Abstract
Computational approaches are becoming increasingly popular for the discovery of drug candidates against a target of interest. Proteins have historically been the primary targets of many virtual screening efforts. While in silico screens targeting proteins has proven successful, other classes of targets, in particular DNA, remain largely unexplored using virtual screening methods. With the realization of the functional importance of many non-cannonical DNA structures such as G-quadruplexes, increased efforts are underway to discover new small molecules that can bind selectively to DNA structures. Here, we describe efforts to build an integrated in silico and in vitro platform for discovering compounds that may bind to a chosen DNA target. Millions of compounds are initially screened in silico for selective binding to a particular structure and ranked to identify several hundred best hits. An important element of our strategy is the inclusion of an array of possible competing structures in the in silico screen. The best hundred or so hits are validated experimentally for binding to the actual target structure by a high-throughput 96-well thermal denaturation assay to yield the top ten candidates. Finally, these most promising candidates are thoroughly characterized for binding to their DNA target by rigorous biophysical methods, including isothermal titration calorimetry, differential scanning calorimetry, spectroscopy and competition dialysis.This platform was validated using quadruplex DNA as a target and a newly discovered quadruplex binding compound with possible anti-cancer activity was discovered. Some considerations when embarking on virtual screening and in silico experiments are also discussed.
Keywords: drug discovery, in silico screening, SURFLEX-DOCK, DNA, G-quadruplex, high-throughput screening
INTRODUCTION
DNA is an underrepresented and underutilized molecular target for small molecule therapeutics. In recent surveys of the biochemical classes of the targets of currently used pharmaceuticals, only 1–2% of known drugs were targeted toward DNA [Drews, 2005; Hopkins et al., 2002; Imming et al., 2006]. Historically, drug discovery has largely focused on proteins, but there is an acute need to find and address alternate non-protein drug targets. A recent critical assessment of potential drug targets concluded that only 10–15% of the human proteome was “druggable”, in which the term is defined as the intersection of sets of proteins that are capable of binding “drug-like” molecules and which are the product of disease modifying genes [Hopkins et al., 2002]. The total number of potentially viable protein drug targets may therefore be surprisingly small [Hopkins et al., 2002; Imming et al., 2006], so it is essential to consider other options for drug discovery that involve other biomolecular targets.
DNA IS AN ATTRACTIVE SMALL MOLECULE TARGET
DNA is a fundamentally attractive drug target. The essence of the “antigene” strategy is that it is advantageous to attack disease targets at their source, at the level of gene expression [Le Doan et al., 1987; Moser et al., 1987]. A protein drug target is the product of a particular gene. At each stage of progression through the central dogma (DNA transcription to RNA, and subsequent translation to protein), the absolute number of target molecules to be hit by a drug inhibitor dramatically increases. A single gene makes multiple copies of mRNA, each of which is translated to make multiple copies of the target protein. The number of target molecules is amplified at each stage in the process. By targeting the single gene, rather than the numerous resultant protein molecules, drug action should become both more selective and efficient. Antigene agents can be either small molecule drug or triplex forming oligonucleotides [Praseuth et al., 1999].
DNA is polymorphic, and adopts a wide variety of secondary and tertiary structures within the genome [Neidle, 1999; Sinden, 1994]. Recent efforts to target DNA were directed toward multistranded triplex and quadruplex structures [Hurley et al., 200, 2006; Mergny et al., 1992;1998; Neidle et al., 2000, 2002]. Using small molecules to target such structures represents a new avenue for drug development, one that is just beginning to be recognized and exploited [Hurley, 2001, 2002; Hurley et al., 2006; Jenkins, 2000; Mergny et al., 1992; 1998, 2001, 2002]. The structures of telomeric or gene promoter G-quadruplexes in particular are diverse and present a variety of groove geometries, stacking arrangements, and loop topologies that offer unique receptor sites for small molecule recognition [Yang & Okamoto, 2010]. Quadruplex structures may be unimolecular, bimolecular or tetramolecular and feature stacked G-quartets, in which four guanine nucleotides are hydrogen bonded to form a square plane [Cuesta et al., 2003]. The high-resolution structure determinations on quadruplexes by NMR and x-ray crystallography have been reviewed recently [Burge et al., 2006; Neidle et al., 2003; Patel et al., 2007]. Targeting quadruplex DNA is important as it is thought to be an integral feature of telomeres [Hurley, 2002; Neidle et al., 2000, 2002, 2005; Cuesta et al., 2003]. Formation and stabilization of quadruplex DNA inhibits telomerase (the enzyme responsible for telomere DNA replication) by rendering its substrate DNA inaccessible for binding [Zahler et al., 1991]. Small molecules that stabilize quadruplex structures within the telomere could effectively inhibit telomerase by blocking its binding to its substrate DNA or by preventing elongation during replication [De Cian et al., 2007]. The observation that telomerase levels are elevated in cancer cells led to concerted attempts to target quadruplex DNA within telomeres as one new avenue of cancer chemotherapy. A second class of functionally important G-quadruplex structures is found in promoter regions of a wide-variety of genes, including a number of oncogenes [Qin & Hurley, 2008]. These quadruplexes are structurally distinct from telomeric quadruplexes, and represent another attractive set of potential drug targets.
The discovery of compounds that bind to quadruplexes has significantly increased interest in this field. Arguably the most studied is the cationic porphyrin, TMPyP4 which binds strongly to quadruplex DNA. Unfortunately, a key limitation appearing almost universally among TMPyP4, and other known small molecules that bind to quadruplexes, is the inability to bind selectively to quadruplex structures compared to other DNA morphologies as well as discriminate between quadruplex DNA structures. In the case of TmPyP4, it appears poorly selective for a number of quadruplex structures, including the C-myc and K-ras quadruplexes [Monchaud et al., 2008]. This poor selectivity is likely because many of these ligands consist of large, planar, aromatic surfaces that appear to stack non-specifically on the G-quartet, which is a conserved feature of G-quadruplex structures. The discovery of ligands that may also interact with the grooves and loop regions, which may impart unique selectivity to a ligand, has proved elusive. This has undoubtedly limited the clinical potential of these small molecules. One possible explanation for the failure to discover new small molecules that bind with high selectivity to quadruplexes is the relatively small number of scaffolds reported. The discovery of new compounds has largely been limited to synthesis of new compounds that are derivatives of known scaffolds as well as isolation of new compounds from natural sources. These drug discovery processes are time consuming, expensive, and "low-throughput" efforts that do not explore much of the enormous chemical diversity that is inherently present in small molecule chemical space. An alternative drug discovery approach that addresses these deficiencies is described here and consists of an integrated in silico screening and in vitro validation approach. This platform is an efficient approach for the discovery of new small molecules with novel scaffolds that may target quadruplex DNA.
IN SILICO VIRTUAL SCREENING STRATEGIES TO TARGET DNA
In silico virtual screening techniques are valuable computational tools for the discovery of new small molecules that can bind to a specific target [Kinnings et al., 2009]. Indeed, computational methods have been integrated into the discovery process for over 50 compounds that are in clinical trials as well as marketed drugs [Jorgensen, 2004]. The benefits of in silico virtual screening are its speed, enrichment rates, and affordability, which circumvent the often laborious, slow and expensive process of experimental screening and library synthesis/collection. Moreover, in silico screening explores a much larger chemical space and increases the chance of discovering new compounds with novel chemical scaffolds. It is estimated that up to 20% of new drugs will be found by virtual screening methods in 2010 [Kapetanovic et al., 2008].
There are several considerations when performing virtual screening including choice of structure or ligand-based approaches as well as the in silico chemical library and computational resources required for the screening studies [Dror et al., 2009]. Structure-based virtual screening methods require the availability of a structure of the target, which is usually acquired through high-resolution NMR or X-Ray crystallography techniques or homology modeling. There are many on-line resources available for structures such as the RSCB protein data bank (PDB) and the Nucleic Acid Database (NDB). These noncommercial databases are useful as the structures can be downloaded directly and prepared for virtual screening experiments. Structure-based virtual screening uses software to screen millions of compounds to determine how well each compound can fit into a active site or inhibit partner interaction on the three dimensional target of interest [Zoete et al., 2009]. Structure-based virtual screening by molecular docking involves two steps; the “docking” of the compounds to the target to determine how well the small molecule fits to the target site as well as “scoring” of the poses which determines which of the top poses produced by the software is “correct” [Rester, 2008]. The “scoring” and ranking of the top poses of each ligand in the binding pocket of the target is one of the most challenging aspects of docking [Kinnings et al., 2009]. Molecular docking using programs such as DOCK, Autodock, Ludi, FlexX and Surflex-Dock have been used to find many lead molecules against a variety of targets, such as thymidylate synthase, retinoic acid receptor, kinases, estrogen receptor and thrombin [Ekins et al., 2007; Schapira et al., 2000; Tondi et al., 1999; Baxter et al., 2000]. The use of molecular docking appears well entrenched in academia and industry and its use will likely increase as virtual databases of small molecules and drug targets continue to expand.
A second type of virtual screening is ligand-based approach which, in contrast to structure-based virtual screening, requires knowledge only of the structure of a biologically active ligand. The structure of the active compound is compared to millions of other chemical compounds to check for chemical and morphological similarity. If the structure of the test compound is similar to that of the known active compound, then the test compound may possess similar biological activity [Rester, 2008]. If multiple small molecules are known to possess similar biological activity, a “pharmacophore” can be constructed that describes the chemical properties that are necessary for a ligand to interact with its target. This “pharmacophore” modeling can be particularly useful to detect a wide number of compounds with diverse chemical features [Dror et al., 2005]. One consideration with ligand-based virtual screening is that it does not require knowledge of the structure of the target. This can sometimes be advantageous particularly when controversy surrounds what target structure is “correct” due to the use of different methods (NMR versus X-Ray crystallography versus in vivo state) for determining the structure. On the other hand, it is disadvantageous in that important interactions of the active compound with the target may not be effectively visualized and assessed due to not being present in the small molecule(s). Ligand-based virtual screening has gained increased acceptance as a method to search for derivatives of known biologically active compounds. This approach has also been used to enrich databases for possible selection of lead compounds [Rester, 2008]. Programs such as FlexS and Surflex-Sim have been previously used with success for ligand-similarity based searches [Dror et al., 2005].
Another aspect of virtual screening is the selection of a database of small molecules for screening experiments. The number of compounds for virtual screening has increased dramatically in recent years, with tens of millions of compounds currently available in multiple databases [Rester, 2008]. In our own experience, one of the ZINC databases used for virtual screening experiment increased from approximately 2.7 million compounds in 2007 to approximately 11.3 million compounds in 2010, the majority of which are available from vendors world-wide. There are other databases with even greater numbers of compounds such as Pubchem that had an estimated 37 million compounds in 2009. The value in having such large compound databases is the expanded chemical space that can be explored. This vastly increases the number of small molecules considered as possible lead candidates that is favorable compared to the relatively few molecules that are evaluated by actual chemical synthesis and other drug discovery techniques. Additionally, many of the in silico libraries have been filtered based on specific criteria to increase the chance that the molecules are “drug-like” in behavior. One such filter is "Lipinski’s Rule of 5", which was derived from a structural and statistical analysis of a large library of drugs that were either currently marketed or in clinical trials. The "Rule of 5" characterizes a small molecule as “drug-like” if it: has ≤5 hydrogen bond donors; has ≤10 hydrogen bond acceptors; is ≤500 daltons molecular weight and has an octanol-water partition coefficient (Log P) of ≤5 [Lipinski et al., 2001]. Taken in total, virtual screening against large databases of compounds is a way to discover new lead candidate small molecules against a target of interest.
Another consideration when performing virtual screening experiments is the computational requirement. Given the sheer size of the in silico libraries and complexity of the virtual screening software, larger computer resources are becoming essential to effectively perform virtual screening experiments. In collaboration with the Kentucky Dataseam Initiative (http://www.kydataseam.com/index2.htm), we use a "grid" of >10,000 computer processors on a network that can be used for in silico screening of large chemical compound libraries. This grid can reduce computational docking time from decades to a few days and is a useful resource for our current drug discovery efforts. Benchmarking of candidate software is another consideration because many of the screening software packages can be operated using a multitude of options that can substantially impact docking accuracy and speed. For example, in some recent docking experiments, the software Surflex-Dock was an order of magnitude faster than Autodock even though the software performance (docking accuracy and ranking) was comparable [Holt et al., 2008]. Thus, a researcher should consider the software, number and speed of available computers and in silico library size when performing virtual screening experiments.
While the use of virtual screening for the discovery of new ligands that target proteins has been well established, very few studies have been performed with nucleic acids [Detering et al., 2004; Hurley, 2001]. This may be partly because almost all virtual screening software has been parameterized for proteins, and may not account for characteristics that are particularly important to nucleic acids, such as their distinct geometrical symmetry, and the electrostatic effects of the phosphate backbone. Moreover, there are few published reports of the use of these programs to target nucleic acids [Chen et al., 1997; Evans et al., 2006]. The most notable attempt at screening against DNA morphologies is an early attempt using the first version of DOCK (1.0) by Kuntz, Kollman et al., [Grootenhuis et al., 1994]. It should be noted that DOCK is now five generations further refined (6.4) and there are now several alternatives available such as Autodock and Surflex. In their pioneering study [Grootenhuis et al., 1994] Kollman et al., used the DOCK algorithm to study the interaction of 10,000 compounds from the Cambridge Crystallographic Database with A-, B-, and Z-form DNA [Kuntz et al., 1982]. Despite this success, perhaps the greatest gap in knowledge in this area is the lack of a systematic study to determine whether docking software can accurately reproduce known structures of ligands bound to nucleic acids and also predict the binding mechanisms of small molecules to nucleic acids. We recently addressed this deficiency for the software Surflex-Dock and Autodock and demonstrated unambiguously that these packages can reproduce known structures of small molecules bound to DNA [Holt et al., 2008]. In total, this work suggests that in silico approaches can be applied to DNA to discover new small molecules that bind to a specific target with a known binding mechanism.
THE DISCOVERY FUNNEL: COMBINING IN SILICO SCREENING AND IN VITRO VALIDATION TO DISCOVER NEW COMPOUNDS
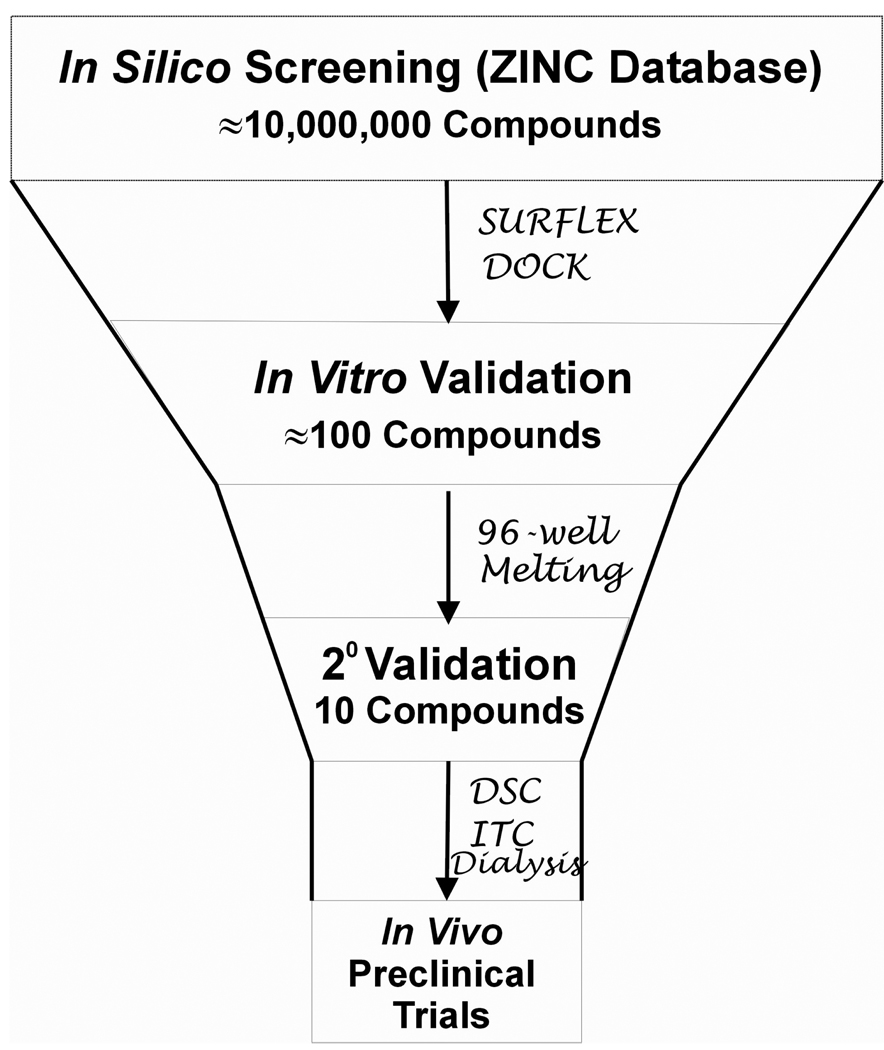
With these in silico strategies and considerations in mind, we have developed an integrated in silico and in vitro screening platform for the purposes of discovering novel small molecules that bind to specific nucleic acid targets (Figure 1). A nucleic acid structure is used as an in silico virtual screening target to probe the binding of virtual libraries of tens of millions of compounds. The virtual screen identifies hundreds of the “best” compounds that will then be tested and validated by a high-throughput 96-well melting assay. The top compounds from the 96-well melting experiments are then subjected to detailed biophysical, molecular dynamics and biological studies. Appropriate feedback loops are included to continuously improve and refine the scoring methods for the virtual screen. As an example of the utility of this platform, we will describe each of the steps in detail and how we integrated this platform to discover a novel compound that binds to the human telomeric quadruplex structure.
Figure 1.
The integrated in silico and in vitro drug discovery platform.
In Silico Screening
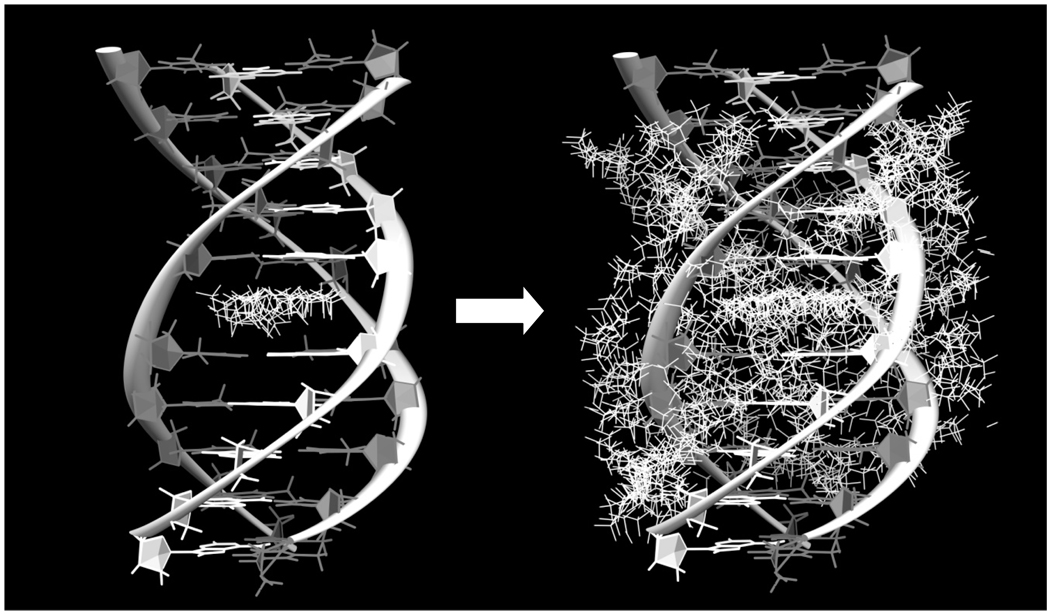
We introduce here a novel in silico approach for discovering new small molecules that bind to a desired DNA target. First is our choice of software. Our previous use and validation of Surflex-Dock have found this software appropriate for targeting DNA [Holt et al., 2008]. It has much faster docking performance (under the conditions previously tested) for large scale virtual screening applications and requires less file preparation than other software. Second is the selection of the in silico library for which we use the ZINC "drug-like" library consisting of approximately 11.3 million compounds. A final component of the in silico screen is the selection of a DNA to target. While the literature shows that many efforts at in silico screening focus on a single site on a target of interest, we believe this is an oversimplification of what potentially occurs in vitro and in vivo. To overcome this deficiency, we have constructed an array of duplex, triplex and quadruplex nucleic acids with variable sequences that represent potential competing binding sites with our target of interest. Additionally, we consider all possible sites on a single target as potential binding sites for a ligand (Figure 2). Figure 2 shows two possible choices for the "protomol" used in ligand docking. In the case shown, one choice (Figure 2A) would be a triplex intercalation site, a choice that would narrow the search but one that would neglect other possible binding modes. Figure 2B shows a better choice, an expanded protomol that encompasses not only the intercalation site, but also several groove binding modes. The relative binding of the small molecule to the site of interest compared to the binding to other sites is used to determine the best "hits." We believe that this in silico setup that evaluates interactions with over a dozen nucleic acid structures is more "representative" of in vitro and in vivo conditions than in silico screening against only a single site and greatly aids in target determining selectivity. This approach was recently validated with our discovery of one of the most highly selective triplex DNA intercalators ever found [Holt et al., 2009]. We have applied this approach to screen for ligands that may bind to a quadruplex structure that is present in the human telomere for the goal of identifying compounds with anti-cancer activity. We screened 11.3 million compounds using Surflex-Dock against the antiparallel "Hybrid-1" quadruplex target (PDB ID: 2HY9) which is one of the proposed quadruplex structures that exists in the human telomere. The top 160 compounds to emerge after scoring for selective binding to this structure were purchased for further evaluation using high-throughput assays.
Figure 2.
The triplex poly dAdTdT is shown with: (Left) the intercalation site targeted by the Surflex-Dock protomol and (Right) all possible binding sites targeted by multiple Surflex-Dock protomols. White bases are adenine and dark grey bases are thymine.
In Vitro Validation
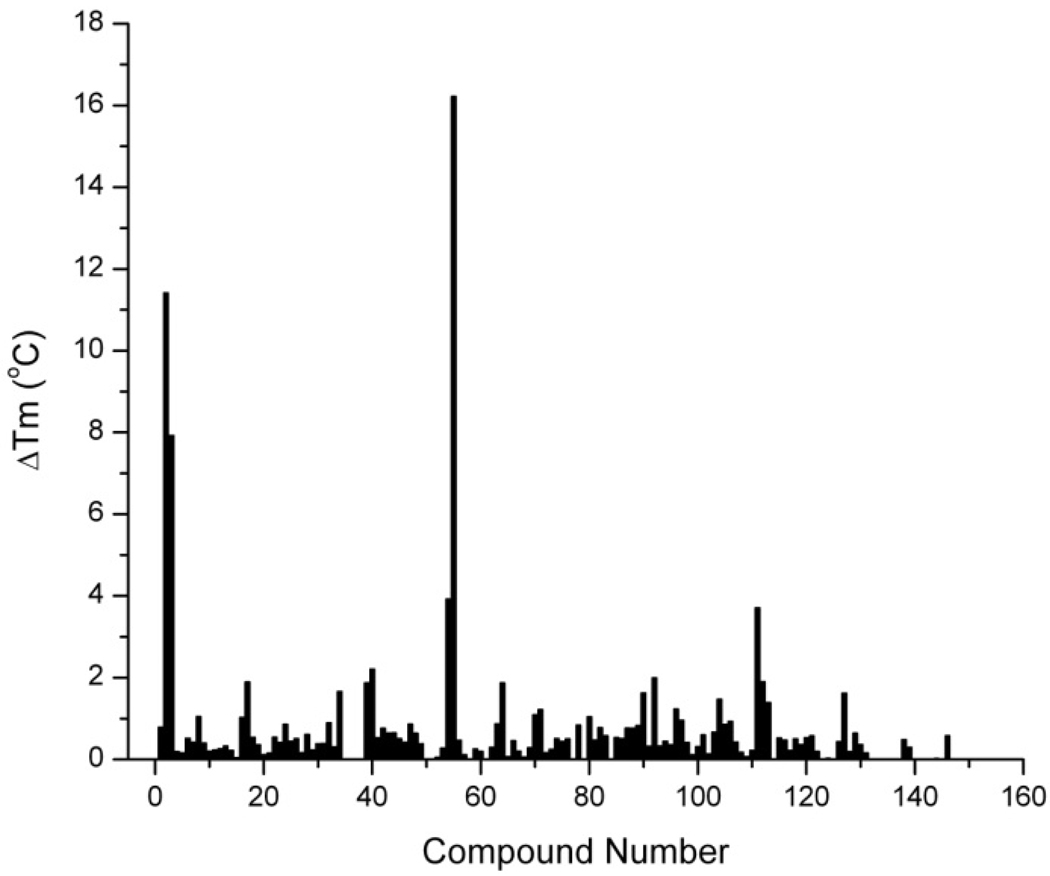
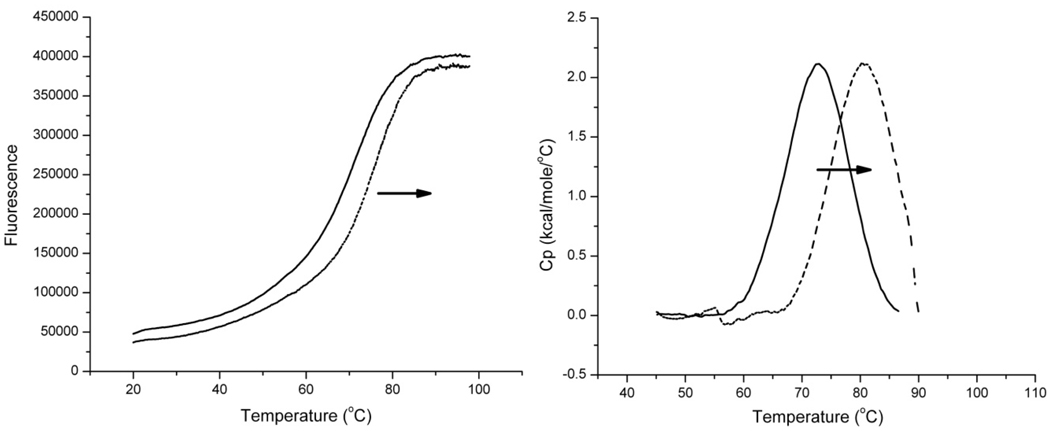
The next step in the testing platform was to develop a high-throughput methodto systemically test the binding of each of the best in silico compounds to the quadruplex target. A recent high-density miniaturized thermal shift assay has proven to be an ideal general strategy for the efficient screening of large libraries of ligands [Pantoliano, et al, 2001]. The assay is based firmly on biophysical principles. Ligand binding to any native macromolecular structure will stabilize that structure against thermal denaturation, and will elevate the melting temperature (Tm). The magnitude of the increase in Tm is a function of binding affinity, stoichiometry and the enthalpies of binding and of denaturation of the native structure [Brandts & Lin, 1990]. We implemented the thermal shift assay using a StepOnePlus™ Real-Time PCR system (Applied Biosystems, Carlsbad, CA) adapted for melting experiments to test the 160 best "hits" that were ordered based on the initial in silico screen. The assay used an antiparallel human telomeric quadruplex structure that incorporated a FRET pair that produced a dramatic increase in fluorescence upon thermal denaturation of the folded form. This melting method is an adaptation of that previously described [Rachwal et al., 2007; Darby et al., 2002; Guedin et al., 2010; Mergny et al., 2003]. The melting assay was chosen due to interest in small molecules that can stabilize the quadruplex structure as shown by an increase in melting temperature of the quadruplex upon addition of a test ligand. While a melting assay was selected for testing purposes here, it should be noted that any number of spectroscopic or fluorescent assays can be adapted to the plate reader to quantify the interaction of small molecules with the target of interest. When the melting assay is adapted to a 96-well plate format, approximately 300 compounds can be screened per day, allowing testing of all of our selected “hit” compounds in a single day. Figure 3 shows the results of the thermal shift assay. After screening the 160 compounds for stabilization of the target quadruplex, it is apparent that five of these stabilize the quadruplex by at least 4°C (Figure 3). Compound 54 was most effective in stabilizing the quadruplex structure, and was chosen for additional studies. Melting curves for the hybrid 1 quadruplex in the presence or absence of compound 54 are shown in Figure 4. The primary data generated from the 96-well instrument are shown in Figure 4A, and the results of secondary screening with differential scanning calorimetry are shown in Figure 4B. These results validate the in silico screen results and the candidates that stabilize the quadruplex the most are selected for further testing.
Figure 3.
In vitro testing by 96-well melting of 160 compounds identified by in silico screening shows that some of the compounds substantially increase the melting temperature of the antiparallel hybrid 1 quadruplex. The ΔTm (the change in melting temperature) is defined as the melting temperature of the quadruplex only substracted from the melting temperature of the quadruplex + ligand.
Figure 4.
96-well melting curves (Left) and DSC Thermograms (Right) for the antiparallel hybrid 1quadruplex without compound 54 (solid black line) and with compound 54 (dashed black line) demonstrating the increase in melting temperature by stabilization of the quadruplex structure.
Secondary Screening
The final step in this integrated platform was testing of the best small molecules from the 96-well melting assay to characterize binding behavior with the quadruplex target. Drug development of lead compounds can be greatly enhanced by detailed knowledge of the thermodynamics of binding to the target [Chaires, 2008]. A number of assays are typically involved in this stage of testing, including rigorous binding studies by calorimetry (isothermal titration calorimetry, differential scanning calorimetry) and spectroscopy [Garbett & Chaires, 2008], competition dialysis studies to validate the structural selectivity of binding [Ragazzon et al, 2007; Ragazzon & Chaires, 2007] structural simulations (Molecular Dynamics) [Trent, 2001] and functional assays (biological assays and tumor inhibition studies). In the case of one of the newly discovered in silico compounds, compound 54, the thermogram generated from DSC shows that once ligand is added to the quadruplex, a right shift in the DSC thermogram occurs, suggesting that the ligand substantially stabilizes the quadruplex (Figure 4B). Again, however, we emphasize that any number of assays can be incorporated into this step to further investigate structural and functional significance of the interaction of a ligand with a DNA target. The totality of work here shows that the in silico and in vitro platform is capable of identifying a compound with quadruplex binding activity.
SUMMARY
DNA continues to be an underrepresented target for new small molecule discovery, despite the potential anti-neoplastic indications for quadruplex stabilizing small molecules. We present here a newly developed in silico and in vitro platform that can be used to discover new small molecules that can bind to therapeutically relevant DNA structures. The platform as outlined here screens over 11.3 million small molecules to a quadruplex structure, performs melting experiments on the top 160 hits and tests the best hits from the melting experiments using other assays to fully characterize structural and functional effects of ligand binding. This platform is powerful for discovering and testing new quadruplex binding ligands, as we demonstrated here but also versatile enough to be tailored to the unique assay expertise in a given laboratory. This approach may be used to discover new compounds with novel scaffolds that may bind to targets that have physiological relevance.
ACKNOWLEDGMENTS
This project was supported by Award Number RO1GM077422 from the National Institutes of Health. The content is solely the responsibility of the authors and does not represent the official views of the NIH.
REFERENCES
- Baxter CA, Murray CW, Waszkowycz B, Li J, Sykes RA, Bone RG, Perkins TD, Wylie W. New approach to molecular docking and its application to virtual screening of chemical databases. J Chem Inf Comput Sci. 2000;40:254–262. doi: 10.1021/ci990440d. [DOI] [PubMed] [Google Scholar]
- Brandts JF, Lin L-N. Study of Strong to Ultratight Protein Interactions Using Differential Scanning Calorimetry. Biochemistry. 1990;29:6927–6940. doi: 10.1021/bi00481a024. [DOI] [PubMed] [Google Scholar]
- Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaires JB. Calorimetry and Thermodynamics in Drug Design. Ann. Rev. Biophysics. 2008;37:135–151. doi: 10.1146/annurev.biophys.36.040306.132812. [DOI] [PubMed] [Google Scholar]
- Chen Q, Shafer RH, Kuntz ID. Structure-based discovery of ligands targeted to the RNA double helix. Biochemistry. 1997;36:11402–11407. doi: 10.1021/bi970756j. [DOI] [PubMed] [Google Scholar]
- Cuesta J, Read MA, Neidle S. The design of G-quadruplex ligands as telomerase inhibitors. Mini Rev Med Chem. 2003;3:11–21. doi: 10.2174/1389557033405502. [DOI] [PubMed] [Google Scholar]
- Darby RA, Sollogoub M, McKeen C, Brown L, Risitano A, Brown N, Barton C, Brown T, Fox KR. High throughput measurement of duplex, triplex and quadruplex melting curves using molecular beacons and a LightCycler. Nucleic Acids Res. 2002;30:e39. doi: 10.1093/nar/30.9.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cian A, Cristofari G, Reichenbach P, De Lemos E, Monchaud D, Teulade-Fichou MP, Shin-Ya K, Lacroix L, Lingner J, Mergny JL. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc Natl Acad Sci U S A. 2007;104:17347–17352. doi: 10.1073/pnas.0707365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detering C, Varani G. Validation of automated docking programs for docking and database screening against RNA drug targets. J Med Chem. 2004;47:4188–4201. doi: 10.1021/jm030650o. [DOI] [PubMed] [Google Scholar]
- Drews J. Drug discovery: a historical perspective. Science. 2000;287:1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- Dror O, Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. Novel approach for efficient pharmacophore-based virtual screening: method and applications. J Chem Inf Model. 2009;49:2333–2343. doi: 10.1021/ci900263d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S, Mestres J, Testa B. In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling. Br J Pharmacol. 2007;152:9–20. doi: 10.1038/sj.bjp.0707305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DA, Neidle S. Virtual screening of DNA minor groove binders. J Med Chem. 2006;49:4232–4238. doi: 10.1021/jm0601957. [DOI] [PubMed] [Google Scholar]
- Garbett NC, Chaires JB. Binding: a polemic and rough guide. Methods Cell Biol. 2008;84:3–23. doi: 10.1016/S0091-679X(07)84001-5. [DOI] [PubMed] [Google Scholar]
- Guedin A, Lacroix L, Mergny JL. Thermal melting studies of ligand DNA interactions. Methods Mol Biol. 2010;613:25–35. doi: 10.1007/978-1-60327-418-0_2. [DOI] [PubMed] [Google Scholar]
- Grootenhuis PD, Roe DC, Kollman PA, Kuntz ID. Finding potential DNA-binding compounds by using molecular shape. Journal of Computer-aided Molecular Design. 1994;8:731–750. doi: 10.1007/BF00124018. [DOI] [PubMed] [Google Scholar]
- Holt PA, Chaires JB, Trent JO. Molecular docking of intercalators and groove-binders to nucleic acids using Autodock and Surflex. J Chem Inf Model. 2008;48:1602–1615. doi: 10.1021/ci800063v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt PA, Ragazzon P, Strekowski L, Chaires JB, Trent JO. Discovery of novel triple helical DNA intercalators by an integrated virtual and actual screening platform. Nucleic Acids Res. 2009;37:1280–1287. doi: 10.1093/nar/gkn1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- Hurley LH. Secondary DNA structures as molecular targets for cancer therapeutics. Biochem Soc Trans. 2001;29:692–696. doi: 10.1042/0300-5127:0290692. [DOI] [PubMed] [Google Scholar]
- Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat Rev Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- Hurley LH, Von Hoff DD, Siddiqui-Jain A, Yang D. Drug targeting of the c-MYC promoter to repress gene expression via a G-quadruplex silencer element. Seminars in oncology. 2006;33:498–512. doi: 10.1053/j.seminoncol.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Hurley LH, Wheelhouse RT, Sun D, Kerwin SM, Salazar M, Fedoroff OY, Han FX, Han H, Izbicka E, Von Hoff DD. G-quadruplexes as targets for drug design. Pharmacol Ther. 2000;85:141–158. doi: 10.1016/s0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- Hurley LH. Secondary DNA structures as molecular targets for cancer therapeutics. Biochem Soc Trans. 2001;29:692–696. doi: 10.1042/0300-5127:0290692. [DOI] [PubMed] [Google Scholar]
- Imming P, Sinning C, Meyer A. Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006;5:821–834. doi: 10.1038/nrd2132. [DOI] [PubMed] [Google Scholar]
- Jenkins TC. Targeting multi-stranded DNA structures. Current Medicinal Chemistry. 2000;7:99–115. doi: 10.2174/0929867003375551. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- Kapetanovic IM. Computer-aided drug discovery and development (CADDD): In silico-chemico-biological approach. Chem Biol Interact. 2008;171:165–176. doi: 10.1016/j.cbi.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnings SL, Jackson RM. LigMatch: a multiple structure-based ligand matching method for 3D virtual screening. J Chem Inf Model. 2009;49:2056–2066. doi: 10.1021/ci900204y. [DOI] [PubMed] [Google Scholar]
- Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. A geometric approach to macromolecule-ligand interactions. J Mol Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- Le Doan T, Perrouault L, Praseuth D, Habhoub N, Decout JL, Thuong NT, Lhomme J, Helene C. Sequence-specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo-[alpha]-thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res. 1987;15:7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- Mergny JL, Duval-Valentin G, Nguyen CH, Perrouault L, Faucon B, Rougee M, Montenay-Garestier T, Bisagni E, Helene C. Triple helix-specific ligands. Science. 1992;256:1681–1684. doi: 10.1126/science.256.5064.1681. [DOI] [PubMed] [Google Scholar]
- Mergny JL, Helene C. G-quadraplex DNA: A target for drug design. Nature Medicine. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- Mergny JL, Lacroix L, Teulade-Fichou MP, Hounsou C, Guittat L, Hoarau M, Arimondo PB, Vigneron JP, Lehn JM, Riou JF, Garestier T, Helene C. Telomerase inhibitors based on quadruplex ligands selected by a fluorescence assay. Proc Natl Acad Sci U S A. 2001;98:3062–3067. doi: 10.1073/pnas.051620698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergny JL, Riou JF, Mailliet P, Teulade-Fichou MP, Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monchaud D, Teulade-Fichou MP. A hitchhiker's guide to G-quadruplex ligands. Org Biomol Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- Moser HE, Dervan PB. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Neidle S. Oxford Handbook of Nucleic Acid Structure. New York: Oxford University Press; 1999. [Google Scholar]
- Neidle S, Parkinson GN. The structure of telomeric DNA. Curr Opin Struct Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat Rev Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- Neidle S, Read MA. G-quadruplexes as therapeutic targets. Biopolymers. 2000;56:195–208. doi: 10.1002/1097-0282(2000)56:3<195::AID-BIP10009>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Neidle S, Thurston DE. Chemical approaches to the discovery and development of cancer therapies. Nat Rev Cancer. 2005;5:285–296. doi: 10.1038/nrc1587. [DOI] [PubMed] [Google Scholar]
- Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5'-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praseuth D, Guieysse AL, Helene C. Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim Biophys Acta. 1999;1489:181–206. doi: 10.1016/s0167-4781(99)00149-9. [DOI] [PubMed] [Google Scholar]
- Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachwal PA, Fox KR. Quadruplex melting. Methods. 2007;43:291–301. doi: 10.1016/j.ymeth.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Ragazzon P, Chaires JB. Use of competition dialysis in the discovery of G-quadruplex selective ligands. Methods. 2007a;43:313–323. doi: 10.1016/j.ymeth.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragazzon PA, Garbett NC, Chaires JB. Competition dialysis: a method for the study of structural selective nucleic acid binding. Methods. 2007b;42:173–182. doi: 10.1016/j.ymeth.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Rester U. From virtuality to reality - Virtual screening in lead discovery and lead optimization: a medicinal chemistry perspective. Curr Opin Drug Discov Devel. 2008;11:559–568. [PubMed] [Google Scholar]
- Schapira M, Raaka BM, Samuels HH, Abagyan R. Rational discovery of novel nuclear hormone receptor antagonists. Proc Natl Acad Sci U S A. 2000;97:1008–1013. doi: 10.1073/pnas.97.3.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden RR. DNA Structure and Function. San Diego: Academic Press; 1994. p. 398. [Google Scholar]
- Tondi D, Slomczynska U, Costi MP, Watterson DM, Ghelli S, Shoichet BK. Structure-based discovery and in-parallel optimization of novel competitive inhibitors of thymidylate synthase. Chem Biol. 1999;6:319–331. doi: 10.1016/S1074-5521(99)80077-5. [DOI] [PubMed] [Google Scholar]
- Trent JO. Advances in DNA Molecular Modeling. An Update. Meth. Enzymol. 2001;340:290–326. doi: 10.1016/s0076-6879(01)40428-9. [DOI] [PubMed] [Google Scholar]
- Yand D, Okamoto K. Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med. Chem. 2010;2:619–646. doi: 10.4155/fmc.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- Zoete V, Grosdidier A, Michielin O. Docking, virtual high throughput screening and in silico fragment-based drug design. J Cell Mol Med. 2009;13:238–248. doi: 10.1111/j.1582-4934.2008.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]