Abstract
Trichophyton tonsurans is the major pediatric pathogen in tinea capitis, causing disparate disease presentations. Little is known about genetic variation, which may ultimately be linked to divergent disease status. This investigation was aimed at identifying genetic variants of T. tonsurans by methods that can facilitate strain discrimination in population-based studies. Ninety-two isolates were acquired from six U.S. microbiology laboratories, and genomic DNA was isolated from mature colonies. The nontranscribed spacer (NTS) was amplified by PCR, and products from isolates with various amplicon sizes were fully sequenced. Nested amplification, targeting a variable internal repeat (VIR) region, allowed assignment of variant type by fragment size. Subvariant type was assigned by a combination of PCR-restriction fragment length polymorphism-based assays. Five variants differing in size (348 to 700 bp) and sequence were identified within the VIR region comprised of several large repeats (104, 140, and 194 bp) arranged in tandem. Seven single-nucleotide polymorphisms (SNPs) were detected across the NTS, with five occurring in the constant regions flanking the VIR region and two occurring in the VIR region. Additionally, a 10-bp insertion and a 14-bp deletion were identified upstream of the VIR region. The combination of SNPs revealed seven haplotype patterns which were stable upon serial passage over 1 year. No sequence variations were identified within the internal transcribed spacer regions. Unique NTS sequences were utilized to develop a duplex PCR assay that discriminated T. tonsurans from other dermatophytes. Of the 92 isolates evaluated, this genotyping scheme distinguished 12 distinct strains, providing evidence of genetic heterogeneity in T. tonsurans.
Cutaneous infections caused by the dermatophyte species remain pervasive in every age group worldwide. Millions of individuals are affected by these pathogens, which contribute to health care costs in excess of $400 million a year for treatment alone (4, 20). In fact, the impact of dermatophyte infections on human disease is so clearly established that a recent white paper by the fungal research community identified Trichophyton rubrum as one of five medically relevant pathogens targeted as initial sequencing candidates by the Fungal Genome Project (2). While T. rubrum is the primary agent of dermatomycoses in adults, Trichophyton tonsurans remains the principal etiologic agent of tinea capitis in the United States and is on the rise in regions of Europe and Japan (5, 16, 19).
Disparate clinical presentations of T. tonsurans tinea capitis are well recognized, with infection spanning the spectrum from an asymptomatic carrier state to a chronic noninflammatory state and a severe acute inflammatory state. While it is likely that interindividual variabilities in both host and fungus work in concert to create these differences, the primary elements that dictate the course of the disease remain to be elucidated. We have recently determined that a significant degree of interstrain variability could be identified in the phenotypic profile of T. tonsurans, with the biochemical expression of proteolytic enzymes spanning several orders of magnitude (1). What remains to be determined is whether this degree of phenotypic difference reflects distinctions in the local microenvironment or is stimulated by factors intrinsic to the fungus (i.e., genetic variability). The relatively limited number of reports attempting to characterize underlying genetic variance within the species prompted us to undertake this investigation aimed at determining whether genetic variability can be elucidated within and between geographically distinct populations of T. tonsurans.
MATERIALS AND METHODS
Specimens.
Ninety-two T. tonsurans cultures, representing scalp isolates from 92 different children, were acquired from six microbiology laboratories across the United States. To ensure purity and to provide a sufficient quantity of fungus for DNA isolation, the organisms were subcultured onto Sabouraud's dextrose agar culture medium with chloramphenicol and cycloheximide (Sab-C; Mycosel; BBL Becton Dickinson, Cockeysville, Md.) and maintained at 25°C for 2 to 4 weeks. Isolates were identified by macroscopic morphology and microscopy at the source institution and by the principal investigator (S. M. Abdel-Rahman). In addition, randomly selected isolates (n = 54) were simultaneously subcultured onto a Sab-C slant for transfer to an independent reference mycology laboratory to confirm species identification. Prototype isolates underwent serial passage every 2 to 4 weeks over 12 months and were reassessed by the methods described below to verify the integrity of the genotyping assays and the stability of the haplotype.
Non-T. tonsurans dermatophytes and nondermatophyte species were evaluated to determine whether related and/or clinically relevant dermatophytes would yield PCR amplification products under the conditions employed for our T. tonsurans samples. This panel was acquired from the American Type Culture Collection (ATCC, Manassas, Va.) and included T. rubrum (ATCC 28188), Trichophyton mentagrophytes (ATCC 28185), Trichophyton violaceum (ATCC 8376), Trichophyton soudanense (ATCC 24583), Trichophyton schoenleinii (ATCC 22775), Trichophyton megninii (ATCC 12106), Trichophyton verrucosum (ATCC 36058), Trichophyton equinum (ATCC 22443), Microsporum audouinii (ATCC 11348), Microsporum canis (ATCC 36299), Microsporum gypseum (ATCC 24102), Microsporum nanum (ATCC 11832), Microsporum persicolor (ATCC 26042), Microsporum ferrugineum (ATCC 42560), Epidermophyton floccosum (ATCC 52066), Candida albicans (ATCC 18804), Aspergillus fumigatus (ATCC 1022), and Malassezia furfur (ATCC 44344). Saccharomyces cerevisiae DNA was purchased from Invitrogen (Carlsbad, Calif.).
DNA isolation.
Isolation of fungal DNA was accomplished by using the DNeasy plant minikit (QIAGEN Inc., Valencia, Calif.) with a slight modification. Fungal mycelia and spores were harvested by flooding the Sab-C plate with phosphate-buffered saline and scraping the colony surface with a sterile loop. The spore suspension was transferred to a 15-ml conical centrifuge tube and centrifuged at 4,000 rpm (1,900 × g) for 25 min. The excess phosphate-buffered saline was decanted, the pellet was transferred to a 1.5-ml microcentrifuge tube, and the sample was lyophilized under vacuum (Speed Vac; Savant, Holbrook, N.Y.).
Approximately 10 to 20 mg of the dried material was suspended in 200 μl of lysis buffer and 2 μl of RNase A. The samples were incubated at 65°C for 30 min, frozen at −70°C for 30 min, and thawed unassisted at room temperature. An additional 200-μl volume of lysis buffer and 2 μl of RNase A were added, and the samples were incubated overnight at 65°C under constant agitation. The column-based DNA purification was carried out according to the manufacturer's protocol, with the exception that the columns were heated to 70°C for 5 min immediately prior to DNA elution. Four separate elutions were performed to maximize the DNA concentration: an initial 30-μl elution, which was discarded, followed by three elutions of 100, 70, and 200 μl. This elution scheme was determined based on preliminary experiments designed to optimize DNA recovery. Elutions 2 through 4 were reserved and stored at 4°C. Elutions were tested for the presence of DNA by amplifying a 368-bp fragment from the ribosomal DNA (rDNA) region containing the internal transcribed spacer region 2 (ITS-2) sequence. (Oligonucleotide primer sequences for all reactions described herein are provided in Table 1.) The reaction was carried out with RedJump Start Taq polymerase (Sigma, St. Louis, Mo.) as recommended by the manufacturer by using primers 5.8S and ITS-4 with 1 μl of genomic DNA in a 15-μl reaction volume. PCR cycling conditions included an initial denaturing step at 94°C for 2 min; 35 cycles of denaturing at 94°C for 10 s, annealing at 54°C for 10 s, and extension at 72°C for 20 s; a hold at 72°C for 5 min; and a final hold at 4°C. All subsequent PCR applications were performed with the elution yielding the strongest product band as assessed by agarose gel electrophoresis.
TABLE 1.
PCR primers employed in the genotyping assaya
| Forward primer | Sequence | Reverse primer | Sequence |
|---|---|---|---|
| 5.8S | 5′ GTA TCG ATG AAG AAC GCA GCG | 5.8SR/2 | 5′ CGC TGC GTT CTT CAT CGA TGC |
| ITS-5 | 5′ AAG TAA AAG TCG TAA CAA GGT TTC CG | ITS-3 | 5′ TAC CAC CAA GAT CTG CAC TAG AGG |
| ITS-5/2 | 5′ TAA AAG TCG TAA CAA GGT TTC CG | ITS-4 | 5′ TCC TCC GCT TAT TGA TAT GC |
| NTS-1 | 5′ GGG TTT AGA CCG TCG TGA GAC AG | NTS-2 | 5′ ATT GCT TAT ACT TAG ACA TGC ATG GC |
| NTS-8 | 5′ GAG CGT CAG CCC GAT CCT TGC C | NTS-6 | 5′ ATA TGA CTA CTG GCA GGA TCA ACC AG |
| NTS-9 | 5′ GGC TGT CTG ACA AGG CAT TGC CG | NTS-8R | 5′ AAG GAT CGG GCT GAC GCT CAG C |
| NTS-13 | 5′ GTA GAA TGC TCC CAA CCA CTC CG | NTS-16 | 5′ GCA TAA AAT ATA TTC TGG CGG CCT C |
| NTS-20 | 5′ GCG TTT GCC CTA GAC GC | NTS-21 | 5′ TCG AAA GTG GGC TTG ACG |
| NTS-24 | 5′ CCT CAC ACG TGA CTA CTA CCA CTC CAA TAT | NTS-22 | 5′ CCT CCA AAT CGA CCG TAG AGC |
| NTS-25 | 5′ CTA CCC GCC GAC TTA TGC | NTS-26 | 5′ AAC GTA GAG AAG ATA CCC TTC GGG TAC |
| NTS-28 | 5′ TGG ACC CTC TGG CCG AG | NTS-27 | 5′ AAC GTA GAG AAG ATA CCC TTC GAG TAC |
| NTS-32 | 5′ TCC GAC AGC TAA GCT TAG G | NTS-29 | 5′ CCA CTC GAT TTT GGA GG |
| NTS-35 | 5′ GCT TGT CGA TTG TTG GCC G) | NTS-33 | 5′ GGA GCA TTC TAC AAC CTC G |
| NTS-40 | 5′ AGT CCG ACA GCT AAG CTT AGG | NTS-36 | 5′ TGC CCG CGG ATC TGG CGT C |
| NTS-37 | 5′ CGG AGT GGT TGG GAG CAT TC |
Partial restriction sites are indicated with boldface text; introduced mismatches are underlined.
PCR amplification of the NTS region.
The nontranscribed spacer (NTS) region was amplified by using primers directed at the highly conserved 5′-flanking 25S (NTS-1) and the 3′-flanking 18S (NTS-2) rRNA gene regions. Each primer produced perfect alignments when we compared them with other species in the rRNA database at http://oberon.rug.ac.be, suggesting that the primers would bind similarly to T. tonsurans. In anticipation of finding fragments between 2 and 5 kb in length, long PCR was performed with Herculase Taq polymerase (Stratagene, La Jolla, Calif.). The reaction mixture composition was that recommended by the manufacturer but supplemented with dimethyl sulfoxide (DMSO) to a final concentration of 5%. One microliter of genomic fungal DNA served as a template for the 12-μl reaction mixtures. PCR conditions included an initial denaturation step (94°C for 2 min); 35 cycles consisting of 94°C for 10 s, annealing at 52°C for 10 s, and extension at 68°C for 3.25 min; and a final hold at 68°C for 5 min with a 4°C hold to complete the reaction. Typically, 1 to 2 μl of product revealed a visible band by gel electrophoresis; however, product yield varied considerably between samples. Hence, a seminested PCR was carried out on a 100- to 2,000-fold-diluted NTS-1-NTS-2 PCR product to generate sufficient sequencing template. Amplification was performed with the original NTS-1 primer paired with a nested primer (NTS-6). PCR cycle conditions were identical to those described above, with the exception that the Eppendorf TripleMaster PCR system with Tuning Buffer (Brinkmann, Westbury, N.Y.) was used at an annealing temperature of 66°C. The NTS-1-NTS-6 product was 19 bp shorter than the NTS-1-NTS-2 product and encompassed the entire NTS region, including approximately 415 and 29 bp of the 25S and 18S rRNA genes, respectively, based on the putative definition of NTS according to a published T. rubrum sequence (GenBank accession no. AF222887). PCR product lengths ranged from 2.5 to 2.9 kb depending on the strain of T. tonsurans. For simplicity, we refer to these PCR fragments containing the NTS as well as the flanking partial 25S and 18S sequences as the NTS region.
PCR amplification of the ITS region.
In order to generate sufficient and clean PCR product for sequence analysis, a 2-kb-long fragment encompassing the entire ITS region along with portions of the 18S and 25S rRNA genes was first amplified (using ITS-5-ITS-3). PCR was performed with 1 μl of genomic DNA in a reaction mixture of 12 μl by using Platinum high-fidelity Taq polymerase (Invitrogen) as recommended by the supplier's protocol. PCR cycling conditions included an initial denaturing step at 94°C for 2 min; 35 cycles of denaturing at 94°C for 10 s, annealing at 64°C for 10 s, and extension at 68°C for 2 min; a hold at 68°C for 5 min; and a final hold at 4°C. The PCR products were assessed by agarose gel electrophoresis and diluted 500- to 2,000-fold with 10 mM Tris (pH 8) for subsequent nested reamplification, with primers ITS-5 and ITS-4 generating a 709-bp amplicon. PCR conditions were identical to those described above with the exception of a 54°C annealing temperature and a 40-s extension.
Sequence analysis of NTS and ITS.
Sequence analysis was performed with PCR products presenting as strong, single bands on agarose gels. Typically, 12 μl of PCR product was treated with 2.5 μl of ExoSAP-IT (Amersham Biosciences, Piscataway, N.J.) and incubated at 37°C for 30 min and at 80°C for 15 min to remove unwanted deoxynucleoside triphosphates and primers. Sequencing was carried out with the DYEnamic ET dye terminator kit for MegaBACE (Amersham Biosciences) with a reaction mixture containing 4 μl of sequencing premix, 4.5 μl of water, 0.5 μl of 10 μM sequencing primer, and 1 μl of ExoSAP-IT-treated PCR product. The reactions were cycle sequenced as recommended, cleaned up with the CleanSEQ magnetic bead system (Agencourt, Beverly, Mass.), and analyzed on a MegaBACE 500 capillary sequencer. Bases were automatically called, uploaded, and analyzed with the Sequencher DNA analysis software (Gene Codes Corp., Ann Arbor, Mich.).
Characterization of the VIR region.
The variable internal repeat (VIR) region within the NTS was characterized initially by sequence analysis and subsequently by PCR-restriction fragment length polymorphism (RFLP) analysis. The diluted initial PCR product (using NTS-1-NTS-2) was reamplified with two nested primers (NTS-13 and NTS-16) by using the Eppendorf TripleMaster PCR system with Tuning Buffer. PCR cycling conditions included an initial denaturing step at 94°C for 2 min; 35 cycles of denaturing at 94°C for 10 s, annealing at 66°C for 10 s, and extension at 68°C for 1.25 min; a hold at 68°C for 5 min; and a final hold at 4°C. Amplicon lengths ranged between 466 bp (variant I) and 818 bp (variant V) and allowed for variant designation based on fragment size. Subsequent digestion with XbaI (Amersham Biosciences) yielded RFLP patterns that were utilized to confirm variant assignment. Finally, the nature of the first repeat 2 element was confirmed by digestion with NaeI (New England Biolabs, Beverly, Mass.) by an assay first developed to detect single-nucleotide polymorphism (SNP) 5 (see below). Amplification of the VIR region directly from genomic DNA was achieved with Herculase Taq polymerase in the presence of 5% DMSO. Cycling conditions were as described above with the exception that the annealing temperature was 64°C.
PCR-RFLP for the detection of NTS SNPs.
SNPs were labeled numerically according to the order in which they were identified. Likewise, the nucleotide presented first (e.g., the T in T→C) refers to the base pair comprising that position for the initial T. tonsurans isolate that was sequenced (variant Ib). As such, it may not reflect the nucleotide occurring with the greatest frequency at that position.
For the SNP 1 (T→C) genotyping assay, a 524-bp fragment was amplified with primers NTS-8 and NTS-37. In the presence of T, BstYI cut the PCR product into 458- and 66-bp fragments, while the product remained uncut in the presence of C. Conversely, a C-carrying PCR product was cut by AvaI, while a PCR product with a T remained uncut.
For SNP 2 (C→T), which is located within the second and third (where applicable) repeat 1 element of the VIR region, an amplicon was generated with primers NTS-28 and NTS-29. Depending on the number of repeats present in the variant, a 177-bp fragment was generated either from one (variants I and II) or two (variants III, IV, and V) repeat loci within the VIR region. TaqαI cut the fragments three times in the presence of C, yielding 85-, 63-, 22-, and 7-bp products. In the presence of T, one TaqαI site was lost, resulting in a band pattern of 107, 63, and 7 bp. While the majority of variants with three repeat 1 elements contained the same nucleotide at both SNP 2 positions, a single isolate of the variant V size was characterized by a “heterozygous” pattern, with a C at one locus and a T at the other, resulting in one occurrence of the 85-, 63-, 22-, and 7-bp band pattern and one occurrence of the 107-, 63-, and 7-bp band pattern. This finding was confirmed with separate DNA isolations from serial passage of the organism.
To detect the T insertion defining SNP 3, PCR was carried out with primers NTS-24 and NTS-21. A partial SspI restriction site was constructed in primer NTS-24, which allowed for digestion of PCR fragments containing the T insertion with SspI. The resulting fragment sizes were 158 and 29 bp. In the absence of the T insertion, the PCR products remained uncut at 186 bp.
SNP 4 (A→G) was detected by amplification with primers NTS-25 and NTS-26 containing a partial KpnI site, allowing for a subsequent cut by the enzyme in the presence of G. Amplification with the same forward primer and NTS-27 containing a partial ScaI site allowed this enzyme to cut in the presence of A. Digests yielded fragments of 253 and 23 bp (KpnI) and 251 and 25 bp (ScaI) or remained uncut at 276 bp in the presence of either A or G, respectively.
SNP 5 (G→C) is located within the first repeat 1 element in the VIR region. Primers NTS-20 and NTS-22 generate products of variable lengths depending on the number of repeat units present (313, 315, or 367 bp). In addition, a uniform 69-bp product was formed from a secondary primer binding site. In the presence of G, NaeI cut once, giving rise to patterns consisting of 219 and 94 bp or 221 and 94 bp when an uninterrupted repeat 2 element was present and 273 and 94 bp when the repeat 2 element was interrupted with the 54-bp motif.
SNPs 6 (T→C) and 7 (T→C) are located in close proximity and were tested with a single amplification product of 361 bp with primers NTS-35 and NTS-36, which was split for restriction digestion. One half was incubated with BslI for SNP 6, cutting fragments once in the presence of T (180 and 181 bp) and twice in the presence of C (181, 131, and 49 bp). The second half was cut with BclI for SNP 7 genotyping, generating 304- and 57-bp fragments in the presence of T and remaining uncut in the presence of C.
All PCRs were performed with a total volume of 15 μl with RedJump Start Taq polymerase (Sigma) as recommended by the manufacturer. Annealing temperatures were 68 and 53°C for SNPs 1 and 2, respectively, 51°C for SNPs 3 and 4, 52°C for SNP 5, and 55°C for SNPs 6 and 7. Extension times ranged from 15 to 35 s depending on the PCR product length. Restriction digestions were incubated for 1 to 3 h at the recommended temperatures (all enzymes were acquired from New England Biolabs), and the fragments were separated on 3 or 4% Synergel agarose gel matrix (Diversified Biotech, Boston, Mass.). All PCRs reported herein were performed with the appropriate positive and negative controls, and the latter controls all tested negative. DNA markers included a 100-bp and a 1-kb ladder (New England Biolabs).
Detection and characterization of the small insertion and deletion.
DNA was amplified with primers NTS-32 and NTS-33. Deletion-negative and insertion-positive DNAs yielded products of 200 and 210 bp, respectively. The 3′ end of NTS-33 binds to nucleotides that are part of the 14-bp deletion; thus, the mismatch prevented amplification from deletion-positive DNA. The SNP 4-detecting primer pair (NTS-25-NTS-26) was added to the PCR to coamplify a 276-bp fragment as an internal control. The PCR was carried out with RedJump Start Taq polymerase as described above, with an annealing temperature of 53°C and a 10-s extension.
Duplex PCR for identifying T. tonsurans.
Two sets of primers were used to set up a duplex reaction that would discriminate T. tonsurans from other dermatophyte species directly from genomic DNA. The reaction mixture was comprised of set 1 primers (NTS-9-NTS-8R) at 0.07 μM, set 2 (T. tonsurans-specific) primers (NTS-40-NTS-37) at 0.2 μM, 5% DMSO, 0.1 mM (each) deoxynucleoside triphosphate, 1× Red JumpStart buffer with MgCl, 0.25 U of Red JumpStart Taq polymerase (Sigma), and 0.75 μl of genomic fungal DNA in a total concentration of 10 μl. PCR cycling included an initial hold for 2 min at 94°C; 35 cycles of 10 s at 94°C, 10 s at 58°C, and 15 s at 72°C; and holds for 5 min at 72 and 4°C. For analysis, 8 μl of PCR mixture was loaded on a 3% Synergel-agarose gel matrix. Primer set 1 yielded a 300-bp product, and primer set 2 yielded a 207-bp product (193 and 217 bp in T. tonsurans strains containing the deletion and insertion, respectively).
PCR-RFLP for the detection of a reported ITS-1 SNP (C→T).
PCR was performed with RedJump Start Taq polymerase according to the manufacturer's recommendations. Cycling included an initial hold for 2 min at 94°C; 35 cycles of 10 s at 94°C, 15 s at 58°C, and 25 s at 72°C; and holds for 5 min at 72 and 4°C. Primers ITS-5/2 and 5.8SR/2 generate an amplicon spanning 49 bp of the 18S gene, the entire ITS1 region (259 bp), and 51 bp of the 5.8S gene. The PCR products were cut twice with NaeI in the presence of C, giving rise to patterns consisting of 241, 67, and 51 bp. In the presence of T, NaeI cut once, giving rise to a 241- and 118-bp band pattern.
Nucleotide sequence accession number.
The nucleic acid sequence for the NTS region (with repeat elements in the VIR region annotated) and partial 25S and partial 18S gene regions has been deposited in the GenBank database under accession no. AY292646.
RESULTS
High-quality DNA was successfully isolated by employing a simple column-based DNA isolation procedure in conjunction with a freeze-thaw technique rather than the labor-intensive mechanical disruption of fungal material in liquid nitrogen followed by a phenol-chloroform extraction as described by other investigators. The DNA preparations were initially assessed by amplifying a 368-bp fragment spanning the ITS-2 region. The highest amplification yields were observed for elution 2 in most cases and for elution 3 when elution 2 produced a weak signal, with products observed for elutions 2 and 3 of all preparations. In no strain was reisolation of DNA required for successful amplification of this region. As such, the PCRs targeting the 2.5- to 2.9-kb NTS region and the approximately 2-kb ITS-containing fragment yielded sufficient PCR product for easy visualization by agarose gel electrophoresis.
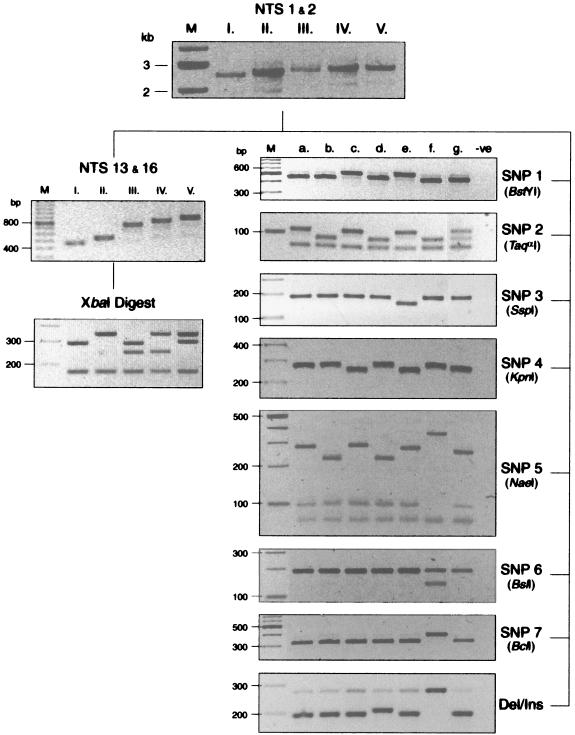
Ninety-two T. tonsurans isolates were acquired from pediatric patients at six different institutions across the United States. We identified five major variants based on the length of the NTS PCR fragment, which ranged from 2.5 to 2.9 kb (Fig. 1, top panel). Complete, contiguous sequences were obtained for all size variants (variant I, n = 6; II, n = 9; III, n = 1; IV, n = 1; V, n = 3), and these data revealed both size and sequence diversity in a VIR region nested within the more constant flanking regions of the NTS (Fig. 2). Sequence data revealed that this region was constituted by the tandem arrangement of two repeat elements (Fig. 2) with smaller subrepeat elements (data not shown) nested internally in the longer of the two repeats (repeat 2). Repeat 1 was characterized by a 104-bp sequence, whereas repeat 2 was defined by a 140-bp sequence which, in variants II, IV, and V, was interrupted by a 54-bp motif (Fig. 3). For routine characterization of the VIR region, a PCR-RFLP-based assay directed at this region was utilized. Restriction digestion of the NTS-13-NTS-16 PCR product, carrying the entire VIR region with XbaI, resulted in specific patterns that allowed easy differentiation of the five variants as shown in the lower left panels of Fig. 1.
FIG. 1.
PCR-RFLP genotyping of T. tonsurans variants and subvariants. Scans of agarose gels are shown in the order of performance (genotyping scheme). Assay details including fragment sizes are provided in Materials and Methods. Lanes are labeled with variant (I to V) or subvariant (a to g). For the SNP 5 panel, only subvariant f remains uncut (evidenced by the absence of the 94-bp band); the remaining subvariants demonstrate an uppermost band at either 219 or 273 bp, depending on whether the first repeat 2 region is uninterrupted or interrupted by the 54-bp motif, respectively. For the deletion-insertion panel, subvariant d indicates the band pattern seen with the 10-bp insertion, and subvariant f indicates the band pattern for isolates possessing the 14-bp deletion. M, molecular size markers; Del, deletion; Ins, insertion. -ve, negative control.
FIG. 2.
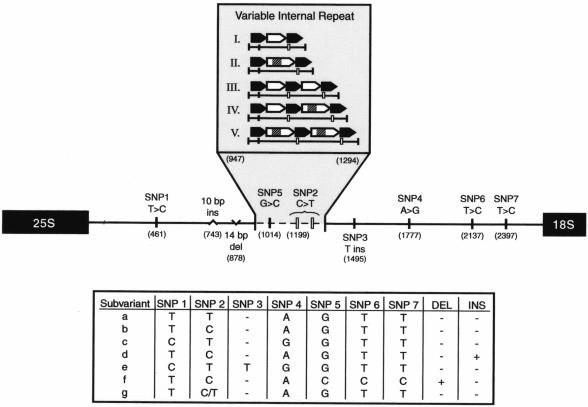
Schematic overview of the VIR region showing the positions of sequence variations within the NTS. The values in parentheses indicate the nucleotide positions of the SNPs relative to the variant I sequence. Resultant haplotype subvariants are shown in the lower panel. The arrows indicate the tandem arrangement of two repeat elements. DEL, deletion; INS, insertion.
FIG. 3.
Full-sequence data for the VIR region by variant (the repeat 1 elements are highlighted in black) and for the insertion and deletion sequences, with repeat motifs underlined.
The sequences of both the repeats and the interruption motif were identical within and among strains, with the exception of two SNPs (SNP 2 and SNP 5) identified in repeat 1 (Fig. 2). In addition, initial sequence analysis of variant IV revealed that the first repeat 2 element contained two additional nucleotides (142 versus 140 bp). These nucleotides were identical in location and sequence to the first 2 bp of the interruption motif; however, the sequence then continued uninterrupted (Fig. 3). Given the difficulty in designing specific primers to target this small sequence variation within the repeat region, the 2-bp insertion was ultimately confirmed for all isolates of the same size (n = 4) by sequence analysis. Aside from variant IV, these two additional nucleotides were not found in any other uninterrupted repeat 2 element that was sequenced (n = 7). Hence, it was presumed to be linked to variant IV, and no additional haplotype designation was assigned to this feature. Overall, more than two dozen smaller repeat elements between 8 and 12 bp in length (not existing in tandem) were found outside of the VIR region throughout the conserved portion of the NTS region. While their significance remains unknown, recognition of such elements was pivotal in primer design.
In addition to interstrain variability in the overall length of the NTS region, seven SNPs were identified: five transitions (SNP 1, T→C; SNP 2, C→T; SNP 4, A→G; SNP 6, T→C; and SNP 7, T→C), one transversion (SNP 5, G→C), and one insertion (SNP 3, T insert). The nucleotide positions for the SNPs relative to the variant I sequence are indicated in Fig. 2. With the exception of SNPs 2 and 5 as noted, the sequence variations occurred in the constant region of the NTS sequence. The restriction pattern for SNP 5 was also informative for additional characterization of the VIR region in isolates whose sizes and XbaI patterns were consistent with those of variant IV. Given that both an interrupted sequence and an uninterrupted repeat 2 sequence were found in the VIR of this variant, the PCR-RFLP band pattern enabled the distinction of whether the interruption motif was located in the first or second of these repeat elements. In all cases, the band pattern suggested that the interruption motif occurred in the second repeat element (Fig. 1), as was confirmed by full sequence data for the region (n = 4).
A 10-bp insertion distinguished several variant I strains. The insertion was positioned immediately downstream of two 10-bp tandem repeats, forming a third repeat unit in the series (Fig. 3). The 14-bp deletion occurred in both variant II and variant V and appeared to share a small (7-bp) repeat motif with the sequence immediately downstream of the deletion (Fig. 3). The constant nature of both the deletion and the insertion between isolates was confirmed by partial sequence analysis for an additional six isolates. While both the deletion and the insertion occurred within a span of approximately 150 nucleotides, no other sequence variations were found within this region. Based on data from these strains, it appears that SNPs 1 and 4 are linked as well as SNPs 5, 6, and 7 and the 14-bp deletion; however, these associations require confirmation in a larger number of isolates.
In combination, the presence or absence of SNPs, the small insertion, and the small deletion yielded seven haplotypes, designated “a” through “g,” which allowed us to further subclassify the existing VIR size variants (i.e., I through V) into a total of 12 genetic subvariants (e.g., Ib, IIc, etc.). We refer to the compound information on VIR size variation and haplotype as the strain's “genotype.” The genotype frequencies are presented overall and by region in Table 2. As indicated in the table, variants IIa (frequency, 0.24) and IIc (frequency, 0.25) along with variant Ib (frequency, 0.22) account for the majority of isolates, irrespective of the region of the country from which the isolates were acquired. By far, the greatest degree of variability was observed in isolates from Columbus, Ohio; Kansas City, Missouri; and Washington, D.C., which is likely due in part to the large number of isolates acquired from the institutions in the last two cities and the relatively large populations of these cities. Aside from this finding, the admittedly limited number of cities evaluated precludes a definitive statement on a trend in population differences, with respect to either variant or subvariant assignment, across the country.
TABLE 2.
Frequency of NTS genetic variants overall and by geographic region of isolationa
| Variant | No. of NTS genetic variants from:
|
||||||
|---|---|---|---|---|---|---|---|
| Washington, D.C. | Columbus, Ohio | Cincinnati, Ohio | Little Rock, Ark. | Kansas City, Mo. | Hollywood, Calif. | Frequency | |
| Ib | 9 | 2 | 1 | 6 | 2 | 0.22 | |
| Id | 5 | 1 | 0.07 | ||||
| IIa | 2 | 2 | 7 | 3 | 5 | 3 | 0.24 |
| IIb | 1 | 1 | 0.02 | ||||
| IIc | 11 | 1 | 2 | 1 | 6 | 2 | 0.25 |
| IIe | 3 | 1 | 2 | 0.07 | |||
| IIf | 1 | 1 | 0.02 | ||||
| IIIb | 1 | 1 | 1 | 1 | 0.04 | ||
| IVa | 1 | 3 | 0.04 | ||||
| Va | 1 | 0.01 | |||||
| Vf | 1 | 0.01 | |||||
| Vh | 1 | 0.01 | |||||
| Total | 33 | 10 | 10 | 4 | 26 | 9 | |
Genetic variants are defined according to (i) the organization of the VIR region and (ii) the haplotype of sequence variations as shown in Fig. 2.
While 90 isolates represented organisms acquired within approximately 6 months of each other, two of the strains received from one of our collaborating institutions had been isolated, and subsequently frozen, approximately 8 to 10 years prior to their acquisition for this study. Of interest was that the genotypes represented by these two strains reflect the most frequently isolated variants (i.e., IIa and IIc), suggesting the relatively constant nature of these sequence variations over the evolutionarily short time frame of a decade.
In attempts to verify that the genetic differences observed between strains reflected actual interstrain variability as opposed to fragment reorganization within a given strain (e.g., rearrangements within an unstable region of tandem repeat sequences), prototype variants underwent serial passage on solid medium every 2 to 4 weeks for 12 months, during which time the genotype was periodically reassessed. Regenotyping assigned the same variant and subvariant classification relative to the parent strain, suggesting that no discernible spontaneous recombination had occurred at the loci evaluated during this time frame.
In addition to the NTS region, the contiguous ITS-1, 5.8S rRNA gene, and ITS-2 regions were also fully sequenced for the prototype variants (variant I, n = 3; II, n = 3; III, n = 1; IV, n = 1; V, n = 2). No interstrain sequence variability was identified, and the consensus sequence was 100% identical to the sequences previously published by a number of investigators (11, 15, 21) and five sequences recently deposited in GenBank by Mochizuki et al. (16). In contrast, Gräser et al. described 12 sequence variations within the ITS regions of four isolates (6). Although the majority of these variations are unique to the findings of these authors, a single SNP (C→T) approximately 18 bp into ITS-1 was also observed by Mochizuki et al. in two of their seven sequences (GenBank accession no. AB094063 and AB094062) (16). We evaluated this putative mutation by PCR-RFLP and were unable to detect this transition in any of our 92 isolates.
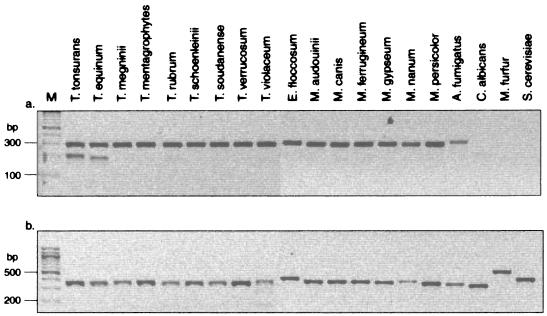
In order to assess the specificity of the genotyping scheme, a panel of non-T. tonsurans dermatophytes was investigated. Successful DNA isolation was again demonstrated by amplification of the ITS-2 fragment; the PCR product yield was equivalent to that observed in T. tonsurans. In contrast, successful amplification of the NTS region was variable and failed completely in T. mentagrophytes, T. schoenleinii, M. audouinii, and M. canis. In attempts to improve this reaction, PCR conditions were modified (e.g., increased extension time and the utilization of other primer combinations) without success. However, amplification of the VIR region (using NTS-16-NTS-13) proceeded only for T. tonsurans regardless of whether a nested amplification from the NTS product or direct amplification from genomic DNA was attempted (data not shown).
While successful amplification of the VIR region appeared to be an indicator of specificity of the genotyping scheme for T. tonsurans and not for related species, the inability to successfully amplify the NTS for all non-T. tonsurans isolates compelled us to develop a duplex PCR to ensure that we could address the issue of species specificity by direct amplification from genomic DNA. Successful amplification with the NTS-9-NTS-8R primer pair confirmed the presence of a dermatophyte (Fig. 4a, upper band), while the NTS-40-NTS-37 primer pair produced an amplicon in T. tonsurans and a less intense and slightly smaller band in T. equinum (Fig. 4a, lower band). The remaining non-T. tonsurans specimens amplified only with the primer pair directed against the more conserved NTS sequence or were negative for both products. Amplification of ITS-2 served as a control to ensure the presence of DNA, and a product was observed for all of the organisms evaluated (Fig. 4b).
FIG. 4.
T. tonsurans-specific duplex PCR. (a) Amplification of a 207-bp T. tonsurans-specific NTS fragment and an ∼300-bp conserved NTS fragment from a panel of dermatophytes and nondermatophyte species; (b) amplification of ITS-2 as DNA quality control for the same samples as are shown in panel a. M, molecular size markers.
DISCUSSION
While dermatophyte genomes remain largely uncharacterized, there have been efforts to understand their genetic diversity, with the majority of work arising from attempts to clarify taxonomy and the phylogenetic relationship among species. However, intraspecies characterization is of particular utility in the epidemiologic evaluation of the endemic anthropophilic organisms that cause chronic persistent infection. Studies employing nonspecific strategies and evaluating limited molecular markers (e.g., nuclear DNA base composition, mitochondrial DNA RFLP, amplified fragment length polymorphism, and PCR fingerprinting) were unable to differentiate strains within species despite the fact that hundreds of isolates collected from various continents over several decades were evaluated (3, 7, 17, 18). These findings led to the general acceptance of the idea that intraspecies polymorphisms in dermatophytes are rare. As a consequence, some investigators maintain that the dermatophyte species have been overclassified, despite their markedly different morphological characteristics, pathogenicity profiles, and host affinities (7, 8, 15).
Approaches directed at specific genes, namely the rRNA gene locus, have been more fruitful at revealing dermatophyte strain variations. Using a Southern blot approach, Jackson et al. were able to identify 14 RFLP band patterns among 50 T. rubrum isolates (12). As only one of the two fragments they observed appeared to be polymorphic, the existence of a repetitive element within the NTS region was hypothesized (12). Subsequently, these investigators cloned and sequenced the NTS region from a single isolate and defined two repeat elements. Based on PCR analysis targeting these repeat regions, it was asserted that a variable number of subrepeat elements accounted for the observed interstrain variations (13). However, the evidence presented was indirect, deduced from variable PCR product lengths, and confirmed by only partial sequence analysis. Moreover, isolates possessing complex pattern types (i.e., numerous PCR products) remained uncharacterized (13).
Given the failure of some of the aforementioned techniques to distinguish T. tonsurans strains (9, 10, 14, 16) and the paucity of sequence information for the species, the rDNA gene locus was selected for this investigation because the flanking 25S, 18S, and 5.8S rDNA sequences are highly conserved among species. In order to unequivocally determine the nature of this locus in T. tonsurans, multiple strains (n = 20), including all those exhibiting length variation in an initial long PCR of the NTS region, were entirely sequenced. This full-sequence approach allowed us to characterize the organization of the VIR region and identify the presence of seven SNPs, a small deletion, and a small insertion. Undoubtedly, a less specific approach would have precluded our ability to discriminate the individual repeat elements, along with the interruption motif, that constitute the VIR region. As such, we could design and subsequently develop a genotyping scheme that differentiates interstrain variants and subvariants by characterizing the internal repeat region and the sequence variations, respectively.
In order to avoid false or unspecific priming of the oligonucleotides elsewhere in the genome, genotyping assays were performed on diluted PCR templates. Despite this, when amplifying the repeat region, we observed multiple additional bands or ladders that were larger than the expected product (data not shown). Since the primers had no secondary binding sites on this template and the ladder formation could be suppressed by using proofreading Taq polymerases such as TripleMaster, we speculate that the nature of the VIR sequence (i.e., repeat 1 and repeat 2 elements, smaller repeat units, and the presence of homo- and heteronucleotide clusters) favor unspecific ladder formation under certain PCR conditions. Interestingly, VIR amplification from genomic DNA would proceed only with Herculase Taq polymerase in the presence of 5% DMSO and failed completely under the conditions used for amplification of the NTS template (data not shown). This implies that the generation of specific products from these variable regions is exquisitely sensitive to PCR conditions and that both multiple-band formation and the failure of amplification need to be interpreted with caution.
The VIR region and the immediate flanking sequences observed are likely specific to T. tonsurans. Database searches with repeat 1 and repeat 2 elements, the interruption motif, and the primers used to amplify the VIR region did not identify any fungal sequences. Furthermore, a comparison of the T. tonsurans NTS region (excluding the flanking 25S and 18S rRNA genes) to that of T. rubrum, the only Trichophyton-derived NTS sequence deposited in GenBank to date, revealed that the overall homology was only 61.5%. The first 71 bp and the last 577 bp were the most similar, at 89 and 87%, respectively, while the central sequence (encompassing the VIR region) exhibited only 53% identity. Finally, the VIR region could be amplified only from genomic T. tonsurans DNA or a long NTS product generated thereof.
In ongoing investigations, we are now utilizing the duplex PCR assay for initial identification of the isolate as T. tonsurans. While the VIR region itself constitutes a more specific target than the region chosen for the duplex reaction, the size variation, the finicky amplification conditions, and the tendency to produce unspecific ladder bands precluded its use. Designing an assay that would allow for the amplification of a small fragment of consistent size within the VIR proved to be impractical because the repeat elements, in combination with the homo- and heteronucleotide motifs, restricted the options for primer design. Therefore, a 207-bp region upstream of the VIR region was targeted. It is sufficiently specific, amplifies well under standard conditions, and is smaller than the nonspecific product in the duplex reaction, ensuring preferential amplification in this reaction. As a result, the presence of only the larger fragment would indicate that a failure to amplify the smaller T. tonsurans-specific product is not due to PCR failure. Among the panel of fungal DNAs tested, only T. equinum DNA revealed a product with the T. tonsurans-specific primers. However, this product may be distinguished from T. tonsurans (including isolates with the small insertion or deletion) by a difference in amplicon size and therefore does not interfere with the interpretation of the results from this assay. Only the yeast species that were evaluated (e.g., C. albicans, M. furfur, and S. cerevisiae) did not yield the approximate 300-bp product from the less-conserved NTS region, despite the presence of a strong band for ITS-2, suggesting sequence heterogeneity within this region. In contrast, all of the dermatophytes, along with A. fumigatus, produced the larger NTS-derived PCR product.
In contrast to the NTS region, no sequence variations were revealed within the ITS regions for any of the prototype strains. Furthermore, the consensus sequence determined in our isolates was identical to those published by several other groups (11, 15, 21). Interestingly, Gräser et al. and Mochizuki et al. reported a combined 14 sequence variations among six isolates of T. tonsurans, with one shared SNP found by both groups of investigators (6, 16). However, we were unable to discover the transition (C→T) in any of our 92 isolates acquired from six cities across the United States, and Mochizuki et al. did not follow up on this SNP in their panel of Japanese isolates. As such, it remains unclear whether the sequence variations observed by Gräser et al. and Mochizuki et al. represent actual genetic variability based on geographic isolation of the strains (which may be the case for the C→T SNP) or simply reflect PCR and/or sequencing artifacts. Irrespective of the discordance, ITS sequence homology in our isolates, each with a different NTS region, supports the idea of the highly conserved nature of this region between strains of T. tonsurans and thus argues that this region constitutes a rather poor candidate for distinguishing variants within the species.
An issue that clearly needs to be addressed and indeed is a prerequisite for any prospective evaluation is the genetic stability of the locus investigated. Using a hybridization technique similar to that of Jackson et al. (12), Gupta and colleagues identified a number of patients in whom multiple RFLP band patterns were noted in the T. rubrum isolates acquired from dermatophyte-infected toenails (10). Unfortunately, they were unable to distinguish whether this finding was the result of concurrent infection with more than one isolate or of genetic instability (e.g., substrain shuffling) within the repetitive regions of this locus (10).
In order to address this issue in our investigation, all prototypes described within this study were continuously passaged over 1 year and their assigned genotypes were confirmed every 2 to 3 months. Of interest was that pleomorphic variation was observed for a number of the prototype isolates that were passaged; however, we found no evidence to suggest that substrain shuffling had occurred. Furthermore, the genotyping assays performed are unequivocally more sensitive than Southern blotting and hybridization of EcoRI-digested DNA, and we are reasonably certain that fragment length differences as small as 10 bp would have been detected in our assays. Spontaneous variations that arise in organisms growing in their natural reservoir may occur with different frequencies than they do in vitro and thus may not have been detected in this investigation. Furthermore, minor sequence variations (e.g., SNPs) could have occurred in areas of the NTS region not covered by our genotyping assays and thus would not have been detected by PCR-RFLP. Although the span of 150 nucleotides in which the small deletion and the insertion reside does not appear to be a hot spot of recombination, this region, along with the VIR, may be more accommodating to such events over longer periods of time. Therefore, we continue to carefully monitor this region.
Since the isolates evaluated represent organisms acquired from children with clinical evidence of infection, the strains that were characterized may not represent the entire spectrum of overall genetic variance in the T. tonsurans population. Subsequent genotypic characterization of isolates acquired from carriers will help to elucidate this matter. Despite this, the genotyping approach presented in this study provides a sufficiently sensitive tool for discriminating differences in T. tonsurans at the genetic level. Given that this investigation was undertaken to evaluate the degree of genetic variance among T. tonsurans isolates, we did not make an attempt to determine whether an association between morphological phenotype and genotype exists. Now that genetic variability has been established using methods that are sensitive, specific, and reproducible, we are in the process of undertaking a broader investigation designed to link genotype with both morphological and biochemical phenotype. Until such data become available, the functional consequences of genotypic variability occurring in this region remain unknown. However, the rDNA locus appears to serve as a suitable surrogate for identifying and discriminating genetic variants of T. tonsurans until such time as additional sequence data from other polymorphic loci become available.
Acknowledgments
This work was supported by a grant from the Katharine B. Richardson Foundation and grant AR46870 from the National Institute of Arthritis, Musculoskeletal, and Skin Diseases, National Institutes of Health.
We gratefully acknowledge the assistance of Brad Schindel for the preparation of samples for sequencing and Joel Bozue for his critical review of the manuscript. We also extend sincere appreciation to the following individuals and institutions for the provision of fungal samples: the Bacteriology Laboratory at Children's Mercy Hospital, Kansas City, Mo.; Mario J. Marcon, Columbus Children's Hospital, Columbus, Ohio; Susan M. Novak, Southern California Permanente Medical Group, North Hollywood, Calif.; Joel Mortensen, Children's Hospital, Cincinnati, Ohio; Laura James, Arkansas Children's Hospital, Little Rock, Ark.; and Joseph M. Campos, Children's National Medical Center, Washington, D.C.
REFERENCES
- 1.Abdel-Rahman, S. M. 2001. Polymorphic exocellular protease expression in clinical isolates of Trichophyton tonsurans. Mycopathologia 150:117-120. [DOI] [PubMed] [Google Scholar]
- 2.Birren B., G. Fink, and E. Lander. 2002. Fungal Genome Initiative: white paper developed by the fungal research community. [Online.] http://www.genome.wi.mit.edu/seq/fgi/FGI_whitepaper_Feb8.pdf. Accessed 30 August 2002.
- 3.Davidson, F. D., D. W. R. MacKenzie, and R. J. Owen. 1980. Deoxyribonucleic acid base composition of dermatophytes. J. Gen. Microbiol. 118:465-470. [DOI] [PubMed] [Google Scholar]
- 4.Drake, L. A., S. M. Dinehart, E. R. Farmer, R. W. Goltz, G. F. Graham, M. K. Hardinsky, C. W. Lewis, D. M. Pariser, J. W. Skouge, S. B. Webster, D. C. Whitaker, B. Butler, B. J. Lowery, B. E. Elewski, M. L. Elgart, P. H. Jacobs, J. L. Lesher, Jr., and R. K. Scher. 1996. Guidelines of care for superficial mycotic infections of the skin: tinea corporis, tinea cruris, tinea faciei, tinea manuum, and tinea pedis. J. Am. Acad. Dermatol. 34:282-286. [DOI] [PubMed] [Google Scholar]
- 5.Fuller, L. C., F. C. Child, G. Midgley, and E. M. Higgins. 2003. Scalp ringworm in south-east London and an analysis of a cohort of patients from a paediatric dermatology department. Br. J. Dermatol. 148:985-988. [DOI] [PubMed] [Google Scholar]
- 6.Gräser, Y., M. El Fari, R. Vilgalys, A. F. Kuijpers, G. S. de Hoog, W. Presber, and H. Tietz. 1999. Phylogeny and taxonomy of the family Arthrodermataceae (dermatophytes) using sequence analysis of the ribosomal ITS region. Med. Mycol. 37:105-114. [PubMed] [Google Scholar]
- 7.Gräser, Y., J. Kuhnisch, and W. Presber. 1999. Molecular markers reveal exclusively clonal reproduction in Trichophyton rubrum. J. Clin. Microbiol. 37:3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gräser, Y., A. F. A. Kuijpers, W. Presber, and G. S. de Hoog. 2000. Molecular taxonomy of the Trichophyton rubrum complex. J. Clin. Microbiol. 38:3329-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gräser, Y., A. F. A. Kuijpers, W. Presber, and G. S. de Hoog. 1999. Molecular taxonomy of Trichophyton mentagrophytes and T. tonsurans. Med. Mycol. 37:315-330. [DOI] [PubMed] [Google Scholar]
- 10.Gupta, A. K., Y. Kohli, and R. C. Summerbell. 2001. Variation in restriction fragment length polymorphisms among serial isolates from patients with Trichophyton rubrum infection. J. Clin. Microbiol. 39:3260-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmsen, D., A. Schwinn, E. B. Brocker, and M. Frosch. 1999. Molecular differentiation of dermatophyte fungi. Mycoses 42:67-70. [DOI] [PubMed] [Google Scholar]
- 12.Jackson, C. J., R. C. Barton, and E. G. V. Evans. 1999. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions. J. Clin. Microbiol. 37:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson, C. J., R. C. Barton, S. L. Kelly, and E. G. V. Evans. 2000. Strain identification of Trichophyton rubrum by specific amplification of subrepeat elements in the ribosomal DNA nontranscribed spacer. J. Clin. Microbiol. 38:4527-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, J. A., K. Takizawa, K. Fukushima, K. Nishimura, and M. Miyaji. 1999. Identification and genetic homogeneity of Trichophyton tonsurans isolated from several regions by random amplified polymorphic DNA. Mycopathologia 145:1-6. [DOI] [PubMed] [Google Scholar]
- 15.Makimura, K., Y. Tamura, T. Mochizuki, A. Hasegawa, Y. Tajiri, R. Hanazawa, K. Uchida, H. Saito, and H. Yamaguchi. 1999. Phylogenetic classification and species identification of dermatophyte strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J. Clin. Microbiol. 37:920-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mochizuki, T., H. Tanabe, M. Kawasaki, H. Ishizaki, and C. J. Jackson. 2003. Rapid identification of Trichophyton tonsurans by PCR-RFLP analysis of ribosomal DNA regions. J. Dermatol. Sci. 32:25-32. [DOI] [PubMed] [Google Scholar]
- 17.Mochizuki, T., S. Watanabe, and M. Uehara. 1996. Genetic homogeneity of Trichophyton mentagrophytes var. interdigitale isolated from geographically distinct regions. J. Med. Vet. Mycol. 34:139-143. [PubMed] [Google Scholar]
- 18.Nishio, K., M. Kawasaki, and H. Ishizaki. 1992. Phylogeny of the genera Trichophyton using mitochondrial DNA analysis. Mycopathologia 117:127-132. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz, R. A., and C. K. Janniger. 1995. Tinea capitis. Cutis 55:29-33. [PubMed] [Google Scholar]
- 20.Smith, E. S., A. B. Fleischer, and S. R. Feldman. 1998. Nondermatologists are more likely than dermatologists to prescribe antifungal/corticosteroid products: an analysis of office visits for cutaneous fungal infections, 1990-1994. J. Am. Acad. Dermatol. 39:43-47. [DOI] [PubMed] [Google Scholar]
- 21.Summerbell, R. C., R. A. Haugland, A. Li, and A. K. Gupta. 1999. rRNA gene internal transcribed spacer 1 and 2 sequences of asexual, anthropophilic dermatophytes related to Trichophyton rubrum. J. Clin. Microbiol. 37:4005-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]