Abstract
Streptococcal fibronectin-binding protein is an important virulence factor involved in colonization and invasion of epithelial cells and tissues by Streptococcus pyogenes. In order to investigate the mechanisms involved in the evolution of sfbI, the sfbI genes from 54 strains were sequenced. Thirty-four distinct alleles were identified. Three principal mechanisms appear to have been involved in the evolution of sfbI. The amino-terminal aromatic amino acid-rich domain is the most variable region and is apparently generated by intergenic recombination of horizontally acquired DNA cassettes, resulting in a genetic mosaic in this region. Two distinct and divergent sequence types that shared only 61 to 70% identity were identified in the central proline-rich region, while variation at the 3′ end of the gene is due to deletion or duplication of defined repeat units. Potential antigenic and functional variabilities in SfbI imply significant selective pressure in vivo with direct implications for the microbial pathogenesis of S. pyogenes.
Streptococcal fibronectin-binding protein I (SfbI), also known as protein F1, is probably the most extensively characterized fibronectin-binding protein in group A streptococcus (GAS; Streptococcus pyogenes). Identified simultaneously in two independent laboratories (15, 39), SfbI is a membrane-anchored surface protein shown to be a major adhesin of GAS (13, 15, 28, 29, 39, 42). In addition, SfbI binds to fibrinogen via a domain in the amino terminus (20) and human immunoglobulin (IgG) via the carboxy-terminal fibronectin-binding repeat domain (25). SfbI is protective in mice (14, 34) and a mucosal adjuvant (26). According to various reports, it is present in 64 to 82% of strains (13, 22, 28). Beside its ability to mediate adherence to host cells via binding to fibronectin as a bridging molecule, SfbI is a potent invasin of epithelial cells (16, 27). While efficient adhesion is mediated through the fibronectin-binding repeat region, internalization into host cells is governed by the fibronectin-binding spacer region in a cooperative manner (41). Recent work demonstrates that SfbI-induced integrin clustering initiates caveola recruitment and subsequent caveola-mediated uptake of the pathogen into epithelial and endothelial cells (33). The interaction between the fibronectin-binding repeat region and the N-terminal F1 modules of fibronectin has been elegantly resolved by defining the structure of the complex that is formed (35). Another important and recently identified function of SfbI is the ability to recruit collagen via prebound fibronectin. SfbI thereby mediates bacterial aggregation, colonization of the collagen matrix, and phagocyte evasion (9). SfbI, like many membrane-associated streptococcal proteins, has a repetitive structure and exhibits decreasing variability from the amino terminus to the carboxy terminus (40). The protein is synthesized as a propeptide with a signal sequence at the amino terminus of the protein which is subsequently cleaved to form the mature protein. The amino terminus of the mature protein contains a domain rich in aromatic amino acids, adjacent to which is a proline-rich domain. The proline-rich domain is made up of two regions: the proline-rich repeats and the proline-rich upstream region encompassing the region previously described as proline-rich repeats 1 and 2 (40). Toward the carboxy terminus lie two functional fibronectin-binding domains: the fibronectin-binding repeat domain and a second fibronectin-binding domain which lies upstream of the fibronectin-binding repeats and which has been designated spacer 2 (40) or the upstream fibronectin-binding domain (36). These domains are highly conserved and display variability only in the number of fibronectin-binding repeats, which may range from one to six (28, 42). Variable numbers of proline-rich repeats have also been reported; however, no correlation between variation in this domain and the numbers of fibronectin-binding repeats has been found (42). The carboxy terminus consists of a wall-associated region, a membrane anchor motif (LPATGD), and a membrane-associated region rich in hydrophobic residues, all of which are typical of surface proteins of gram-positive cocci (11).
Several groups have described the arrangement of the genomic region containing the gene encoding SfbI (6, 30). This locus has been designated the fibronectin-collagen-T antigen (FCT) region (6) since it may contain genes encoding the fibronectin-binding protein SfbI and/or protein F2, a collagen-binding protein (Cpa), and the T antigen. Incorporation of sequences from the recently completed M3 (4), M5 (http://www.sanger.ac.uk/Projects/S_pyogenes), and M18 (37) genomes reveals a complex gene arrangement in this region which is indicative of a zone of frequent genetic rearrangement.
Mosaic alleles are made up of distinct DNA cassettes of different phylogenic origins which have recombined to produce novel chimeric genes. While gene mosaic structures for genes encoding streptokinase (19), hyaluronidase (24), streptococcal superantigen (32), and a novel collagen-like protein called SclA (31) have been reported in GAS, the classic example in GAS are members of the M-protein family encoded by genes within the Vir (or mga) regulon (43). Horizontal transfer and intergenic recombination have played major roles in the evolution of the emm-like family, contributing to both sequence variation and variation in the overall architecture of the Vir regulon (44). Since different members of the M-protein family exhibit different protein-binding capabilities, this rearrangement may have direct ramifications for the virulence and tropisms of the various strains. This is supported by the observation that different architectural arrangements (emm pattern types) have been associated with preferences for specific tissue sites (5).
Although evaluation of the accumulation of mutations is a valuable tool for assessment of strain relatedness, transfer of whole cassettes of DNA between strains complicates such analyses. Multilocus sequence typing with several presumably selection-neutral housekeeping genes has been used to determine the extent of intergenic recombination in bacterial genomes. The lack of congruence between maximum-likelihood trees for each of the “neutral” genes in GAS was indicative of very high rates of recombination, and it was concluded that this was sufficient to “obliterate the phylogenetic signal in [individual] gene trees” (10).
Recombination at distal sites such as the Vir regulon and the FCT region is most likely independent. Evidence to support this theory includes the fact that T types do not always conform to M types, with several T types reported for isolates of the same M type and vice versa (2, 17). In addition, while the distribution of sfbI within M types (28) or Vir types (VTs) (13) is generally consistent, there is evidence of heterogeneity of sfbI species within M types, as the number of fibronectin-binding repeats detected in M8 and M28 serotype strains is variable (28). Significant heterogeneity in sfbI sequences encoding the amino-terminal half of the aromatic amino acid-rich domain from M4, M12, M15, and M18 strains has also been reported (20).
The aim of this study was to examine precisely the genetic variability in the entire sfbI in order to determine possible mechanisms involved in the evolution of this important streptococcal adhesin and invasin. Additionally, we have examined whether particular sfbI sequence types (STs) are associated with the clinical source of the isolates.
(Part of this work was presented at the XVth Lancefield International Symposium on Streptococci and Streptococcal Disease, October 2002, Goa, India [R. Towers, M. J. Walker, and G. S. Chhatwal, Lancefield Int. Symp. Streptococci Streptococcal Dis. abstr. O5.3, 2002].)
MATERIALS AND METHODS
Bacterial strains.
Isolates were obtained from the Menzies School of Health Research streptococcal collection. This collection contains isolates from patients of the Royal Darwin Hospital and the surrounding Aboriginal communities. In addition, an M13 GAS reference strain from the United Kingdom (Cathy13) and the homologous sfbII strain (strain 75401) from Germany were included (22, 38). Background information regarding clinical details, VTs (12), and emm STs (3), when available, are included in Table 1. All streptococcal strains were grown on agar plates containing 5% sheep blood (Sigma) at 37°C overnight.
TABLE 1.
Summary of sfbI STs
| sfbI ST | Strain | Clinical source | Diseasea | VT | emm ST | Aro type | Pro typeb | No. of Pro rptsc | No. of Fn rptsd |
|---|---|---|---|---|---|---|---|---|---|
| 1 | NS678 | Throat | N | 33.2 | NDe | 1 | A1 | 2 | 1 |
| 1 | NS704 | Throat | N | 33.2 | ND | 1 | A1 | 2 | 1 |
| 2 | NS1045 | Skin | N | 89 | emm60 | 1 | A1 | 1 | 2 |
| 2 | NS1053 | Blood | I | 89 | emm60 | 1 | A1 | 1 | 2 |
| 3 | NS1036 | Skin | N | 33.1 | emm110 | 1 | A1 | 1 | 4 |
| 4 | BSB19 | Skin | N | 6 | stbsb19 | 2 | A1 | 5 | 3 |
| 5 | NS473 | Skin | N | 37.1 | stns554 | 2 | A1 | 1 | 4 |
| 6 | NS539 | Skin | N | 1 | emm22 | 2 | A1 | 3 | 4 |
| 7 | NS35 | Skin | N | 22 | emm58 | 2 | A1 | 2 | 4 |
| 7 | NS351 | Skin | N | 22 | emm58 | 2 | A1 | 2 | 4 |
| 7 | NS474 | Skin | N | 22 | emm58 | 2 | A1 | 2 | 4 |
| 7 | NS687 | Blood | I | 22 | emm58 | 2 | A1 | 2 | 4 |
| 8 | BL16 | Skin | N | 25 | emm85 | 2 | A2 | 4 | 1 |
| 9f | D471 | Throat | N | ND | (M6)g | 3 | A1 | 1 | 5 |
| 10 | NS691 | Skin | N | 57 | emm69 | 4 | A1 | 1 | 4 |
| 10 | NS930 | Skin | I | 57 | emm69 | 4 | A1 | 1 | 4 |
| 10 | NS931 | Blood | I | 57 | emm69 | 4 | A1 | 1 | 4 |
| 11 | NS16 | Skin | N | 37.1 | stns554 | 5 | B1 | 1 | 5 |
| 11 | NS240 | Blood | I | 116 | st2904 | 5 | B1 | 1 | 5 |
| 12 | NS190 | Skin | I | 19 | emm74 | 5 | B1 | 1 | 4 |
| 12 | DRX5 | Skin | N | 30 | ND | 5 | B1 | 1 | 4 |
| 13 | NS210 | Blood | I | 34 | emm22 | 5 | B1 | 2 | 3 |
| 14 | NS1120 | Throat | N | 105 | ND | 5 | B3 | 5 | 3 |
| 15 | NS179 | Pus | I | 7.2 | emm9.1 | 6 | B3 | 11 | 2 |
| 16 | NS192 | Blood | I | 3.2 | emm100 | 6 | B2 | 4 | 4 |
| 17 | CATHY13 | Unknown | U | 2.1 | emm13 | 6 | B1 | 4 | 1 |
| 17 | NS297 | Skin | N | 3.1 | emm44/61 | 6 | B1 | 4 | 1 |
| 17 | NS495 | Skin | N | 2.2 | emm90 | 6 | B1 | 4 | 1 |
| 17 | NS730 | Pus | I | 2.2 | emm90 | 6 | B1 | 4 | 1 |
| 17 | NS731 | Pus | I | 2.2 | emm90 | 6 | B1 | 4 | 1 |
| 17 | NS732 | Aspirate | I | 2.2 | emm90 | 6 | B1 | 4 | 1 |
| 17 | NS755 | Blood | I | 2.2 | emm90 | 6 | B1 | 4 | 1 |
| 17 | NS989 | Skin | N | 2.2 | emm90 | 6 | B1 | 4 | 1 |
| 18 | NS226 | Blood | I | 14.1 | emm42 | 7 | A1 | 5 | 4 |
| 18 | NS244 | Skin | N | 14.1 | stns244 | 7 | A1 | 5 | 4 |
| 18 | NS415 | Skin | N | 14.1 | stns244 | 7 | A1 | 5 | 4 |
| 19 | NS14 | Skin | N | 96 | emm102 | 7 | A1 | 4 | 4 |
| 20 | NS488 | Throat | N | 52 | emm12 | 8 | A2 | 5 | 4 |
| 20 | NS880 | Throat | N | 52 | emm12 | 8 | A2 | 5 | 4 |
| 20 | A735 | Throat | N | ND | (M12) | 8 | A2 | 5 | 4 |
| 21 | 75401 | Throat | N | ND | (M75) | 9 | B1 | 2 | 3 |
| 22 | NS101 | Blood | I | 33.1 | emm110 | 10 | B1 | 1 | 3 |
| 22 | NS516 | Skin | N | 33.1 | emm110 | 10 | B1 | 1 | 3 |
| 23 | BSA39 | Skin | N | 7.1 | emm63 | 10 | B1 | 1 | 5 |
| 24 | NS1132 | Skin | N | 104 | emm25 | 11 | B1 | 1 | 3 |
| 25 | DRV1 | Skin | N | 18 | emm55 | 12 | B1 | 1 | 4 |
| 26 | NS216 | Blood | I | 122 | ND | 12 | B1 | 4 | 4 |
| 27 | EF1949 | Unknown | U | ND | (M15) | 13 | A2 | 5 | 4 |
| 28 | NS424 | Skin | N | 61 | emm14 | 13 | A2 | 6 | 4 |
| 28 | NS483 | Skin | N | 61 | emm14 | 13 | A2 | 6 | 4 |
| 28 | NS501 | Blood | I | 61 | emm14 | 13 | A2 | 6 | 4 |
| 28 | NS506 | Skin | N | 61 | emm14 | 13 | A2 | 6 | 4 |
| 29 | NS534 | Blood | I | 37.1 | stns554 | 13 | A2 | 3 | 1 |
| 30 | DRY5 | Skin | N | 37.1 | stns554 | 13 | A2 | 1 | 2 |
| 31 | DSM2071 | Unknown | N | ND | (M23) | 14 | A1 | 2 | 4 |
| 32 | NS90 | Blood | I | 52 | stns90 | 15 | B1 | 1 | 4 |
| 33 | NS195 | Blood | I | 120 | ND | 15 | B1 | 1 | 5 |
| 34 | NS1042 | Skin | N | 77 | stck401 | 15 | B1 | 1 | 2 |
Invasive or noninvasive disease: I, invasive; N, noninvasive; U, unknown.
Pro type, SfbI proline-rich region sequence subtype.
Pro rpts, “true” proline-rich repeats.
Fn rpts, fibronectin-binding repeats.
ND, not determined.
Boldface indicates information published previously.
M serotype, as stated in relevant publications.
PCR.
The template for PCR was prepared by using the InstaGene matrix (Bio-Rad) according to the instructions of the manufacturer, except that due to the small size of the streptococcal colonies, 10 to 20 streptococcal colonies were used. The sequencing template was amplified with primers sfbI-F1 and sfbI-R4. Each 50-μl PCR mixture contained 5 μl of InstaGene preparation. The PCR parameters used were an initial denaturing step at 94°C for 2 min and then 35 cycles of denaturation at 94°C for 1 min, annealing at 49°C for 1 min, and extension at 72°C for 2 min 30 s, with a final extension step of 72°C for 5 min.
Screening for sfbI and rofA.
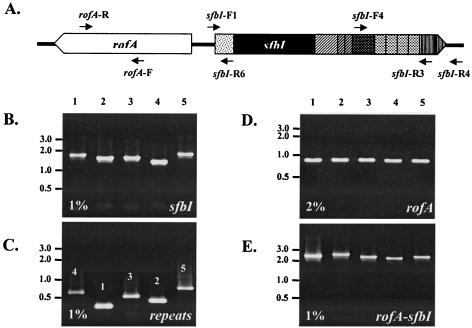
Many of the data on the distribution of sfbI in the GAS strains used in this study have been published elsewhere (8, 13). For the purposes of this study all strains were retested for the presence of sfbI by PCR, and the results were compared with previous results. PCR screening was performed with primers sfbI-F1 and sfbI-R3. The number of fibronectin-binding repeats was determined with primers sfbI-F4 and sfbI-R3. Isolates were also screened for the presence of rofA with primers rofA-F and rofA-R. The arrangement of rofA, relative to that of sfbI, was determined with primers rofA-R and sfbI-R6. All primer sequences are shown in Table 2, while their positions relative to those of the respective genes are shown in Fig. 1A.
TABLE 2.
Primer sequences used in this study
| Primer direction and primer | Sequence |
|---|---|
| Forward | |
| sfbI-F1 | 5′-GTCTTTCTTGACAATAACGTGGTAAGCTC-3′ |
| sfbI-F2 | 5′-GTAGCCTATGCTGCCGATGAGAAG-3′ |
| sfbI-F3 | 5′-TTTGTACCAGAAAATCCCCCTAAACCTG-3′ |
| sfbI-F4 | 5′-GACTTACCTATTGAAGATCCTCGTTATGAG-3′ |
| sfbI-F5 | 5′-GCATGCGCGGGTGCTATCG-3′ |
| sfbI-F7 | 5′-GTACAGAATATGTACAAGATAATCC-3′ |
| sfbI-S1F | 5′-GTGCTGAATATGTACCTGATAGTCC-3′ |
| PCRP1 | 5′-TATCAAAATCTTCTAAGTGCTGAG-3′ |
| rofA-F | 5′-GCCAATAACTGAGGTAGC-3′ |
| Reverse | |
| sfbI-R1 | 5′-CTCATAACGAGGATCTTCAATAGGTAAGTC-3′ |
| sfbI-R2 | 5′-CACTCCTGGCTCTTTCGTATCTTCTG-3′ |
| sfbI-R3 | 5′-GTATCTTCAACAATGGTCACTGTTTCACTG-3′ |
| sfbI-R4 | 5′-CAGTTATCCACTATTCAGCATATTTGCGC-3′ |
| sfbI-R5 | 5′-CGATAGCACCCGCGCATGC-3′ |
| sfbI-R6 | 5′-GCAGCATAGGCTACTTGACCAAAAC-3′ |
| sfbI-R7 | 5′-GGATTATCTTGTACATATTCTGTAC-3′ |
| sfbI-R8 | 5′-CAGGTCTAGGGGGATCTTCTGGTACAAA-3′ |
| sfbI-S1R | 5′-GGACTATCAGGTACATATTCAGCAC-3′ |
| PCRP1R | 5′-CTCAGCACTTAGAAGATTTTG-3′ |
| rofA-R | 5′-GGTTTTGCTCTTTTAGGT-3′ |
FIG. 1.
(A) Positions of primers relative to the two genes. The shading of the SfbI domains is as described in the legend to Fig. 2. The schematic is not drawn to scale. (B to E) Ethidium bromide-stained 1 or 2% agarose gels showing the results of PCR screening for sfbI and rofA for five representative strains. (B) Screening for sfbI with primers sfbI-F1 and sfbI-R3; (C) determining the number of fibronectin-binding repeats in SfbI with primers sfbI-F4 and sfbI-R3; (D) screening for rofA with primers rofA-F and rofA-R; (E) determining arrangement of rofA relative to that of sfbI with primers rofA-R and sfbI-R6. DNA marker (GeneRuler DNA Ladder Mix, MBI Fermentas) sizes (in kilobases) are shown at the left. Lanes 1 to 5, GAS strains DSM2071, NS732, NS1132, DRY5, and BSA39, respectively.
DNA sequence analysis.
The PCR product was used as the template for sequencing reactions. This template was generated with primers sfbI-F1 and sfbI-R4 or sfbI-R3. A total of 5 μl of the PCR product was visualized in an ethidium bromide-stained 1% agarose gel to determine its purity and relative concentration. The remainder was purified with QIAquick columns (Qiagen), eluted in 30 μl of sterile distilled water, and stored at −20°C. The primers used for the sequence reactions are shown in Table 2.
Sequence alignment was performed with a range of DNASTAR programs. Applied Biosystems electropherograms were aligned to generate DNA contigs by using SeqManII, and then DNA contigs were translated to amino acid sequences with the MegAlign program and were subsequently aligned by the Clustal V method. The alignment was then modified by eye before being converted back to the DNA sequence alignment.
Nucleotide sequence accession numbers.
The nucleotide sequences for the 54 sfbI genes can be found in the GenBank database under accession numbers AJ347791 to AJ347844.
RESULTS
Screening for sfbI and rofA.
The presence of sfbI was confirmed by PCR for 54 GAS isolates. Representative PCR results are shown in Fig. 1B. PCR was also used to determine the number of fibronectin-binding repeats as an initial indicator of variation (Fig. 1C). In addition, the relative distribution and arrangement of rofA with respect to those of sfbI were established by PCR. All strains were tested for the presence of rofA with primers rofA-F and rofA-R (30) (Fig. 1D), while primers rofA-R and sfbI-R6 were used to detect adjacent rofA and sfbI genes (Fig. 1E). rofA was present in all 54 strains and was situated immediately upstream of sfbI, but in an orientation opposite that shown in Fig. 1A.
Sequence analysis of sfbI.
The entire sfbI gene was amplified by PCR for use as the sequencing template from most strains by using primers sfbI-F1 and sfbI-R4. Initially, the sfbI PCR products from 20 strains were sequenced in their entirety in both directions. It was determined from these data that the 3′ end of sfbI was highly conserved, apart from the various numbers of proline-rich and fibronectin-binding repeats. As the number of fibronectin-binding repeats could be readily determined by PCR (Fig. 1C) and these results were consistent with preliminary DNA sequence data, no attempt was made to sequence the fibronectin-binding repeat domains for the additional 34 strains examined in this study, and conclusions as to the relatedness of the genes were drawn from the sequence and PCR results combined.
The nucleotide sequences were aligned with published sfbI sequences, including those encoding full-length SfbI (GenBank accession no. X67947) (2); protein F1 (GenBank accession no. L10919) (20); protein F1.12 (GenBank accession no. AF447492) (6); protein F15 and 12 partial sfbI sequences (GenBank accession nos. AF009908 to AF009920) (20); and gfbA, an sfbI homolog from GGS (GenBank accession no. U131115) (21). The nucleotide sequence alignment can be found at GenBank alignment accession number ALIGN_000212 (data not shown).
Source of variation in sfbI.
Figure 2 shows an overall schematic of the variation observed in the sfbI sequences. As noted previously (40), variation in sfbI decreases in the 5′-to-3′ direction. The variation at the 3′ end of the gene is due to intragenic recombination, i.e., rearrangement of repetitive elements within genes through homologous recombination. The deletion or duplication of repeat units (narrower bars in Fig. 2) has resulted in a variable number of proline-rich and fibronectin-binding repeats. There is no relationship between the numbers of proline-rich repeats and the numbers of fibronectin-binding repeats, nor is there an association between the numbers of repeats and the upstream sequence variation. Therefore, it can be assumed that intragenic recombination is a frequent phenomenon within these regions.
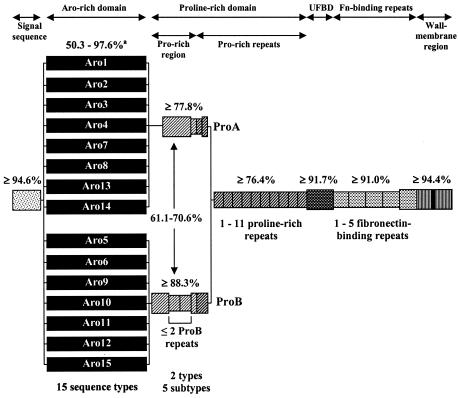
FIG. 2.
Overall schematic of variation seen in SfbI. Decreasing variability of SfbI occurs from the amino-terminal aromatic amino acid-rich domain (solid black) through the proline-rich region (thin diagonal stripe) and to the proline-rich repeats (thick diagonal stripe) and the functional fibronectin (Fn)-binding domains (stippled). Narrower bars indicate the deletion or duplication of repeat units. The percent identities observed in pairwise DNA alignments is shown above each domain. Gaps introduced to aid alignment have been treated as single base changes. The diagram is not drawn to scale. UFBD, upstream fibronectin-binding domain.
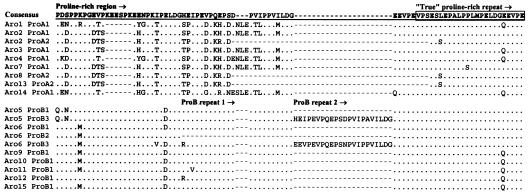
Two distinct and divergent STs which exhibited 61 to 70% identity in pairwise alignments of the DNA sequences were identified within the central proline-rich region. These have been designated ProA and ProB. In addition, defined internal sequences have been duplicated or deleted, thereby generating two to three subtypes within each proline-rich region type (Fig. 2 and 3). In the case of ST ProB, in which up to two copies of the internal sequences were present, these repetitive structures were designated the ProB repeats.
FIG. 3.
Alignment of amino acid sequences corresponding to the proline-rich domain of SfbI. The consensus sequence is shown at the top in boldface with underlining. The proline-rich region has two STs, ProA and ProB. Deletion or duplication events within these STs define the proline-rich region subtypes. Dots represent identical nucleotides relative to the consensus sequence, and hyphens indicate the positions of insertions found in particular isolates. The ProB repeats are indicated, and the first true proline-rich repeat is boxed.
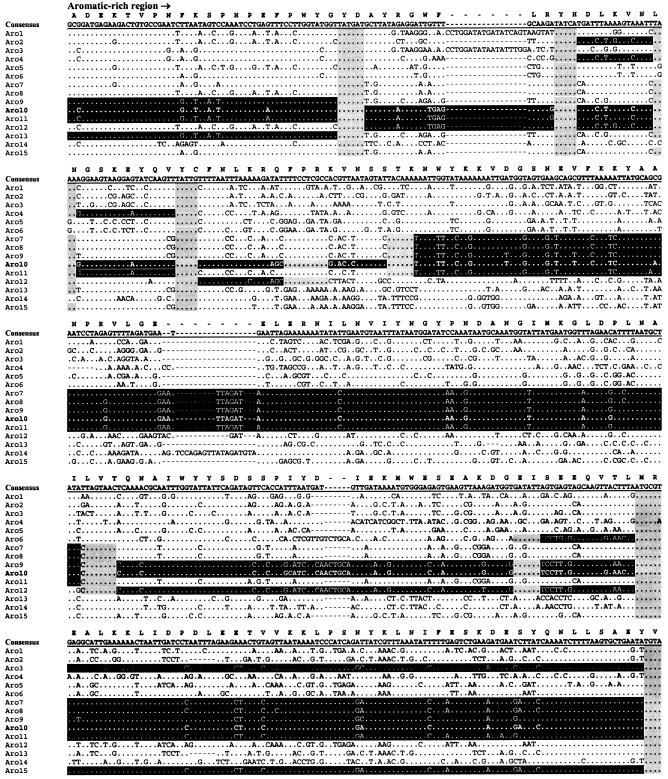
The 5′ end of sfbI, which encodes the aromatic amino acid-rich domain, is by far the most variable region of the gene. Fifteen distinct aromatic amino acid-rich domain sequences types (Aro types), designated Aro1 to Aro15, were identified. In pairwise alignments, a minimum 98.8% identity was seen within Aro types, while 50.3 to 97.6% identity was observed between types. While some of this variation is due to point mutations, which are easily identifiable as single nucleotide changes within otherwise conserved regions, the majority appear to be due to intergenic recombination. Similar to the emm gene, this intergenic DNA transfer has generated a gene mosaic structure in sfbI. The overall effects at the protein level can be seen in the amino acid sequence alignments of the various aromatic amino acid-rich domains (Fig. 4). Aro10 has been used to illustrate how distinct DNA cassettes, flanked by highly conserved junction sequences, are shared between otherwise divergent STs. This type of mosaic allele can be generated only by horizontal gene transfer between strains.
FIG. 4.
Alignment of nucleotide sequences corresponding to the aromatic amino acid-rich domain STs. The overall consensus sequence is shown in boldface with underlining above the alignment, and the translated protein sequence is also shown. Dots represent identical nucleotides relative to the consensus sequence, and hyphens indicate the positions of insertions found in particular isolates. The gene mosaic structure is illustrated by using Aro10 as an example (boldface), putative horizontally transferred DNA cassettes are shaded in black, while predicted junctions are shaded in gray.
Conserved structure of sfbI.
Despite the high degree of variation observed, the overall structural properties of SfbI were conserved. Within the aromatic amino acid-rich domain, the distribution of aromatic amino acids remained relatively unchanged, and similarly, the distribution of proline residues within the proline-rich domain remained relatively unchanged. Over the entire length of the deduced amino acid sequence, hydrophobicity, average charge, and surface probability plots were generally conserved among the SfbI proteins from different strains. By restriction of comparisons to the aromatic amino acid- and proline-rich domains, the positively charged regions remained clustered in the aromatic amino acid-rich domain, despite sequence variation, with a few peaks in the proline-rich region before the negatively charged proline-rich repeats (Fig. 4). The periodicity of the repeat sequences is apparent in hydrophobicity, charge, and surface probability plots (data not shown). The theoretical pI of the putative SfbI proteins ranged between 4.11 and 4.81 (mean ± standard deviation, 4.51 ± 0.13), indicating that SfbI would have a net negative charge at physiological pH.
Summary of sfbI STs.
Thirty-five individual sfbI STs were identified on the basis of combinations of aromatic amino acid- and proline-rich domain STs and the numbers of proline-rich and fibronectin-binding repeats (Table 1). The strains within some VTs (VT2.2, VT14.1, VT22, VT33.1, VT33.2, VT52, VT57, and VT61) appeared to be clonal and were of the same emm and sfbI STs. M12 strain A735 (6) is also likely a clone of VT52, since all three strains are M12 and have identical sfbI genes. By contrast, VT37.1 was highly heterogeneous and included strains exhibiting sfbI STs 5, 11, 30, and 31. Conversely, other strains had virtually identical sfbI genes but belonged to different VTs; for example, sfbI ST18 included strains of VT2.1, VT2.2, and VT3.1. Some sfbI STs were identical except in the variable numbers of repeats; for example, sfbI ST30 and sfbI ST31 were identical except that the former has three proline-rich repeats and one fibronectin-binding repeat, while the latter has one and two repeats, respectively. This demonstrates that while the presence of sfbI is associated with VT, the species of the sfbI allele may be independent.
No correlation between the relative numbers of proline-rich and fibronectin-binding repeats was observed. There was also no apparent relationship between the numbers of repeats and aromatic amino acid- or proline-rich STs; for example, compare sfbI STs 4 to 8 within Aro2 (Table 1). Proline-rich domain STs were associated with particular aromatic amino acid-rich domain STs; however, several proline-rich domain sequence subtypes could be seen to be associated with a single aromatic amino acid-rich domain ST. For example, within Aro7, sfbI ST16, sfbI ST17, and sfbI ST18 exhibit proline-rich subtypes ProB3, ProB2, and ProB1, respectively.
Phylogenic analysis of sfbI.
The aromatic amino acid-rich domain was considered to be the greater indicator of relatedness, since the high degree of variability of this domain appeared to have been generated by many individual mutations, while the variability in the numbers of repeats was due to a smaller number of mutation events that occurred at a relatively high frequency but that involved large fragments of highly conserved DNA. This is probably best illustrated by the Aro6 group, the sequences of which are highly conserved in the aromatic amino acid-rich domain but which contain 0, 1, or 2 internal ProB repeats; 4 or 11 proline-rich repeats; and 1, 2, or 4 fibronectin-binding repeats. This suggests that the repetitive units are frequently shuffled and that this has little or no true bearing on the phylogeny of the strain. Variation in sfbI has arisen through genetic exchange evolution of different alleles and is not independent. Therefore, a phylogenic tree was not generated, since the principal assumption of such analysis is that the alleles have evolved independently.
Is the sfbI ST associated with the clinical source of the isolate?
Information on the clinical sources of the isolates was available. Clonal distribution indicated that the skin was the principal reservoir for invasive disease isolates in the Northern Territory of Australia. Some sfbI STs were exclusively throat isolates (1, 9, 14, 21, 22); however, this association did not extend to Aro or proline-rich types. Further investigation revealed that all throat isolates were cultured from samples from nonindigenous people; therefore, this observation was probably due to the differences in the strains circulating in the two populations rather than to the tissue tropism that might be attributed to the SfbI variants. Overall, there was no common trend to indicate that particular or related STs were associated with the clinical sources of the isolates.
DISCUSSION
Components of both the Vir regulon and the FCT region have undergone frequent genetic rearrangement. These regions encode important virulence factors, principally adhesins with affinities for different human proteins such as fibronectin, plasminogen, collagen, fibrinogen, and immunoglobulins. Horizontal gene transfer has played a major role in the generation of variability within both regions, resulting in the mosaic alleles seen in emm/enn and sfbI.
The advantages of increased variability through horizontal gene transfer include the generation of antigenic variation and variable binding capabilities. As a mechanism of generating genetic diversity, horizontal gene transfer has the unique advantage of allowing incorporation of cassettes or domains which have already been checked for fitness in another genetic context. This bypasses the less efficient trial-and-error strategy and is particularly useful in altering the functional capabilities of a protein in a relatively short time frame.
The mosaic structure of the genes encoding the M-protein family has resulted in a divergent group of proteins exhibiting variable binding abilities and serotype-specific epitopes. The role that such variation plays in the pathogenesis of GAS is purely speculative; however, clear links have been shown between Vir regulon arrangements (emm patterns) and tissue tropism (5). We have shown that sfbI also has a gene mosaic structure especially apparent in the amino-terminal aromatic amino acid-rich domain and have identified 35 distinct sfbI STs. The biological function of the aromatic amino acid-rich domain is not known. To date, the reported binding functions of this domain include fibrinogen binding (20) and a putative carbohydrate-binding motif (40) which, from our data, appears to be highly conserved (Fig. 3A). The conserved distribution of aromatic amino acids within this domain, as well as the apparent intragenomic translocation of a highly homologous domain into Cpa (30), implies that the domain may have a significant function. In addition, the fact that the entire domain is present as a defined unit in Cpa implies that the function(s) and/or properties of the aromatic amino acid-rich domain require the whole domain. Despite the high degree of amino acid sequence variability, this domain remains positively charged. Since the proline-rich domain is predominantly negatively charged, we speculate that interaction between the two domains may be pivotal to the overall structure or folding of the native protein.
The other possibility is that variability has arisen in response to selective pressure from the immune system. Anti-M-protein antibodies are protective yet are serotype specific (23), as the protective epitopes reside in the variable amino terminus of the M protein (1, 7, 18). SfbI is a protective antigen when used in animal infection models (14); however, unlike M protein the protective epitope(s) resides in the conserved carboxy terminus and can provide protection from heterologous challenge (34). Natural infection in individuals from populations in which streptococcal infections are endemic also elicits significant levels of anti-SfbI IgG antibodies (13). Since these individuals are still infected, despite the presence of anti-SfbI IgG antibodies, it appears that anti-SfbI antibodies either are not protective in humans or are not present in significantly high enough concentrations to be effective. While the fibronectin-binding domain is protective and conserved, variability in the amino terminus may represent a mechanism of immune evasion, thereby directing the immune response away from the carboxy terminus.
Recombination within selection-neutral genes in GAS is common to the point that it masks phylogenic signals (10). As a consequence, phylogenic analyses of sfbI do not clarify evolutionary relationships between strains but clarify only the similarities between sfbI alleles. In looking at possible correlations between the sfbI STs and the clinical sources of the isolates, we did not find any associations. As with T-agglutination patterns, specific sfbI STs were consistently associated with emm STs; however, like T types, they were not restricted to singular emm STs. This seems to indicate that emm and sfbI and not closely linked. Given that the tee and sfbI genes both reside in the FCT region, it would be interesting to determine if T types and sfbI STs are more closely associated with each other than with emm STs. Such an analysis would require determination of the T types of the 54 GAS strains examined in this study.
The 34 individual sfbI STs identified in this study seem to have evolved through a combination of different evolutionary mechanisms working independently on distinct regions of the sfbI gene. The majority of divergence in the Aro domain appears to be due to intergenic recombination of horizontally transferred DNA cassettes, resulting in a genetic mosaic; two distinct and divergent STs are apparent in the central proline-rich region of sfbI; and the deletion or duplication of repeat units has resulted in a variable number of ProB, proline-rich, and fibronectin-binding repeats.
SfbI is an important virulence factor involved in colonization and internalization in fibronectin-rich epithelial tissues such as the upper respiratory mucosa and injured skin epithelium. Without clear associations between SfbI variants and function or tissue tropism, it is impossible to draw conclusions regarding the selective factors influencing these evolutionary mechanisms; however, the variability observed implies significant selective pressure on this locus in vivo. In light of the role played by intergenic transfer in the generation of variability in ligand-binding capacities in members of the M-protein family, the implications for microbial pathogenesis of possible functional and antigenic variability in SfbI warrant further investigation.
Acknowledgments
R.J.T. received postgraduate awards from the University of Wollongong and the German Centre for Biotechnology (Braunschweig, Germany). This work was partially supported by the National Health and Medical Research Council of Australia (grant 9936609) and BMBF Network Pathogenomics.
We thank those people who have contributed to the accumulation of data on isolates in the Northern Territory Streptococcal Collection, including M. Hibble, D. E. Bessen, J. Carapetis, A. Delvecchio, M. L. Edwards, and A. M. Goodfellow.
REFERENCES
- 1.Beachey, E. H., J. M. Seyer, J. B. Dale, W. A. Simpson, and A. H. Kang. 1981. Type-specific protective immunity evoked by synthetic peptide of Streptococcus pyogenes M protein. Nature 292:457-459. [DOI] [PubMed] [Google Scholar]
- 2.Beall, B., R. R. Facklam, J. A. Elliott, A. R. Franklin, T. Hoenes, D. Jackson, L. Laclaire, T. Thompson, and R. Viswanathan. 1998. Streptococcal emm types associated with T-agglutination types and the use of conserved emm gene restriction fragment patterns for subtyping group A streptococci. J. Med. Microbiol. 47:893-898. [DOI] [PubMed] [Google Scholar]
- 3.Beall, B., R. Facklam, and T. Thompson. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bessen, D. E., M. W. Izzo, T. R. Fiorentino, R. M. Caringal, S. K. Hollingshead, and B. Beall. 1999. Genetic linkage of exotoxin alleles and emm gene markers for tissue tropism in group A streptococci. J. Infect. Dis. 179:627-636. [DOI] [PubMed] [Google Scholar]
- 6.Bessen, D. E., and A. Kalia. 2002. Genomic localization of a T serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infect. Immun. 70:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale, J. B., and E. H. Beachey. 1986. Localization of protective epitopes of the amino terminus of type 5 streptococcal M protein. J. Exp. Med. 163:1191-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delvecchio, A., B. J. Currie, J. D. McArthur, M. J. Walker, and K. S. Sriprakash. 2002. Streptococcus pyogenes prtFII, but not sfbI, sfbII or fbp54, is represented more frequently among invasive-disease isolates of tropical Australia. Epidemiol. Infect. 128:391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinkla, K., M. Rohde, W. T. M. Jansen, J. R. Carapetis, G. S. Chhatwal, and S. R. Talay. 2003. Streptococcus pyogenes recruits collagen via surface bound fibronectin: a novel colonisation and immune evasion mechanism. Mol. Microbiol. 47:861-869. [DOI] [PubMed] [Google Scholar]
- 10.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1980. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 12.Gardiner, D., J. Hartas, B. Currie, J. D. Mathews, D. J. Kemp, and K. S. Sriprakash. 1995. Vir typing: a long-PCR typing method for group A streptococci. PCR Methods Appl. 4:288-293. [DOI] [PubMed] [Google Scholar]
- 13.Goodfellow, A. M., M. Hibble, S. R. Talay, B. Kreikemeyer, B. J. Currie, K. S. Sriprakash, and G. S. Chhatwal. 2000. Distribution and antigenicity of fibronectin binding proteins (SfbI and SfbII) of Streptococcus pyogenes clinical isolates from the Northern Territory, Australia. J. Clin. Microbiol. 38:389-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzmán, C. A., S. R. Talay, G. Molinari, E. Medina, and G. S. Chhatwal. 1999. Protective immune response against Streptococcus pyogenes in mice after intranasal vaccination with the fibronectin-binding protein SfbI. J. Infect. Dis. 179:901-906. [DOI] [PubMed] [Google Scholar]
- 15.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 89:6172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jadoun, J., V. Ozeri, E. Burstein, E. Skutelsky, E. Hanski, and S. Sela. 1998. Protein F1 is required for efficient entry of Streptococcus pyogenes into epithelial cells. J. Infect. Dis. 178:147-158. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, D. R., and E. L. Kaplan. 1995. A review of the correlation of T-agglutination patterns and M-protein typing and opacity factor production in the identification of group A streptococci. J. Med. Microbiol. 38:311-315. [DOI] [PubMed] [Google Scholar]
- 18.Jones, K. F., B. N. Manjula, K. H. Johnston, S. K. Hollingshead, J. R. Scott, and V. A. Fischetti. 1985. Location of variable and conserved epitopes among the multiple serotypes of streptococcal M protein. J. Exp. Med. 161:623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapur, V., S. Kanjilal, M. R. Hamrick, L. L. Li, T. S. Whittam, S. A. Sawyer, and J. M. Musser. 1995. Molecular population genetic analysis of the streptokinase gene of Streptococcus pyogenes: mosaic alleles generated by recombination. Mol. Microbiol. 16:509-519. [DOI] [PubMed] [Google Scholar]
- 20.Katerov, V., A. Andreev, C. Schalen, and A. A. Totolian. 1998. Protein F, a fibronectin-binding protein of Streptococcus pyogenes, also binds human fibrinogen: isolation of the protein and mapping of the binding region. Microbiology 144:119-126. [DOI] [PubMed] [Google Scholar]
- 21.Kline, J. B., S. Xu, A. L. Bisno, and C. M. Collins. 1996. Identification of a fibronectin-binding protein (GfbA) in pathogenic group G streptococci. Infect. Immun. 64:2122-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreikemeyer, B., S. R. Talay, and G. S. Chhatwal. 1995. Characterization of a novel fibronectin-binding surface protein in group A streptococci. Mol. Microbiol. 17:137-145. [DOI] [PubMed] [Google Scholar]
- 23.Lancefield, R. C. Current knowledge of the type specific M antigens of group A streptococci. J. Immunol. 89:307-313. [PubMed]
- 24.Marciel, A. M., V. Kapur, and J. M. Musser. 1997. Molecular population genetic analysis of a Streptococcus pyogenes bacteriophage-encoded hyaluronidase gene: recombination contributes to allelic variation. Microb. Pathog. 22:209-217. [DOI] [PubMed] [Google Scholar]
- 25.Medina, E., G. Molinari, M. Rohde, B. Haase, G. S. Chhatwal, and C. A. Guzman. Fc-mediated nonspecific binding between fibronectin-binding protein I of Streptococcus pyogenes and human immunoglobulins. J. Immunol. 163:3396-3402. [PubMed]
- 26.Medina, E., S. R. Talay, G. S. Chhatwal, and C. A. Guzmán. 1998. Fibronectin-binding protein I of Streptococcus pyogenes is a promising adjuvant for antigens delivered by mucosal route. Eur. J. Immunol. 28:1069-1077. [DOI] [PubMed] [Google Scholar]
- 27.Molinari, G., S. R. Talay, P. Valentin-Weigand, M. Rohde, and G. S. Chhatwal. 1997. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect. Immun. 65:1357-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natanson, S., S. Sela, A. E. Moses, J. M. Musser, M. G. Caparon, and E. Hanski. 1995. Distribution of fibronectin-binding proteins among group A streptococci of different M types. J. Infect. Dis. 171:871-878. [DOI] [PubMed] [Google Scholar]
- 29.Okada, N., A. P. Pentland, P. Falk, and M. G. Caparon. 1994. M protein and protein F act as important determinants of cell-specific tropism of Streptococcus pyogenes in skin tissue. J. Clin. Investig. 94:965-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Podbielski, A., M. Woischnik, B. A. Leonard, and K. H. Schmidt. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051-1064. [DOI] [PubMed] [Google Scholar]
- 31.Rasmussen, M., A. Eden, and L. Bjorck. 2000. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect. Immun. 68:6370-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reda, K. B., V. Kapur, D. Goela, J. G. Lamphear, J. M. Musser, and R. R. Rich. 1996. Phylogenetic distribution of streptococcal superantigen SSA allelic variants provides evidence for horizontal transfer of ssa within Streptococcus pyogenes. Infect. Immun. 64:1161-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohde, M., E. Müller, G. S. Chhatwal, and S. R. Talay. 2003. Host cell caveolae act as an entry-port for group A streptococci. Cell. Microbiol. 5:323-342. [DOI] [PubMed] [Google Scholar]
- 34.Schulze, K., E. Medina, S. R. Talay, R. J. Towers, G. S. Chhatwal, and C. A. Guzman. 2001. Characterization of the domain of fibronectin-binding protein I of Streptococcus pyogenes responsible for elicitation of a protective immune response. Infect. Immun. 69:622-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarz-Linek, U., J. M. Werner, A. Pickford, S. Gurusiddappa, J. H. Kim, E. S. Pilka, J. A. Briggs, T. S. Gough, M. Hook, I. D. Campbell, and J. R. Potts. 2003. Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature 423:177-181. [DOI] [PubMed] [Google Scholar]
- 36.Sela, S., A. Aviv, A. Tovi, I. Burstein, M. G. Caparon, and E. Hanski. 1993. Protein F: an adhesin of Streptococcus pyogenes binds fibronectin via two distinct domains. Mol. Microbiol. 10:1049-1055. [DOI] [PubMed] [Google Scholar]
- 37.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talay, S. R., E. Ehrenfeld, G. S Chhatwal, and K. N. Timmis. 1991. Expression of the fibronectin-binding components of Streptococcus pyogenes in Escherichia coli demonstrates that they are proteins. Mol. Microbiol. 5:1727-1734. [DOI] [PubMed] [Google Scholar]
- 39.Talay, S. R., P. Valentin-Weigand, P. G. Jerlstrom, K. N. Timmis, and G. S. Chhatwal. 1992. Fibronectin-binding protein of Streptococcus pyogenes: sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infect. Immun. 60:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Talay, S. R., P. Valentin-Weigand, K. N. Timmis, and G. S. Chhatwal. 1994. Domain structure and conserved epitopes of Sfb protein, the fibronectin-binding adhesin of Streptococcus pyogenes. Mol. Microbiol. 13:531-539. [DOI] [PubMed] [Google Scholar]
- 41.Talay, S. R., A. Zock, M. Rohde, G. Molinari, M. Oggioni, G. Pozzi, C. A. Guzman, and G. S. Chhatwal. 2000. Cooperative binding of human fibronectin to SfbI protein triggers streptococcal invasion into respiratory epithelial cells. Cell. Microbiol. 2:521-535. [DOI] [PubMed] [Google Scholar]
- 42.Valentin-Weigand, P., S. R. Talay, A. Kaufhold, K. N. Timmis, and G. S. Chhatwal. 1994. The fibronectin binding domain of the Sfb protein adhesin of Streptococcus pyogenes occurs in many group A streptococci and does not cross-react with heart myosin. Microb. Pathog. 17:111-120. [DOI] [PubMed] [Google Scholar]
- 43.Whatmore, A. M., and M. A. Kehoe. 1994. Horizontal gene transfer in the evolution of group A streptococcal emm-like genes: gene mosaics and variation in Vir regulons. Mol. Microbiol. 11:363-374. [DOI] [PubMed] [Google Scholar]
- 44.Whatmore, A. M., V. Kapur, J. M. Musser, and M. A. Kehoe. 1995. Molecular population genetic analysis of the enn subdivision of group A streptococcal emm-like genes: horizontal gene transfer and restricted variation among enn genes. Mol. Microbiol. 15:1039-1048. [DOI] [PubMed] [Google Scholar]