_2380.
A second function for MMAR_2380 could be as a regulatory protein in the biosynthesis of LAM and related lipoglycans. The biosynthesis pathway of LAM has recently become better characterized. Not only glycosyltransferases directly involved in the addition of mannose and arabinose have been discovered, but also proteins with a role in the regulation of this pathway. One of these proteins is lipoprotein LpqW, which directs PIM4 into the biosynthesis route of LAM (Crellin et al., 2008; Kovacevic et al., 2006). Furthermore, the production of PIM2 and PIM6 has been shown to be compartmentalized in separate cell domains, a plasma membrane domain (PMf) and a plasma membrane–cell wall domain (PM–CW), respectively, each containing the enzymes required specifically for their biosynthesis (Morita et al., 2005). Hence, as mutation of MMAR_2380 resulted in a small quantitative decrease of [1,2-14C]acetate incorporation in mutant LAM as well as LM and PIM6 (Fig. 4), MMAR_2380 may also have a regulatory function in the metabolic pathway of these lipoglycans, e.g. in the recruitment of specific enzymes to the different membrane/cell wall fractions. In addition, as it is a regulatory protein, mutation of MMAR_2380 could be the cause of the higher branching degrees of the mutant LAM, which could subsequently affect the LAM-capping activity in the mutant strain. Regarding the potential regulatory role of MMAR_2380, it has been reported that for certain enzymes additional processing such as acetylation is essential for proper transport and localization or for the activity of the enzyme (Banaei et al., 2009; Fukuda et al., 2002; Tschumi et al., 2009), and MMAR_2380 may function in this way.
Finally, considering the altered growth of the mutant strain (Fig. 5), it cannot be excluded that mutation of MMAR_2380 has a pleiotropic effect on the organization of the whole cell wall, and that it thus disturbs the compartmentalized biosynthesis of the higher-order PIM6, LAM and LM, and/or the transport of lower-order PIM2 from the PMf domain to the PM–CW fraction as precursor for the other lipoglycans, possibly resulting in the inability of the mutant strain to produce LAM with a mannose cap.
Summarizing, we have identified a novel gene involved, possibly indirectly, in the biosynthesis of the mannose cap on LAM, MMAR_2380, which is the M. marinum orthologue of Rv1565c. The mutant LAM was not only devoid of the mannose cap, but also displayed a higher degree of branching in both the mannan core and the arabinan domain as compared with wild-type LAM. In addition, mutation of MMAR_2380 caused small quantitative differences in [1,2-14C]acetate incorporation in LAM, LM and PIMs, while all the different acylated forms of the PIMs seen in the wild-type strain were still present. Furthermore, another effect was seen: M. marinum ΔMMAR_2380 showed increased aggregation in liquid cultures as compared with the wild-type strain, a phenotypic trait which was not observed for the previous mutant M. marinum ΔMMAR_2439, which produced ‘capless’ LAM. Hence, in all likelihood, MMAR_2380 encodes an acyltransferase, although neither the length of the acyl chain transferred nor the acceptor can be predicted. Acylation activity by MMAR_2380 may be important for the biosynthesis of the mannose cap in a way which has yet to be determined.
Acknowledgments
B. J. A. acknowledges support to R. U. from the EU (FP6 grant: ImmunovacTB 37388). G. S. B. acknowledges support in the form of a Personal Research Chair from Mr James Bardrick, Royal Society Wolfson Research Merit Award, as a former Lister Institute-Jenner Research Fellow, the Medical Research Council and The Wellcome Trust (081569/Z/06/Z). We thank Jeroen Geurtsen, Nicole N. van der Wel, Eveline Weerdenburg and Shomara Willems for technical assistance and helpful advice.
Abbreviations
Ara6, hexaarabinofuranoside
Araf, arabinofuranosyl unit
AraLAM, LAM with linked arabinosyl units
BCA, bicinchoninic acid
CE, capillary electrophoresis
LAM, lipoarabinomannan
LM, lipomannan
ManLAM, mannose-capped LAM
Manp, mannopyranosyl residue
MPI, mannosylphosphatidylinositol
PIMs, phosphatidylinositol mannosides
PIM2, phosphatidylinositol dimannosides
PIM4, phosphatidylinositol tetramannosides
PIM6, phosphatidylinositol hexamannosides
PM–CW, plasma membrane–cell wall domain
Footnotes
Six supplementary figures and a supplementary table, with supplementary references, are available with the online version of this paper.
References
- Abdallah, A. M., Verboom, T., Hannes, F., Safi, M., Strong, M., Eisenberg, D., Musters, R. J., Vandenbroucke-Grauls, C. M., Appelmelk, B. J. & other authors (2006). A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol 62, 667–679. [DOI] [PubMed] [Google Scholar]
- Appelmelk, B. J., den Dunnen, J., Driessen, N. N., Ummels, R., Pak, M., Nigou, J., Larrouy-Maumus, G., Gurcha, S. S., Movahedzadeh, F. & other authors (2008). The mannose cap of mycobacterial lipoarabinomannan does not dominate the Mycobacterium–host interaction. Cell Microbiol 10, 930–944. [DOI] [PubMed] [Google Scholar]
- Banaei, N., Kincaid, E. Z., Lin, S. Y., Desmond, E., Jacobs, W. R., Jr & Ernst, J. D. (2009). Lipoprotein processing is essential for resistance of Mycobacterium tuberculosis to malachite green. Antimicrob Agents Chemother 53, 3799–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle, J. T. & Brennan, P. J. (1989). Chemical basis of rough and smooth variation in mycobacteria. J Bacteriol 171, 3465–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle, J. T., Klaczkiewicz, K., Brennan, P. J., Jacobs, W. R. & Inamine, J. M. (1993). Rough morphological variants of Mycobacterium avium. Characterization of genomic deletions resulting in the loss of glycopeptidolipid expression. J Biol Chem 268, 10517–10523. [PubMed] [Google Scholar]
- Besra, G. S. (1998). Preparation of cell-wall fractions from mycobacteria. In Methods in Molecular Biology: Mycobacteria Protocols, vol. 101, pp. 91–107. Edited by Parish, T. & Stoker, N. G.. Totowa, NJ. : Humana Press. [DOI] [PubMed] [Google Scholar]
- Besra, G. S., Morehouse, C. B., Rittner, C. M., Waechter, C. J. & Brennan, P. J. (1997). Biosynthesis of mycobacterial lipoarabinomannan. J Biol Chem 272, 18460–18466. [DOI] [PubMed] [Google Scholar]
- Briken, V., Porcelli, S. A., Besra, G. S. & Kremer, L. (2004). Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response. Mol Microbiol 53, 391–403. [DOI] [PubMed] [Google Scholar]
- Burguière, A., Hitchen, P. G., Dover, L. G., Kremer, L., Ridell, M., Alexander, D. C., Liu, J., Morris, H. R., Minnikin, D. E. & other authors (2005). LosA, a key glycosyltransferase involved in the biosynthesis of a novel family of glycosylated acyltrehalose lipooligosaccharides from Mycobacterium marinum. J Biol Chem 280, 42124–42133. [DOI] [PubMed] [Google Scholar]
- Chatterjee, D. & Khoo, K. H. (1998). Mycobacterial lipoarabinomannan: an extraordinary lipoheteroglycan with profound physiological effects. Glycobiology 8, 113–120. [DOI] [PubMed] [Google Scholar]
- Ciucanu, I. & Kerek, F. (1984). A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res 131, 209–217. [Google Scholar]
- Crellin, P. K., Kovacevic, S., Martin, K. L., Brammananth, R., Morita, Y. S., Billman-Jacobe, H., McConville, M. J. & Coppel, R. L. (2008). Mutations in pimE restore lipoarabinomannan synthesis and growth in a Mycobacterium smegmatis lpqW mutant. J Bacteriol 190, 3690–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffé, M., Mcneil, M. & Brennan, P. J. (1991). Novel type-specific lipooligosaccharides from Mycobacterium tuberculosis. Biochemistry 30, 378–388. [DOI] [PubMed] [Google Scholar]
- Dinadayala, P., Kaur, D., Berg, S., Amin, A. G., Vissa, V. D., Chatterjee, D., Brennan, P. J. & Crick, D. C. (2006). Genetic basis for the synthesis of the immunomodulatory mannose caps of lipoarabinomannan in Mycobacterium tuberculosis. J Biol Chem 281, 20027–20035. [DOI] [PubMed] [Google Scholar]
- Dobson, G., Minnikin, D. E., Minnikin, S. M., Parlett, J. H., Goodfellow, M., Ridell, M. & Magnusson, M. (1985). Systematic analysis of complex mycobacterial lipids. In Chemical Methods in Bacterial Systematics, vol. 1, pp. 237–265. Edited by Goodfellow, M. & Minnikin, D. E.. London. : Academic Press. [Google Scholar]
- Fukuda, A., Matsuyama, S., Hara, T., Nakayama, J., Nagasawa, H. & Tokuda, H. (2002). Aminoacylation of the N-terminal cysteine is essential for Lol-dependent release of lipoproteins from membranes but does not depend on lipoprotein sorting signals. J Biol Chem 277, 43512–43518. [DOI] [PubMed] [Google Scholar]
- Gadikota, R. R., Callam, C. S., Appelmelk, B. J. & Lowary, T. L. (2003). Synthesis of oligosaccharide fragments of mannosylated lipoarabinomannan appropriately functionalized for neoglycoconjugate preparation. J Carbohydr Chem 22, 149–170. [Google Scholar]
- Geijtenbeek, T. B., van Vliet, S. J., Koppel, E. A., Sanchez-Hernandez, M., Vandenbroucke-Grauls, C. M., Appelmelk, B. & Van Kooyk, Y. (2003). Mycobacteria target DC-SIGN to suppress dendritic cell function. J Exp Med 197, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleron, M., Bala, L., Brando, T., Vercellone, A. & Puzo, G. (2000). Mycobacterium tuberculosis H37Rv parietal and cellular lipoarabinomannans. Characterization of the acyl- and glyco-forms. J Biol Chem 275, 677–684. [DOI] [PubMed] [Google Scholar]
- Gilleron, M., Lindner, B. & Puzo, G. (2006). MS/MS approach for characterization of the fatty acid distribution on mycobacterial phosphatidyl-myo-inositol mannosides. Anal Chem 78, 8543–8548. [DOI] [PubMed] [Google Scholar]
- Hoffmann, C., Leis, A., Niederweis, M., Plitzko, J. M. & Engelhardt, H. (2008). Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci U S A 105, 3963–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, S. T., Rhoades, E., Recht, J., Pang, X. H., Alsup, A., Kolter, R., Lyons, C. R. & Byrd, T. F. (2006). Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology 152, 1581–1590. [DOI] [PubMed] [Google Scholar]
- Jackson, M., Crick, D. C. & Brennan, P. J. (2000). Phosphatidylinositol is an essential phospholipid of mycobacteria. J Biol Chem 275, 30092–30099. [DOI] [PubMed] [Google Scholar]
- Kang, P. B., Azad, A. K., Torrelles, J. B., Kaufman, T. M., Beharka, A., Tibesar, E., DesJardin, L. E. & Schlesinger, L. S. (2005). The human macrophage mannose receptor directs Mycobacterium tuberculosis lipoarabinomannan-mediated phagosome biogenesis. J Exp Med 202, 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, D., Berg, S., Dinadayala, P., Gicquel, B., Chatterjee, D., McNeil, M. R., Vissa, V. D., Crick, D. C., Jackson, M. & other authors (2006). Biosynthesis of mycobacterial lipoarabinomannan: role of a branching mannosyltransferase. Proc Natl Acad Sci U S A 103, 13664–13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, D., Obregon-Henao, A., Pham, H., Chatterjee, D., Brennan, P. J. & Jackson, M. (2008). Lipoarabinomannan of Mycobacterium: mannose capping by a multifunctional terminal mannosyltransferase. Proc Natl Acad Sci U S A 105, 17973–17977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J. D., Kocincova, D., Westman, E. L. & Lam, J. S. (2009). Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun 15, 261–312. [DOI] [PubMed] [Google Scholar]
- Kolk, A. H., Ho, M. L., Klatser, P. R., Eggelte, T. A., Kuijper, S., de Jonge, S. & van Leeuwen, J. (1984). Production and characterization of monoclonal antibodies to Mycobacterium tuberculosis, M. bovis (BCG) and M. leprae. Clin Exp Immunol 58, 511–521. [PMC free article] [PubMed] [Google Scholar]
- Korduláková, J., Gilleron, M., Puzo, G., Brennan, P. J., Gicquel, B., Mikusova, K. & Jackson, M. (2003). Identification of the required acyltransferase step in the biosynthesis of the phosphatidylinositol mannosides of Mycobacterium species. J Biol Chem 278, 36285–36295. [DOI] [PubMed] [Google Scholar]
- Kovacevic, S., Anderson, D., Morita, Y. S., Patterson, J., Haites, R., McMillan, B. N., Coppel, R., McConville, M. J. & Billman-Jacobe, H. (2006). Identification of a novel protein with a role in lipoarabinomannan biosynthesis in mycobacteria. J Biol Chem 281, 9011–9017. [DOI] [PubMed] [Google Scholar]
- Lamichhane, G., Tyagi, S. & Bishai, W. R. (2005). Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect Immun 73, 2533–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R. E., Brennan, P. J. & Besra, G. S. (1998). Synthesis of β-d-arabinofuranosyl-1-monophosphoryl polyprenols: examination of their function as mycobacterial arabinosyl transferase donors. Bioorg Med Chem Lett 8, 951–954. [DOI] [PubMed] [Google Scholar]
- Ludwiczak, P., Brando, T., Monsarrat, B. & Puzo, G. (2001). Structural characterization of Mycobacterium tuberculosis lipoarabinomannans by the combination of capillary electrophoresis and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem 73, 2323–2330. [DOI] [PubMed] [Google Scholar]
- Maeda, N., Nigou, J., Herrmann, J. L., Jackson, M., Amara, A., Lagrange, P. H., Puzo, G., Gicquel, B. & Neyrolles, O. (2003). The cell surface receptor DC-SIGN discriminates between Mycobacterium species through selective recognition of the mannose caps on lipoarabinomannan. J Biol Chem 278, 5513–5516. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., Fong, J. H., Geer, L. Y., Geer, R. C., Gonzales, N. R. & other authors (2009). CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res 37, D205–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, A. K., Alderwick, L. J., Rittmann, D., Wang, C., Bhatt, A., Jacobs, W. R., Jr, Takayama, K., Eggeling, L. & Besra, G. S. (2008). Identification of a novel α(1→6) mannopyranosyltransferase MptB from Corynebacterium glutamicum by deletion of a conserved gene, NCgl1505, affords a lipomannan- and lipoarabinomannan-deficient mutant. Mol Microbiol 68, 1595–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, Y. S., Velasquez, R., Taig, E., Waller, R. F., Patterson, J. H., Tull, D., Williams, S. J., Billman-Jacobe, H. & McConville, M. J. (2005). Compartmentalization of lipid biosynthesis in mycobacteria. J Biol Chem 280, 21645–21652. [DOI] [PubMed] [Google Scholar]
- Nigou, J., Vercellone, A. & Puzo, G. (2000). New structural insights into the molecular deciphering of mycobacterial lipoglycan binding to C-type lectins: lipoarabinomannan glycoform characterization and quantification by capillary electrophoresis at the subnanomole level. J Mol Biol 299, 1353–1362. [DOI] [PubMed] [Google Scholar]
- Nigou, J., Gilleron, M., Rojas, M., Garcia, L. F., Thurnher, M. & Puzo, G. (2002). Mycobacterial lipoarabinomannans: modulators of dendritic cell function and the apoptotic response. Microbes Infect 4, 945–953. [DOI] [PubMed] [Google Scholar]
- Nigou, J., Gilleron, M. & Puzo, G. (2003). Lipoarabinomannans: from structure to biosynthesis. Biochimie 85, 153–166. [DOI] [PubMed] [Google Scholar]
- Pitarque, S., Herrmann, J. L., Duteyrat, J. L., Jackson, M., Stewart, G. R., Lecointe, F., Payre, B., Schwartz, O., Young, D. B. & other authors (2005). Deciphering the molecular bases of Mycobacterium tuberculosis binding to the lectin DC-SIGN reveals an underestimated complexity. Biochem J 392, 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitarque, S., Larrouy-Maumus, G., Payre, B., Jackson, M., Puzo, G. & Nigou, J. (2008). The immunomodulatory lipoglycans, lipoarabinomannan and lipomannan, are exposed at the mycobacterial cell surface. Tuberculosis (Edinb) 88, 560–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttinaowarat, S., Thompson, K., Lilley, J. & Adams, A. (1999). Characterization of Mycobacterium spp. isolated from fish by pyrolysis mass spectrometry (PyMS) analysis. AAHRI Newslett 8, 4–8. [Google Scholar]
- Rubin, E. J., Akerley, B. J., Novik, V. N., Lampe, D. J., Husson, R. N. & Mekalanos, J. J. (1999). In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc Natl Acad Sci U S A 96, 1645–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti, C. M., Boyd, D. H. & Rubin, E. J. (2001). Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci U S A 98, 12712–12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger, L. S., Hull, S. R. & Kaufman, T. M. (1994). Binding of the terminal mannosyl units of lipoarabinomannan from a virulent strain of Mycobacterium tuberculosis to human macrophages. J Immunol 152, 4070–4079. [PubMed] [Google Scholar]
- Sonnhammer, E. L., Heijne, G. & Krogh, A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, pp. 175–182. Edited by J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff & C. Sensen. Menlo Park, CA: AAAI Press. [PubMed]
- Tschumi, A., Nai, C., Auchli, Y., Hunziker, P., Gehrig, P., Keller, P., Grau, T. & Sander, P. (2009). Identification of apolipoprotein N-acyltransferase (Lnt) in mycobacteria. J Biol Chem 284, 27146–27156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welin, A., Winberg, M. E., Abdalla, H., Sarndahl, E., Rasmusson, B., Stendahl, O. & Lerm, M. (2008). Incorporation of Mycobacterium tuberculosis lipoarabinomannan into macrophage membrane rafts is a prerequisite for the phagosomal maturation block. Infect Immun 76, 2882–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, Y., Danelishvili, L., Wu, M., Hidaka, E., Katsuyama, T., Stang, B., Petrofsky, M., Bildfell, R. & Bermudez, L. E. (2006a). The ability to form biofilm influences Mycobacterium avium invasion and translocation of bronchial epithelial cells. Cell Microbiol 8, 806–814. [DOI] [PubMed] [Google Scholar]
- Yamazaki, Y., Danelishvili, L., Wu, M., Macnab, M. & Bermudez, L. E. (2006b). Mycobacterium avium genes associated with the ability to form a biofilm. Appl Environ Microbiol 72, 819–825. [DOI] [PMC free article] [PubMed] [Google Scholar]