Abstract
Human papillomaviruses (HPVs) are important in the development of human cancers, including cervical and oral tumors. However, most existing methods for HPV typing cannot routinely distinguish among the more than 100 distinct types of HPV or the natural HPV intratypic variants that have also been documented. To address this problem, we developed a novel method, general primer-denaturing high-performance liquid chromatography (GP-dHPLC), for the detection and typing of genital HPV using an automated 96-well plate format. GP-dHPLC uses general primer PCR (GP-PCR) to amplify the viral DNA and then analyzes the GP-PCR products by denaturing high-performance liquid chromatography (dHPLC). A number of different primer pairs with homology to most known genital HPV types were tested, and the L1C1-L1C2M pair specific for the L1 region of the viral genome was chosen. A set of HPV standard control patterns, consisting of those for HPV types 16, 18, 31, 33, 39, 45, 51, 52, 56, 58, 59, 6, and 11, was established for genital HPV typing. One hundred eighty-six frozen and formalin-fixed cervical cancer tissue samples were analyzed for the presence of HPV and the HPV type by this method, and 95.8% of them were found to contain HPV DNA. GP-dHPLC accurately discriminated among HPV variants that differed by as little as one nucleotide. Several new variants of HPV types 16, 18, 39, 45, 52, and 59 were identified. Moreover, multiple HPV infections were detected in 26.6% of the samples. Our results indicate that HPV typing by GP-dHPLC permits discrimination of common genital HPV types, detection of multiple HPV infections, and identification of HPV variants in clinical samples.
Human papillomaviruses (HPVs) form a heterogeneous group whose genome is an 8-kb double-stranded circle of DNA. They are the most diverse group of DNA viruses involved in human disease. More than 100 putative HPV types have been described to date, including 85 that have been cloned and officially designated and others that have been identified from the sequences of PCR products. More than 40 types of HPV are sexually transmitted; these are called genital HPVs. The genital HPVs can be grouped on the basis of their clinical associations and in vitro transforming capabilities into high-risk types (e.g., HPV types 16 and 18), intermediate-risk types (e.g., HPV types 31 and 33), and low-risk types (e.g., HPV types 6 and 11) (8, 16, 20). HPV is considered the major etiological agent of cervical cancer and its precursor lesions throughout the world (3). The majority of invasive cervical cancers and high-grade lesions are associated with high-risk and intermediate-risk HPV infections, while most low-grade dysplastic lesions and benign condylomata acuminata are associated with low-risk HPV types (3, 8, 16, 20, 22, 34).
Nucleic acid sequencing has revealed the existence of many natural sequence variations among HPV types. HPV genomes that vary from that of the prototype by ≤2% are defined as “variants,” whereas those that diverge by ≥10% from the genome of any described HPV type qualify as independent types. Isolates whose genomes differ from that of an established type by 2 to 10% are considered subtypes (30). A number of HPV variants for high-risk, intermediate-risk, and low-risk types have been described to date (11). More than 40 naturally occurring variants have been classified (10, 11, 13); their biological and biochemical properties differ in vitro (26), and their oncogenicities also differ (13, 19, 36). Several studies have indicated that viral persistence and the development of high-grade lesions and invasive carcinomas are closely associated with the presence of specific HPV variants (1, 2, 4-7, 10, 15, 18, 21, 31, 40).
Given the different oncogenic potentials of the various HPV types and variants, HPV genotyping is becoming increasingly important as a diagnostic aid and prognostic tool. Recently, HPV detection has been proposed as an alternative to cytological screening for cervical cancer (24, 32, 33).
Two main approaches are used for HPV detection and typing: (i) general primer PCR (GP-PCR) (also referred to as degenerate or consensus primer PCR), which amplifies a conserved region in the virus followed by HPV type-specific identification, and (ii) the hybrid capture assay, which detects and types HPV by direct hybridization with type-specific RNA probes (25). A variety of general primers are available for the detection of HPV through GP-PCR (9, 23, 28, 37, 38). These primer sets are designed to match the short regions of homology among the HPV genotypes. For example, GP5+-GP6− (28), MY11-MY09 (23), and L1C1-L1C2 (37) from the L1 region and pU-1M-pU-2R (38) from the E6/E7 region can detect a broad spectrum of known and unknown HPV DNAs. Because of similarities between the GP-PCR products, however, it remains difficult to accurately differentiate among the genital HPV types. A variety of techniques, including DNA hybridization with type-specific probes (12, 28), type-specific PCR (27), multiplex PCR with type-specific antisense primers (14), single-strand conformation polymorphism analysis (39), restriction fragment length polymorphism analysis (9, 37), enzyme-linked immunosorbent assay (17), and direct sequence analysis (29), have been used in combination with GP-PCR to facilitate HPV typing. Each detection method has advantages and disadvantages, but most are unable to discriminate among multiple HPV types and variants.
Denaturing high-performance liquid chromatography (dHPLC) separates PCR products by size and sequence. It detects the sequence divergence of DNA fragments, including single base substitutions and short deletions and insertions. This technology is sufficiently sensitive for the reliable detection of nearly 100% of DNA sequence variations at optimized partially denaturing temperatures (35). We therefore developed a novel method for the detection and typing of genital HPV that uses a combination of GP-PCR and dHPLC (referred as general primer-dHPLC [GP-dHPLC]). The method allows us to discriminate among common genital HPV types, to detect multiple HPV infections, and to identify HPV variants from clinical samples. Moreover, samples can be processed in a 96-well format and continuously loaded on the column for analysis.
MATERIALS AND METHODS
HPV controls.
We used HPV controls to evaluate the sensitivities of the GP-PCR primers and to establish the gradient conditions for dHPLC to establish the optimal conditions and standard pattern for HPV typing. Subclones of HPV genomes were kindly provided by Lou Laimins at Northwestern University (HPV types 6, 11, 16, 18, 31, 33, 35, and 42), E.-M. de Villiers at Deutsches Krebsforschungszentrum in Heidelberg, Germany (HPV types 45 and 51), and Wayne D. Lancaster at Wayne State University School of Medicine (HPV type 52). Controls for HPV types 39, 56, 58, and 59 were identified from cervical cancer tissue samples. All HPV controls were confirmed by bidirectional sequencing of their PCR products.
Cervical cancer tissue samples.
Cervical cancer tissue samples were obtained from pathological specimens from patients with invasive cervical carcinomas. Thirty-six samples were snap-frozen in liquid nitrogen, 169 were embedded in Optimal Cutting Temperature compound (Sakura Finetek Inc., Torrance, Calif.) and frozen in isobutane, and 41 were in formalin-fixed, paraffin-embedded tissue blocks. All cervical tissue specimens were reviewed by a gynecologic pathologist (P.C.H. or D.L.) to confirm the diagnosis of invasive cervical cancer. Four to eight 10-μm sections from each tumor were placed in a sterile microcentrifuge tube. The microtome blade was changed for the preparation of each tumor. HPV-negative control tissue from normal human spleen and ovary tissues were used to verify that no cross-contamination occurred between samples. DNA was extracted by the standard sodium dodecyl sulfate-proteinase K procedure, purified with phenol-chloroform, resuspended in TE (Tris-EDTA) buffer (pH 8.0), and then quantitated. The study was approved by the Washington University Human Studies Committee.
PCRs.
Several general primer sets were tested (Table 1). The PCR mixture contained 50 mM KCl, 10 mM Tris-Cl (pH 8.4), 1.0 mM MgCl2, 100 μM each deoxynucleoside triphosphate, 1 μM each primer, and a mixture (9:1; vol/vol) of 5 U of AmpliTaq (Applied Biosystems, Foster City, Calif.) per μl with 2.5 U of Pfu (Stratagene, La Jolla, Calif.) per μl and 40 ng of genomic DNA (or 40 pg of plasmid HPV DNA) in a final volume of 20 μl. The general PCR program was 94°C for 2 min, 94°C for 30 s, 40 to 47°C (1°C increase each cycle) for 30 s, and 72°C for 1 min for 8 cycles and then 94°C for 30 s, 48°C for 30 s, and 72°C for 1 min for 35 cycles on a PCR Express thermal cycler (Hybaid, Franklin, Mass.). The presence of PCR products was confirmed by 2% agarose gel electrophoresis and ethidium bromide staining.
TABLE 1.
General primer sets
| Primer name | ORFa of viral genome | Primer sequence (5′-3′) | Primer locationb | PCR product size (bp) | Reference(s) |
|---|---|---|---|---|---|
| L1C1 | L1 | CGTAAACGTTTTCCCTATTTTTTT | 5609-5632 | 243-262 | 9, 37 |
| L1C2 | TACCCTAAATACTCTGTATTG | 5841-5861 | |||
| (L1C2M) | (TACCCTAAATACCCTATATTG) | (5841-5861) | |||
| GP5+ | L1 | TTTGTTACTGTGGTAGATAC | 6624-6643 | 139-149 | 25 |
| GP6− | GAAAAATAAACTGTAAATCA | 6746-6765 | |||
| L1S-L | L1 | TTTAATAAACCATATTGGTTACA | 6555-6577 | 89 | —c |
| L1S-R | GTATCCACAACAGTAACAAA | 6624-6643 | |||
| PU-1M | E6/E7 | TGTCAAAAACCGTTGTGTCC | 419-438 | 205-271 | 38 |
| (pU-1M-L) | (TGTCAAAAACCGTTGTGTCCAGAAGAAAA) | (419-447) | |||
| pU-2R | GAGCTGTCGCTTAATTGCTC | 637-656 | |||
| (pU-2R-N) | (TCTGAGTCGCTTAATTGCTC) | (637-656) | |||
| E6-L | E6 | ACCGAAAACGGTTCATATAAA | 50-70 | 192-212 | —c |
| E6-R | AAATGCAAATTCATATACCTC | 224-244 |
ORF, open reading frame.
Primer location refers to the sequence of HPV type 16 (GenBank accession no. K02718).
—, new primer design.
HPV typing by dHPLC.
Genotyping of the amplified GP-PCR products was performed by dHPLC with the WAVE Nucleic Acid Fragment Analysis System (Wave system; Transgenomic, Inc., San Jose, Calif.). The reagents used were buffer A (0.1 M triethylammonium acetate [TEAA], 0.025% [vol/vol] acetonitrile) and buffer B (0.1 M TEAA, 25% [vol/vol] acetonitrile); all reagents were purchased from Transgenomic, Inc.
dHPLC uses the principles of reverse-phase ion-paired chromatography to separate PCR products. The chromatographic column (DNASep column) in the Wave system is packed with C18 alkylated polystyrene-divinylbenzene polymeric beads. A positively charged ion-pairing reagent (TEAA) allows the negatively charged DNA to interact with the hydrophobic DNASep column matrix. An increasing proportion of organic mobile phase (acetonitrile) is used to elute the DNA fragments from the matrix in a size-dependent manner (for nondenaturing conditions) and a sequence-dependent manner (for partially and fully denaturing conditions), and the fragments are detected by UV analysis. For our analysis, we set the temperature high enough to partially denature DNA duplexes. GP-PCR products with sequence divergence have different retention times on the column. Identical HPV types elute as a single peak, whereas different HPV types or variants elute in double peaks or in a single peak containing a shoulder or having a retention time different from that of the control sample.
We optimized the temperature, gradient conditions, and flow rates on the Wave system to differentiate among the most common high- and intermediate-risk genital HPV types. HPV genotypes were determined by mixing each sample with an HPV control and comparing the results with those for standard HPV controls. PCR products whose peaks differed from those of the HPV controls were sequenced. Multiple peaks were isolated for sequencing by using the Fragment Collector of the Wave system. The samples were also automatically quantitated by the Wave system according to peak area.
Sequencing.
A 5-μl aliquot from the PCR mixture was treated by use of the ExoSAP-IT kit (U.S. Biochemical Corporation, Cleveland, Ohio), according to the instructions of the manufacturer, and was then used as the sequencing template without further purification. Direct sequencing was performed with the BigDye Terminator sequencing kit (version 3.0; Applied Biosystems). Sequencing reaction products were run on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). The DNA sequences were compared by using DNAStar software (DNAStar, Inc., Madison, Wis.).
RESULTS
Selection of general primer set for GP-PCR.
Several general primer sets are available for the detection of HPV in clinical specimens. They are designed to match the short regions of conserved homology among the HPV genotypes. The primer sets evaluated in this study are presented in Table 1. We also designed and tested two additional sets: L1S-L and L1S-R from the L1 region and E6-L and E6-R from the E6 region (Table 1). We compared mismatches between the general primer sets and genital HPV DNA templates. The results of the comparisons are displayed in Table 2.
TABLE 2.
Comparison of mismatches between general primer sets and HPVs DNAa
| HPV type (associated riskb) | GenBank accession No. | No. of nucleotide mismatches
|
||||||
|---|---|---|---|---|---|---|---|---|
| GP5+/GP6− | L1C1/L1C2 | L1C1/L1C2M | pU-1M/pU-2R | pU-1M-L/pU-2R-N | L1S-L/L1S-R | E6-L/E6-R | ||
| 16 (HR) | K02718 | 2/0 | 2/3 | 2/3 | 3/2 | 4/7 | 2/1 | 3/2 |
| 18 (HR) | X05015 | 0/1 | 1/2 | 1/2 | 4/5 | 5/0 | 0/2 | 2/1 |
| 31 (IR) | J04353 | 0/2 | 4/3 | 4/1 | 1/2 | 1/7 | 2/2 | 3/3 |
| 33 (IR) | A12360 | 0/2 | 1/4 | 1/2 | 2/1 | 4/6 | 2/2 | 2/1 |
| 35 (IR) | M74117 | 2/0 | 5/0 | 5/3 | 1/3 | 3/8 | 1/2 | 3/3 |
| 39 (IR) | M62849 | 4/2 | 1/5 | 1/3 | 3/8 | 4/3 | 3/2 | 3/0 |
| 45 (IR) | X74479 | 3/0 | 1/5 | 1/3 | 6/6 | 7/1 | 1/2 | 2/2 |
| 51 (IR) | M62877 | 6/3 | 2/1 | 2/1 | 6/10 | 7/5 | 4/5 | 1/5 |
| 52 (IR) | X74481 | 4/1 | 1/4 | 1/4 | 4/2 | 5/7 | 2/2 | 3/5 |
| 53 (IR) | X74482 | 3/4 | 2/5 | 1/3 | 4/9 | 6/4 | 3/1 | 1/3 |
| 56 (IR) | X74483 | 1/3 | 1/2 | 1/0 | 5/10 | 7/5 | 2/2 | 1/4 |
| 58 (IR) | D90400 | 2/2 | 1/5 | 1/3 | 2/2 | 3/7 | 3/3 | 3/2 |
| 59 (IR) | X77858 | 4/2 | 1/1 | 1/3 | 5/7 | 8/4 | 2/4 | 1/2 |
| 66 (IR) | U31794 | 2/4 | 1/3 | 1/3 | 7/10 | 9/5 | 2/0 | 0/5 |
| 68 (IR) | M73258 | 3/3 | 3/4 | 3/4 | 5/8 | 8/3 | 5/1 | 2/1 |
| 70 (IR) | U21941 | 3/2 | 1/3 | 1/1 | 4/9 | 5/4 | 3/3 | 2/0 |
| 82 (IR) | AF293961 | 6/2 | 2/4 | 2/2 | 4/10 | 8/5 | 2/5 | 1/5 |
| 6b (LR) | X00203 | 0/1 | 3/3 | 3/3 | 4/3 | 5/8 | 2/2 | 6/6 |
| 11 (LR) | M14119 | 0/0 | 3/3 | 3/3 | 4/3 | 6/8 | 2/2 | 6/5 |
| 26 (LR) | X74472 | 5/3 | 3/4 | 3/4 | 5/11 | 5/6 | 2/6 | 2/4 |
| 34 (LR) | X74476 | 3/5 | 3/3 | 3/3 | 3/10 | 3/5 | 3/3 | 4/3 |
| 40 (LR) | X74478 | 3/1 | 9/1 | 9/1 | 5/9 | 8/4 | 5/3 | 2/9 |
| 42 (LR) | M73236 | 3/4 | 4/3 | 4/5 | 5/11 | 9/6 | 0/4 | 3/10 |
| 43 (LR) | U12504 | —/—c | —/— | —/— | 7/— | 9/— | —/3 | 3/8 |
| 44 (LR) | U31788 | 1/3 | 4/6 | 4/4 | 5/9 | 6/4 | 4/1 | 5/3 |
| 54 (LR) | NC_001676 | 4/1 | 3/4 | 2/2 | 6/8 | 9/4 | 2/4 | 2/6 |
| 73 (LR) | X94165 | 3/1 | 5/4 | 5/4 | 3/11 | 6/6 | 2/3 | 2/1 |
The published primer sequence was compared to HPV DNA sequences obtained from GenBank (http://www.ncbi.nlm.nih.gov) and the HPV sequence database at Los Alamos National Laboratory (http://hpv-web.lanl.gov).
HR, high risk; IR, intermediate risk; LR, low risk.
—, for HPV type 43, only a partial sequence is available from the indicated sources.
The L1C1-L1C2M, L1S-L-L1S-R, pU-1M-pU-2R, and E6-L-E6-R primer sets were further tested for their abilities to amplify HPV by using plasmids with HPV type 16, 18, 31, 33, 6, and 11 DNA. All the primer sets worked well with all of these major HPV types. The smallest amount of each virus that was detectable depended on the number of mismatches between the primer and the template. HPV types with fewer mismatches were easier to amplify. Primer sets containing a mismatch of >5 nucleotides did not consistently amplify HPV DNA (data not shown). We chose the L1C1-L1C2M primer set for use with clinical samples because its sequence had few mismatches when it was used with high- and intermediate-risk HPV types (Table 2). Also, the size of the product permitted good amplification from samples embedded in paraffin and separation by dHPLC.
Establishment of a set of HPV standard control patterns.
The resolution of DNA fragments by dHPLC depends on several factors, including the denaturing temperature, the gradient concentrations of buffer A and buffer B, flow rate, slope, and run time. All these factor need to be optimized to distinguish among the GP-PCR products of the common genital HPV types.
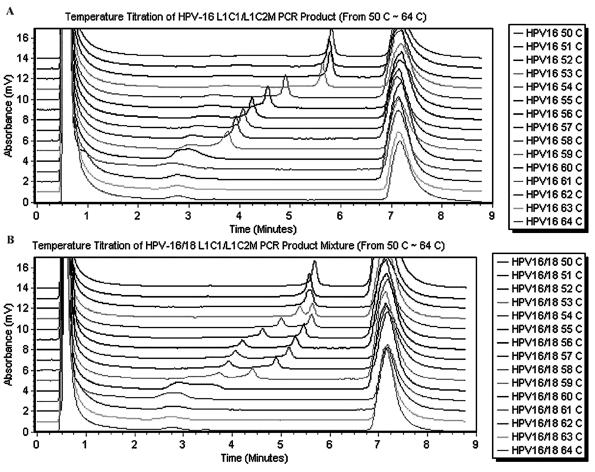
To determine the temperature at which the different HPV PCR products could be distinguished, we performed a temperature titration of the GP-PCR products from the most commonly encountered HPV types, types 16, 18, 31, and 33, using a temperature range of 50 to 64°C. Samples containing individual HPV types as well as mixtures of different types were tested. Samples containing just one HPV type gave a single peak over the entire temperature range (Fig. 1A), whereas samples containing two HPV types displayed two clearly separated peaks only between 56 and 58°C (Fig. 1B). We therefore selected the temperature range 56 to 58°C for further testing.
FIG. 1.
Temperature titration of individual GP-PCR products and mixtures of GP-PCR products. GP-PCR products were amplified by using primers L1C1-L1C2M. HPV controls were run through a temperature gradient of 50 to 64°C. (A) Temperature titration of HPV type 16 GP-PCR product. The eluted peak for HPV is between 3 and 6 min; the large peak at 7 min is what was washed off between two samples. (B) Temperature titration of a mixture of GP-PCR products from both HPV type 16 and HPV type 18. The two separate well between 53 and 59°C.
The temperature titration revealed that each HPV type had a specific retention time at a given temperature (data not shown). Therefore, we mixed the most common genital HPV types together in order to establish an HPV standard control pattern for future HPV typing. After optimizing the denaturing temperature, gradient concentration, flow rate, slope, and run time, we established a set of standard control patterns for HPV typing on the Wave system. The set included the general standard pattern and standard patterns for groups I to IV. The conditions used to obtain these standard control patterns are listed in Tables 3 and 4.
TABLE 3.
dHPLC conditions for HPV genotyping
| Groupa | Run time (min) | Pump flow rate (ml/min) | Slope (% buffer B/min) | Oven temp (°C) |
|---|---|---|---|---|
| General | 13.2 | 0.9 | 1.2 | 57 |
| I | 10.1 | 0.6 | 1.2 | 57 |
| II | 10.8 | 0.5 | 1.2 | 58 |
| III | 10.1 | 0.6 | 1.2 | 58.9 |
| IV | 17.6 | 0.4 | 0.12 | 57.4 |
See Fig. 3 for the HPV composition of each group.
TABLE 4.
Five different gradient conditions for HPV genotypinga
| Step | General
|
Group I
|
Group II
|
Group III
|
Group IV
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (min) | % A | % B | Time (min) | % A | % B | Time (min) | % A | % B | Time (min) | % A | % B | Time (min) | % A | % B | |
| Loading | 0.0 | 49 | 51 | 0.0 | 49 | 51 | 0.0 | 49 | 51 | 0.0 | 48 | 52 | 0.0 | 47 | 53 |
| Start gradient | 0.5 | 48 | 52 | 0.5 | 48 | 52 | 0.5 | 48 | 52 | 0.5 | 47 | 53 | 0.5 | 46 | 54 |
| Stop gradient | 7.3 | 45 | 55 | 5.0 | 47 | 53 | 5.0 | 47 | 53 | 5.0 | 46 | 54 | 9.0 | 45 | 55 |
| Start clean | 7.4 | 0 | 100 | 5.1 | 0 | 100 | 5.1 | 0 | 100 | 5.1 | 0 | 100 | 9.1 | 0 | 100 |
| Stop clean | 7.9 | 0 | 100 | 5.6 | 0 | 100 | 5.6 | 0 | 100 | 5.6 | 0 | 100 | 9.5 | 0 | 100 |
| Start equilibrate | 8.0 | 49 | 51 | 5.7 | 49 | 51 | 5.7 | 49 | 51 | 5.7 | 48 | 52 | 9.6 | 47 | 55 |
| Stop equilibrate | 11.0 | 49 | 51 | 6.8 | 49 | 51 | 6.8 | 49 | 51 | 6.8 | 48 | 52 | 12.6 | 47 | 55 |
See Fig. 3 for the HPV composition of each group. A and B refer to buffers A and B, respectively, as defined in the text.
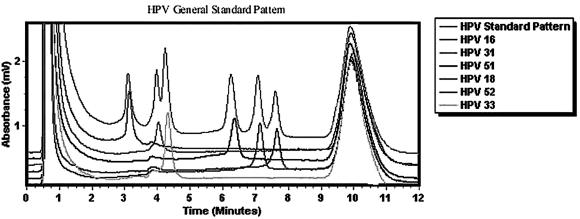
The general standard pattern included six of the most common high-risk and intermediate-risk HPV types (HPV types 16, 31, 33, 51, 52, and 18). The HPV types are displayed from left to right in the chromatograph in Fig. 2. This general standard pattern was used as the preliminary typing step. It was not precise enough to distinguish among all the common genital HPV types.
FIG. 2.
HPV general standard pattern. The general standard pattern is that obtained with a mixture of six common high-risk and intermediate-risk HPV types: types 16, 31, 33, 51, 52, and 18. The PCR products for these controls are mixed together and are shown on the top elution run. The retention time is unaffected by mixing, as can be seen by the run for each individual sample below. The gradient conditions for this general standard pattern are presented in Tables 3 and 4.
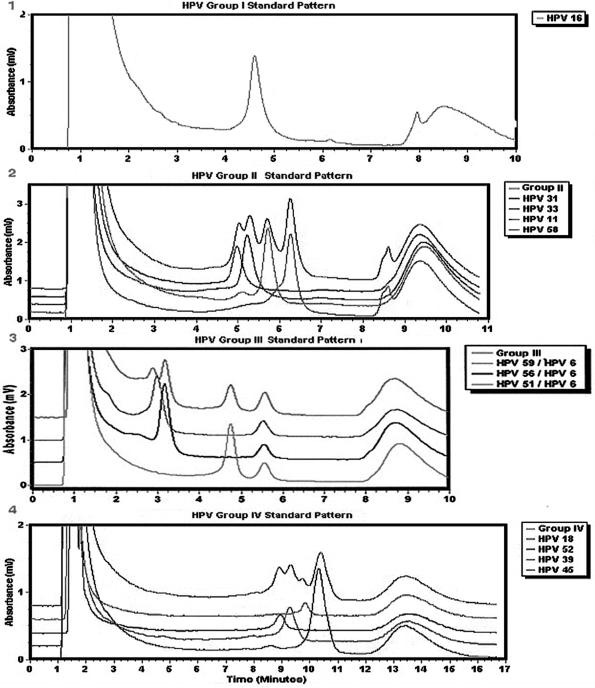
The final identification of HPV types was achieved by mixing each sample with an HPV control, running the mixture under one of the four subgroup conditions, and then sequencing the products with novel peaks, i.e., variants and novel types (Fig. 3). Each subgroup contains one to four HPV types, and the 13 HPV types in Fig. 3 comprise over 95% of the viruses to be identified. The subgroups were determined by comparing the retention times of the most common viral types through the column.
FIG. 3.
Standard patterns for groups I to IV. After the unknown sample is mixed with the control with which it is most likely to match, the mixed sample is run under one of four conditions: panel 1, the group I standard pattern is for HPV type 16 only; panel 2, the group II standard pattern includes HPV types 31, 33, 11, and 58; panel 3, the group III standard pattern includes HPV types 59, 56, 51, and 6; panel 4, the group IV standard pattern includes HPV types 18, 52, 39, and 45. The gradient condition for each standard pattern is presented in Tables 3 and 4.
The precise compositions of buffers A and B are key factors affecting the retention times and resolutions of the peaks. Thus, HPV peaks shifted slightly between batches of buffer, creating the possibility of assigning a sample to the wrong control. To catch such errors, we ran HPV standard controls after about every 10 to 15 samples were run.
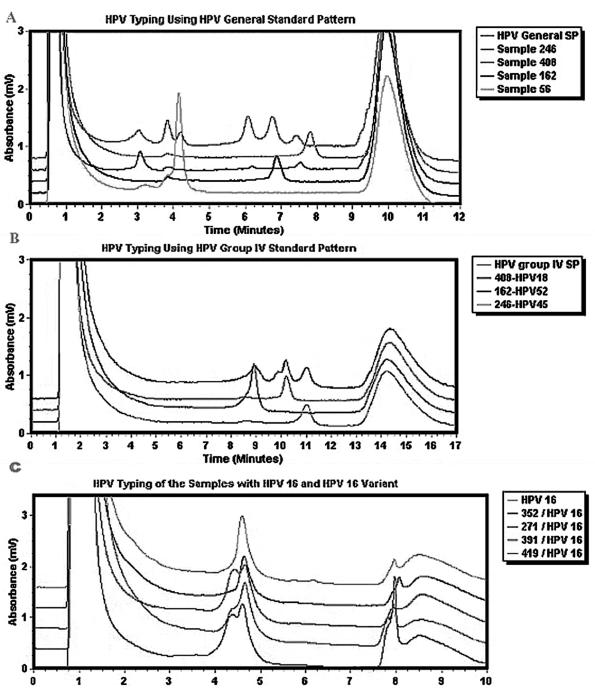
Use of GP-dHLPC to detect and type clinical specimens.
Detection and typing of HPV in clinical specimens were performed in three steps. First, HPV was detected by GP-PCR with the L1C1-L1C2M general primer set. The presence and size of the PCR products were ascertained by agarose gel electrophoresis. Second, each GP-PCR product was analyzed with the Wave system. The general standard pattern condition was used to compare the retention time of the DNA sample to that of the control HPV and to assign each sample to one of four groups (Fig. 4A). Finally, each GP-PCR product (or a single DNA peak collected from the column) was mixed with an equal amount of the control that displayed a similar retention time or with the most prevalent HPV type encountered within its group. The mixtures were analyzed on the Wave system by using a group-specific standard pattern condition (Fig. 4B and C). Thus, samples assigned to group I were mixed with HPV type 16, samples assigned to group II were mixed with HPV type 31 or 33 (depending on the sample's position during the first run), samples assigned to group III were mixed with HPV type 51, and samples assigned to group IV were mixed with HPV type 18, 52, or 45. If the mixture produced a single peak with the same retention time as that for the known HPV control, the HPV genotype of the sample was confirmed. If the mixture revealed a shoulder or double peak (Fig. 4C, samples 271, 391, and 419), it was assumed that a different HPV type or variant was present. In this case, the DNA product was sequenced from both ends to determine the genotype.
FIG. 4.
HPV genotyping of cervical cancer tissue specimens. (A) A first run of the HPV general standard pattern followed by the PCR product of four unknown samples. (B) The second run mixes the unknown samples with HPVs from group IV. Because each sample in the mixture produces a sharp peak, it is easy to deduce that the sequence of sample 408 is identical to that of HPV type 18, that the sequence of sample 162 matches that of HPV type 52, and that the sequence of sample 246 matches that of HPV type 45. (C) A second run that mixes unknown samples with HPV type 16 by using group I conditions. The peaks produced by samples 271, 391, and 419 contain a shoulder; sequencing of the PCR product revealed an HPV type 16 variant.
We analyzed 186 cervical tissue samples for the presence and type of HPV. More than 95% of the samples were infected with the virus. The validity of HPV genotyping was obtained by direct sequencing of 12 PCR products from several HPV types. In addition, all samples that displayed a peak different from that displayed by the HPV control were sequenced, and in all cases a different type or variant was identified. We discovered 25 HPV variants in all: 12 HPV type 16 variants; 8 HPV type 18 variants; 2 HPV type 59 variants; and 1 variant each of HPV types 45, 52, and 39 (Table 5). Every variant was confirmed by bidirectional sequencing. The sequences of the HPV variants differed from the HPV prototype recognized in GenBank by 1 to 5 bp. Because of the degeneracy of the genetic code, not all of the HPV variants had altered amino acid sequences.
TABLE 5.
HPV variants identified in this study
| HPV type and variant | No. of basepair mismatches | Mutation locationa | Codon change | Protein sequence changeb | GenBank accession no. of known variant | No. of cases |
|---|---|---|---|---|---|---|
| 16 variant 1 | 1 | 5696 | AAG-AAA | Lys(46)-Lys(46) | U37217 | 8 |
| 16 variant 2 | 2 | 5637 | AGT-AGC | Ser(33)-Ser(33) | None | 2 |
| 5696 | AAG-AAA | Lys(46)-Lys(46) | ||||
| 18 variant 1 | 1 | 5701 | CCC-CGC | Pro(91)-Arg(91) | None | 4 |
| 18 variant 2 | 2 | 5619 | TTG-ATG | Leu(64)-Met(64) | None | 3 |
| 5701 | CCC-CGC | Pro(91)-Arg(91) | ||||
| 18 variant 3 | 5 | 5648 | TAT-TAC | Tyr(73)-Tyr(73) | None | 1 |
| 5684 | ACC-ACT | Thr(85)-Thr(85) | ||||
| 5701 | CCC-CGC | Pro(91)-Arg(91) | ||||
| 5729 | AGC-AGT | Ser(100)-Ser(100) | ||||
| 5789 | AAG-AAA | Lys(120)-Lys(120) | ||||
| 59 variant | 1 | 5683 | GAG-GAT | Glu(26)-Asp(26) | None | 2 |
| 39 variant | 1 | 5778 | GTA-GTG | Val(45)-Val(45) | None | 1 |
| 45 variant | 1 | 5675 | AGC-AAC | Ser(49)-Asn(49) | None | 1 |
| 52 variant | 1 | 5828 | AAA-AAG | Lys(88)-Lys(88) | None | 1 |
The number indicates the location of the mutation in the complete HPV viral genome given in GenBank. The GenBank accession numbers are as follows: HPV type 16, K02718; HPV type 18, X05015; HPV type 59, X77858; HPV type 39, M62849; HPV type 45, X74479; HPV type 52, X74481.
The numbers in parentheses represent the locations of the amino acid in the HPV L1 gene.
Multiple HPV infections were detected in 26.6% of the cervical cancer tissue samples. GP-dHLPC could identify multiple HPV infections easily and reliably by detecting multiple peaks (Fig. 4A, sample 408). The HPV types in the majority of samples infected with multiple HPV types could be determined by mixing the PCR product with multiple HPV controls, particularly if two different groups were used in the second run (Fig. 4A and B). When a new peak was noted for a sample containing more than one HPV variant or type, we used the Wave system's Fragment Collector to isolate the peaks before sequencing the fragments.
DISCUSSION
We have developed a novel method for genotyping genital HPV that uses GP-PCR followed by dHPLC on the Wave system. The Wave system is a commercially available automated technology for detecting variations in DNA sequences. The PCR products can be processed in a 96-well format and continuously loaded on the column for analysis without any initial purification. This technique allows reliable discrimination of the most common genital HPV types detected by the consensus PCR primers as well as identification of HPV variants whose sequences diverge by as little as 1 nucleotide in a ∼250-bp segment of the HPV L1 region. It can also detect and type multiple HPV types infecting patients in an easy and potentially more accurate way than hybridization techniques, which may have difficulty sorting multiple types due to cross-hybridization of signals. Finally, this method has the potential to quantify HPV according to individual HPV peak areas if proper internal standards are used. These combined advantages make GP-dHPLC a valuable methodology for HPV genotyping.
Sequencing has detected many HPV variants with variations in the E6 (4, 6, 7, 18, 19, 21, 36), E7 (5, 6, 21), LCR (13), and E2 (10) regions. Previous studies have documented a close association between these HPV variants and progression to invasive cervical cancer (11). At present, such variants are identified by PCR with direct sequencing of the PCR products, but this may be difficult without a subcloning step if samples are infected with multiple HPV types. GP-dHPLC is a powerful tool for identifying HPV variants, especially those found in samples with multiple HPV types. In the present study, a number of HPV variants with sequence variations within the L1 gene were identified. We are unaware of any relationship between these variants and the recognized variants with variations in the E6, E7, LCR, and E2 regions. PCR with consensus or HPV-specific primers with dHPLC has the ability to detect any sequence variations within any HPV region. We have designed several primer sets that can reliably distinguish a sequence divergence of only 1 bp within the E6 and E7 genes of HPV type 16 (data not shown).
There are some limitations in the detection and genotyping of genital HPV by GP-dHPLC. First, as is common with all GP-PCRs, the L1C1-L1C2M primers cannot consistently amplify all genital HPV types, especially those with more than five mismatches, such as HPV type 35. For samples that test negative for HPV with the L1C1-L1C2M primer set, use of a second general primer set, such as pU-1M-pU-2R, would improve the sensitivity. Second, we have developed conditions for detecting only about 20 of the more than 40 genital HPV types reported (although these 20 types account for over 95% of the most frequently occurring types), although the specific retention times of additional HPV-positive controls could be determined. Third, the retention times of the eluted peaks change slightly with each new batch of buffer as well as during long sample runs; they also vary according to which model of the Wave system is used. Therefore, in addition to optimizing gradient conditions, it is critical to run HPV-positive controls periodically between runs with unknown samples.
We have demonstrated that HPV detection and typing by GP-PCR in combination with analysis by dHPLC are both reliable and accurate, validating the results obtained by direct sequencing of the GP-PCR products. As clinical indications for the detection and typing of HPV expand, GP-dHPLC offers a new and accurate methodology for HPV analysis.
Acknowledgments
The work was supported by grants from ACS (grant RSG-96-088-08-CCE) to J.S.R. and NIH (grant CA094141) to D.S.G. The Wave system is supported by the BJC Foundation and the Department of Obstetrics and Gynecology at Washington University School of Medicine.
REFERENCES
- 1.Berumen, J., R. M. Ordonez, E. Lazcano, J. Salmeron, S. C. Galvan, R. A. Estrada, E. Yunes, A. Garcia-Carranca, G. Gonzalez-Lira, and A. Madrigal-de la Campa. 2001. Asian-American variants of human papillomavirus 16 and risk for cervical cancer: a case-control study. J. Natl. Cancer Inst. 93:1325-1330. [DOI] [PubMed] [Google Scholar]
- 2.Bible, J. M., C. Mant, J. M. Best, B. Kell, W. G. Starkey, K. Shanti Raju, P. Seed, C. Biswas, P. Muir, J. E. Banatvala, and J. Cason. 2000. Cervical lesions are associated with human papillomavirus type 16 intratypic variants that have high transcriptional activity and increased usage of common mammalian codons. J. Gen. Virol. 81:1517-1527. [DOI] [PubMed] [Google Scholar]
- 3.Bosch, F. X., M. M. Manos, N. Munoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, K. V. Shah, et al. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87:796-802. [DOI] [PubMed] [Google Scholar]
- 4.Buonaguro, F. M., M. L. Tornesello, I. Salatiello, P. Okong, L. Buonaguro, E. Beth-Giraldo, B. Biryahwaho, S. D. Sempala, and G. Giraldo. 2000. The Uganda study on HPV variants and genital cancers. J. Clin. Virol. 19:31-41. [DOI] [PubMed] [Google Scholar]
- 5.Chan, P. K., C. W. Lam, T. H. Cheung, W. W. Li, K. W. Lo, M. Y. Chan, J. L. Cheung, and A. F. Cheng. 2002. Association of human papillomavirus type 58 variant with the risk of cervical cancer. J. Natl. Cancer Inst. 94:1249-1253. [DOI] [PubMed] [Google Scholar]
- 6.Chan, P. K., C. W. Lam, T. H. Cheung, W. W. Li, K. W. Lo, M. Y. Chan, J. L. Cheung, L. Y. Xu, and A. F. Cheng. 2002. Human papillomavirus type 16 intratypic variant infection and risk for cervical neoplasia in southern China. J. Infect. Dis. 186:696-700. [DOI] [PubMed] [Google Scholar]
- 7.Da Costa, M. M., C. J. Hogeboom, E. A. Holly, and J. M. Palefsky. 2002. Increased risk of high-grade anal neoplasia associated with a human papillomavirus type 16 E6 sequence variant. J. Infect. Dis. 185:1229-1237. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs, P. G., F. Girardi, and H. Pfister. 1988. Human papillomavirus DNA in normal, metaplastic, preneoplastic and neoplastic epithelia of the cervix uteri. Int. J. Cancer 41:41-45. [DOI] [PubMed] [Google Scholar]
- 9.Fujinaga, Y., M. Shimada, K. Okazawa, M. Fukushima, I. Kato, and K. Fujinaga. 1991. Simultaneous detection and typing of genital human papillomavirus DNA using the polymerase chain reaction. J. Gen. Virol. 72:1039-1044. [DOI] [PubMed] [Google Scholar]
- 10.Giannoudis, A., M. Duin, P. J. Snijders, and C. S. Herrington. 2001. Variation in the E2-binding domain of HPV 16 is associated with high-grade squamous intraepithelial lesions of the cervix. Br. J. Cancer 84:1058-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giannoudis, A., and C. S. Herrington. 2001. Human papillomavirus variants and squamous neoplasia of the cervix. J. Pathol. 193:295-302. [DOI] [PubMed] [Google Scholar]
- 12.Gravitt, P. E., C. L. Peyton, R. J. Apple, and C. M. Wheeler. 1998. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 36:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kammer, C., M. Tommasino, S. Syrjanen, H. Delius, U. Hebling, U. Warthorst, H. Pfister, and I. Zehbe. 2002. Variants of the long control region and the E6 oncogene in European human papillomavirus type 16 isolates: implications for cervical disease. Br. J. Cancer 86:269-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazarus, P., and S. Caruana. 1996. Typing of common human papilloma virus strains by multiplex PCR. Anal. Biochem. 243:198-201. [DOI] [PubMed] [Google Scholar]
- 15.Londesborough, P., L. Ho, G. Terry, J. Cuzick, C. Wheeler, and A. Singer. 1996. Human papillomavirus genotype as a predictor of persistence and development of high-grade lesions in women with minor cervical abnormalities. Int. J. Cancer 69:364-368. [DOI] [PubMed] [Google Scholar]
- 16.Lorincz, A. T., G. F. Temple, R. J. Kurman, A. B. Jenson, and W. D. Lancaster. 1987. Oncogenic association of specific human papillomavirus types with cervical neoplasia. JNCI 79:671-677. [PubMed] [Google Scholar]
- 17.Lungu, O., X. W. Sun, T. C. Wright, Jr., A. Ferenczy, R. M. Richart, and S. Silverstein. 1995. A polymerase chain reaction-enzyme-linked immunosorbent assay method for detecting human papillomavirus in cervical carcinomas and high-grade cervical cancer precursors. Obstet. Gynecol. 85:337-342. [DOI] [PubMed] [Google Scholar]
- 18.Luxton, J., C. Mant, B. Greenwood, N. Derias, R. Nath, P. Shepherd, and J. Cason. 2000. HPV16 E6 oncogene variants in women with cervical intraepithelial neoplasia. J. Med. Virol. 60:337-341. [PubMed] [Google Scholar]
- 19.Matsumoto, K., H. Yoshikawa, S. Nakagawa, X. Tang, T. Yasugi, K. Kawana, S. Sekiya, Y. Hirai, I. Kukimoto, T. Kanda, and Y. Taketani. 2000. Enhanced oncogenicity of human papillomavirus type 16 (HPV16) variants in Japanese population. Cancer Lett. 156:159-165. [DOI] [PubMed] [Google Scholar]
- 20.Pfister, H. 1987. Relationship of papillomaviruses to anogenital cancer. Obstet. Gynecol. Clin. N. Am. 14:349-361. [PubMed] [Google Scholar]
- 21.Radhakrishna Pillai, M., S. Sreevidya, B. H. Pollock, P. G. Jayaprakash, and B. Herman. 2002. Human papillomavirus type 16 E6 and E7 gene variations in Indian cervical cancer. Gynecol. Oncol. 87:268-273. [DOI] [PubMed] [Google Scholar]
- 22.Reid, R., M. Greenberg, A. B. Jenson, M. Husain, J. Willett, Y. Daoud, G. Temple, C. R. Stanhope, A. I. Sherman, G. D. Phibbs, et al. 1987. Sexually transmitted papillomaviral infections. I. The anatomic distribution and pathologic grade of neoplastic lesions associated with different viral types. Am. J. Obstet. Gynecol. 156:212-222. [DOI] [PubMed] [Google Scholar]
- 23.Resnick, R. M., M. T. Cornelissen, D. K. Wright, G. H. Eichinger, H. S. Fox, J. ter Schegget, and M. M. Manos. 1990. Detection and typing of human papillomavirus in archival cervical cancer specimens by DNA amplification with consensus primers. J. Natl. Cancer Inst. 82:1477-1484. [DOI] [PubMed] [Google Scholar]
- 24.Schiffman, M., R. Herrero, A. Hildesheim, M. E. Sherman, M. Bratti, S. Wacholder, M. Alfaro, M. Hutchinson, J. Morales, M. D. Greenberg, and A. T. Lorincz. 2000. HPV DNA testing in cervical cancer screening: results from women in a high-risk province of Costa Rica. JAMA 283:87-93. [DOI] [PubMed] [Google Scholar]
- 25.Schiffman, M. H., N. B. Kiviat, R. D. Burk, K. V. Shah, R. W. Daniel, R. Lewis, J. Kuypers, M. M. Manos, D. R. Scott, M. E. Sherman, et al. 1995. Accuracy and interlaboratory reliability of human papillomavirus DNA testing by hybrid capture. J. Clin. Microbiol. 33:545-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoppler, M. C., K. Ching, H. Stoppler, K. Clancy, R. Schlegel, and J. Icenogle. 1996. Natural variants of the human papillomavirus type 16 E6 protein differ in their abilities to alter keratinocyte differentiation and to induce p53 degradation. J. Virol. 70:6987-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van den Brule, A. J., C. J. Meijer, V. Bakels, P. Kenemans, and J. M. Walboomers. 1990. Rapid detection of human papillomavirus in cervical scrapes by combined general primer-mediated and type-specific polymerase chain reaction. J. Clin. Microbiol. 28:2739-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Brule, A. J., P. J. Snijders, R. L. Gordijn, O. P. Bleker, C. J. Meijer, and J. M. Walboomers. 1990. General primer-mediated polymerase chain reaction permits the detection of sequenced and still unsequenced human papillomavirus genotypes in cervical scrapes and carcinomas. Int. J. Cancer 45:644-649. [DOI] [PubMed] [Google Scholar]
- 29.van den Brule, A. J., P. J. Snijders, P. M. Raaphorst, H. F. Schrijnemakers, H. Delius, L. Gissmann, C. J. Meijer, and J. M. Walboomers. 1992. General primer polymerase chain reaction in combination with sequence analysis for identification of potentially novel human papillomavirus genotypes in cervical lesions. J. Clin. Microbiol. 30:1716-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Ranst, M., R. Tachezy, and R. D. Burk. 1996. In C. Lacey (ed.), Papillomavirus review. Leeds University Press, Leeds, United Kingdom.
- 31.Villa, L. L., L. Sichero, P. Rahal, O. Caballero, A. Ferenczy, T. Rohan, and E. L. Franco. 2000. Molecular variants of human papillomavirus types 16 and 18 preferentially associated with cervical neoplasia. J. Gen. Virol. 81:2959-2968. [DOI] [PubMed] [Google Scholar]
- 32.Wright, T. C., Jr., J. T. Cox, L. S. Massad, L. B. Twiggs, and E. J. Wilkinson. 2002. 2001 consensus guidelines for the management of women with cervical cytological abnormalities. JAMA 287:2120-2129. [DOI] [PubMed] [Google Scholar]
- 33.Wright, T. C., Jr., L. Denny, L. Kuhn, A. Pollack, and A. Lorincz. 2000. HPV DNA testing of self-collected vaginal samples compared with cytologic screening to detect cervical cancer. JAMA 283:81-86. [DOI] [PubMed] [Google Scholar]
- 34.Wright, T. C., Jr., and R. M. Richart. 1990. Role of human papillomavirus in the pathogenesis of genital tract warts and cancer. Gynecol. Oncol. 37:151-164. [DOI] [PubMed] [Google Scholar]
- 35.Xiao, W., and P. J. Oefner. 2001. Denaturing high-performance liquid chromatography: a review. Hum. Mutat. 17:439-474. [DOI] [PubMed] [Google Scholar]
- 36.Xin, C. Y., K. Matsumoto, H. Yoshikawa, T. Yasugi, T. Onda, S. Nakagawa, M. Yamada, S. Nozawa, S. Sekiya, Y. Hirai, K. Shiromizu, T. Fujii, and Y. Taketani. 2001. Analysis of E6 variants of human papillomavirus type 33, 52 and 58 in Japanese women with cervical intraepithelial neoplasia/cervical cancer in relation to their oncogenic potential. Cancer Lett. 170:19-24. [DOI] [PubMed] [Google Scholar]
- 37.Yoshikawa, H., T. Kawana, K. Kitagawa, M. Mizuno, H. Yoshikura, and A. Iwamoto. 1990. Amplification and typing of multiple cervical cancer-associated human papillomavirus DNAs using a single pair of primers. Int. J. Cancer 45:990-992. [DOI] [PubMed] [Google Scholar]
- 38.Yoshikawa, H., T. Kawana, K. Kitagawa, M. Mizuno, H. Yoshikura, and A. Iwamoto. 1991. Detection and typing of multiple genital human papillomaviruses by DNA amplification with consensus primers. Jpn. J. Cancer Res. 82:524-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zehbe, I., J. F. Sallstrom, M. Evander, K. Edlund, E. Rylander, G. Wadell, and E. Wilander. 1996. Nonradioisotopic detection and typing of human papillomaviruses by use of polymerase chain reaction and single-strand conformation polymorphism. Diagn. Mol. Pathol. 5:206-213. [DOI] [PubMed] [Google Scholar]
- 40.Zehbe, I., E. Wilander, H. Delius, and M. Tommasino. 1998. Human papillomavirus 16 E6 variants are more prevalent in invasive cervical carcinoma than the prototype. Cancer Res. 58:829-833. [PubMed] [Google Scholar]