Abstract
Rett syndrome (RTT) is a neurodevelopmental autism spectrum disorder that affects girls due primarily to mutations in the gene encoding methyl-CpG binding protein 2 (MECP2). The majority of RTT patients carry missense and nonsense mutations leading to a hypomorphic MECP2, while null mutations leading to the complete absence of a functional protein are rare. MECP2 is an X-linked gene subject to random X-chromosome inactivation resulting in mosaic expression of mutant MECP2. The lack of human brain tissue motivates the need for alternative human cellular models to study RTT. Here we report the characterization of a MECP2 mutation in a classic female RTT patient involving rearrangements that remove exons 3 and 4 creating a functionally null mutation. To generate human neuron models of RTT, we isolated human induced pluripotent stem (hiPS) cells from RTT patient fibroblasts. RTT-hiPS cells retained the MECP2 mutation, are pluripotent and fully reprogrammed, and retained an inactive X-chromosome in a nonrandom pattern. Taking advantage of the latter characteristic, we obtained a pair of isogenic wild-type and mutant MECP2 expressing RTT-hiPS cell lines that retained this MECP2 expression pattern upon differentiation into neurons. Phenotypic analysis of mutant RTT-hiPS cell-derived neurons demonstrated a reduction in soma size compared with the isogenic control RTT-hiPS cell-derived neurons from the same RTT patient. Analysis of isogenic control and mutant hiPS cell-derived neurons represents a promising source for understanding the pathogenesis of RTT and the role of MECP2 in human neurons.
INTRODUCTION
Rett syndrome [RTT (MIM 312750)] is a neurodevelopmental disorder affecting roughly 1 in 10 000 live female births (1). RTT girls develop normally until 6–18 months of age when they enter a stage of developmental arrest. Clinical features include microcephaly, characteristic hand wringing, autistic features, loss of language, and mental retardation (2). Genetically, over 95% of classic RTT patients harbour a loss-of-function mutation in an X-linked gene encoding the methyl-CpG binding protein 2 (MECP2) (3). MECP2 functions as a transcriptional regulator, both as an activator and repressor, by binding to methylated CpG dinucleotides of target genes via its methyl-CpG binding domain (MBD) and recruiting chromatin remodelling proteins via its transcriptional repression domain (TRD) (4–8).
Most mutations in MECP2 are de novo from the paternal germline involving a C-to-T mutation at CpG hotspots (9,10). In North America, ∼39 and ∼35% of RTT patients are due to missense and nonsense mutations in MECP2, respectively, while large deletions are relatively rare (∼6%) (11–15). Furthermore, as MECP2 is X-linked, it is subject to the effect of X-chromosome inactivation (XCI) in female cells. XCI occurs during female development when one of the two X-chromosomes is randomly inactivated such that approximately half the cells inactivate the maternally derived X-chromosome, while the other half inactivates the paternally derived X-chromosome (16). Therefore, RTT patients are mosaic where half of their cells express wild-type (WT) MECP2, while the other half express mutant MECP2. However, although XCI is random in most cases, it can occasionally be nonrandom which could lead to phenotypic variability in RTT patients depending on the extent of favourable XCI skewing (17).
Most of our understanding of RTT and MECP2 has been attributed to the study of Mecp2 mutant mouse models as access to patient neurons, such as postmortem tissues, is severely limited and may not accurately reflect early pathogenesis of RTT (18). Although, Mecp2 mutant mouse models recapitulate key characteristics associated with RTT patients including an initial phase of apparently normal development followed by severe neurodevelopmental dysfunction, there is evidence that mouse models are an underrepresentation of the human condition (19–21). Mecp2−/y mice are viable, whereas the equivalent mutation in human males is associated with severe congenital encephalopathy and early death (22). Disease onset in RTT mouse models is during adulthood, whereas disease onset in RTT patients is during childhood (2). This could be due to the fact that RTT mouse models exhibit unbalanced XCI favouring the expression of WT Mecp2, whereas in RTT patients, balanced XCI is the norm (23–25). For these reasons, a method that will allow for the generation of large numbers of affected neurons directly from RTT patients will be advantageous to further understand the role of MECP2 and pathogenesis of RTT in human neurons.
Human induced pluripotent stem (hiPS) cells, which are similar to human embryonic stem (hES) cells molecularly and functionally, can be derived from adult somatic cell types via the introduction of defined transcription factors (26–28). The generation of patient-specific hiPS cells has major implications for translational medicine, such as disease phenotyping, drug screens, and cell therapy. Indeed, hiPS cells have been generated from a variety of diseases where specific phenotypes have been observed in vitro and proof-of-principle drug screens have been performed (29–37). More recently, it has been observed that female hiPS cells retain an inactive X-chromosome in a nonrandom pattern (38), in contrast to their mouse counterparts which reactivate the inactive X-chromosome thus carrying two active X-chromosomes and exhibit random XCI upon differentiation (39). This pattern of XCI in female hiPS cells provide prospects to isolate isogenic control and experimental hiPS cell lines for heterozygous X-linked diseases, such as RTT.
Here we report the characterization of a functionally null mutation in MECP2 attributable to rearrangements removing exons 3 and 4 (Δ3–4) in a classic RTT patient. We generated hiPS cells from this patient, predicted to carry a severe mutation, and demonstrate that these hiPS cells are pluripotent and fully reprogrammed. Taking advantage of the fact that female hiPS cells retain an inactive X-chromosome in a nonrandom pattern, we obtained hiPS cell lines with alternative parental X-chromosomes inactivated. Directed differentiation of hiPS cells into neurons demonstrated that MECP2 expression follows the XCI pattern, allowing the generation of a pair of isogenic control (expressing WT MECP2) and experimental (expressing mutant MECP2) hiPS cell lines which have important prospects for downstream applications.
RESULTS
Characterization of the Δ3–4 MECP2 mutation
The majority of RTT patients carry missense and nonsense mutations in the MECP2 gene leading to a hypomorphic protein with partial function, while null mutations leading to the complete absence of a functional protein are relatively rare (21,40,41). Preliminary screening for mutations in a 6-year-old classic RTT patient using multiplex ligation-dependent probe amplification indicated that she carries a deletion involving exons 3 and 4 of MECP2 (data not shown). She has growth and developmental delay, inability to walk without assistance, ataxia, nonverbal, has no hand use and constant repetitive hand motions, some tremor, has had epileptic seizures and significant abnormal electroencephalogram, teeth grinding, some sleep difficulties, and breath holding and hyperventilation.
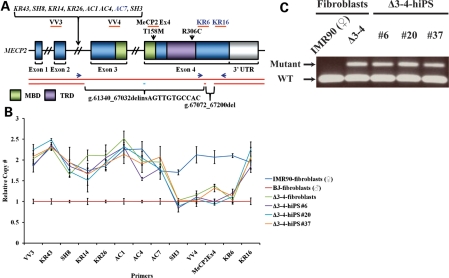
To precisely map the Δ3–4 MECP2 mutation of this RTT patient, a skin punch biopsy was obtained, fibroblasts expanded and DNA harvested for genotyping. Quantitative polymerase chain reaction (qPCR) with primers spanning the MECP2 locus was performed on Δ3–4-fibroblasts to determine copy number variations (Fig. 1A and B). The region of the breakpoints was identified, allowing primers to be designed that span the Δ3–4 MECP2 mutation and amplify the mutant Δ3–4 MECP2 allele (Fig. 1C). Sequencing of the mutant Δ3–4 MECP2 allele revealed a pair of deletions, g.61340_67032delinsAGTTGTGCCAC and g.67072_67200del, where the larger deletion was also associated with an 11 bp insertion, ultimately removing the entire exon 3 and the 5′ end of exon 4, including the MBD and TRD (Fig. 1A). Further analysis of the Δ3–4 MECP2 mutation revealed genetic features associated with genomic copy number variations (Supplementary Material, Fig. S1a). We detected an AluSx element spanning the 5′ end of the larger deletion (g.61340_67032delinsAGTTGTGCCAC) (Supplementary Material, Fig. S1b), which could potentially trigger Alu recombination-mediated deletions (42–44). We were not able to determine the origin of the insertion associated with the larger deletion as an 11 bp motif is insufficient to accurately perform a genome search (Supplementary Material, Fig. S1c). However, the co-occurrence of an insertion with a deletion has been observed previously and the insertion is thought to originate from an inversion of the local genomic sequence (44,45). On the other hand, we detected a 3 bp microhomology sequence flanking the smaller deletion (g.67072_67200del) (Supplementary Material, Fig. S1d), which could potentially trigger microhomology-mediated processes such as microhomology-mediated end joining or microhomology-mediated break-induced replication (43–45). Therefore, it appears that the two deletions may have been caused by two separate mechanisms. Together, these data demonstrate the identification of a complex mutation comprising a pair of deletions, where the larger deletion is also associated with an insertion, within the MECP2 gene, removing the two important domains, in a classic RTT patient.
Figure 1.
Mapping of the Δ3–4 MECP2 mutation. (A) Schematic of the MECP2 locus. Primers used for analysis of copy number variations and their approximate amplifying regions are indicated, where applicable, with orange bars. Primers in italics belong to intron 2. Primers in dark blue were used for amplification of WT (KR6-Fwd and KR16-Rev) and mutant Δ3–4 (AC7-Fwd and KR16-Rev) MECP2 alleles in (C) and their approximate location are indicated with dark blue arrows. The two alleles of MECP2 are indicated with red bars. There are two deletions that comprise the Δ3–4 MECP2 mutation, g.61340_67032delinsAGTTGTGCCAC and g.67072_67200del. The pair of deletions is indicated by the absence of one of the red bars indicative of the deletion of one of the two alleles creating a heterozygous deletion. The insertion associated with the larger deletion is indicated with a light blue bar. The nomenclature of the mutations relates to the genomic DNA sequence with position 1 defined as the first nucleotide of NG_007107.1 (NCBI: reference sequence). The approximate location of the p.T158M and p.R306C mutations is indicated. MBD, methyl-CpG binding domain; TRD, transcriptional repression domain; UTR, untranslated region. (B) qPCR analysis of copy number variations along the MECP2 locus. Data are expressed as mean ± SEM. (C) PCR using WT and mutant-specific primers to amplify the WT and Δ3–4 MECP2 allele, respectively.
Generation and characterization of RTT-hiPS cells
The predicted severity of the Δ3–4 MECP2 mutation prompted us to generate hiPS cells from this patient. Δ3–4-fibroblasts were transduced with OCT4, SOX2, KLF4 and c-MYC retroviral vectors and the EOS lentiviral vector that reports pluripotency as previously described (30,46). Three Δ3–4-hiPS cell lines propagated robustly under puromycin selection driven by the EOS-pluripotent reporter and were subjected to further characterization. Δ3–4-hiPS cells were pluripotent and fully reprogrammed as indicated by the expression of pluripotency-associated markers, including bona fide hiPS cell markers REX1, ABCG2, DNMT3B and TRA1-60 (47) (Fig. 2A and B, Supplementary Material, Fig. S2a). Their ability to differentiate into the three germ layers was shown in vitro via embryoid body formation (Fig. 2C, Supplementary Material, Fig. S2b) and in vivo via teratoma formation by injection into immunodeficient mice (Fig. 2D, Supplementary Material, Fig. S2c). DNA fingerprinting by short tandem repeat analysis indicated that all RTT-hiPS cells came from their fibroblast of origin and not from contaminating hES cells within the laboratory (Fig. 2E). Δ3–4-hiPS cells were fully reprogrammed, as retroviral transgenes were largely silenced and have simultaneously reactivated the endogenous loci of the reprogramming factors similar to previously published patient-specific hiPS cell lines (29,31,32) (Fig. 2F). Δ3–4-hiPS cells maintained a normal female karyotype (Fig. 2G, Supplementary Material, Fig. S2d). Finally, we confirmed that the Δ3–4-hiPS cells carried the Δ3–4 MECP2 mutant and WT allele present in the parental fibroblasts (Fig. 1).
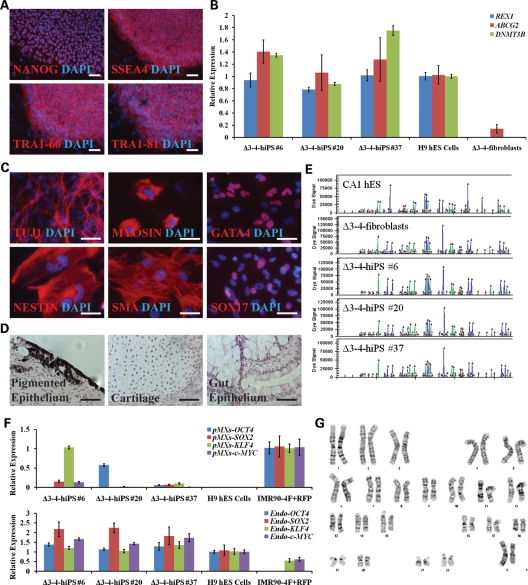
Figure 2.
Δ3–4-hiPS cells are pluripotent and fully reprogrammed. (A) Δ3–4-hiPS #37 express pluripotency markers by immunocytochemistry. Scale bars, 100 µm. (B) Δ3–4-hiPS cells express bona fide hiPS cell markers by qRT–PCR. Data are expressed as mean ± SEM. (C) Δ3–4-hiPS #37 differentiate into the three germ layers, ectoderm (TUJ1, NESTIN), mesoderm [MYOSIN, SMA (SMOOTH MUSCLE ACTIN)] and endoderm (GATA4, SOX17), in vitro via embryoid body formation. Scale bars, 50 µm. (D) Δ3–4-hiPS #37 differentiates into the three germ layers, ectoderm (pigmented epithelium), mesoderm (cartilage) and endoderm (gut epithelium), in vivo via teratoma formation by injection into immunodeficient mice. Scale bars, 50 µm. (E) Δ3–4-hiPS cells carry identical short tandem repeat profile as their parental fibroblast of origin and are distinct from CA1 hES cells in the laboratory. (F) qRT–PCR shows that Δ3–4-hiPS cells have largely silenced the reprogramming vectors in comparison to IMR90-4F+RFP and have reactivated the endogenous loci of the reprogramming factors similar to H9 hES cells. ‘pMXs' and ‘Endo' refers to primers specifically recognizing the exogenous reprogramming factors and endogenous loci, respectively. Data are expressed as mean ± SEM. (G) Δ3–4-hiPS #37 carries a normal female karyotype by G-banding analysis.
In addition, we acquired additional primary fibroblasts from the Coriell Cell Repository which carried a common RTT-associated p.T158M point mutation in MECP2 for reprogramming. Three T158M-hiPS cell lines were generated and were shown to be pluripotent and fully reprogrammed using a similar panel of characterization assays and carried the p.T158M mutation (Supplementary Material, Figs S3 and S4). From these data, we conclude that the RTT-hiPS cells carry a RTT-associated mutation and are pluripotent and fully reprogrammed.
RTT-hiPS cells retain an inactive X-chromosome in a nonrandom pattern
It has been recently demonstrated that female hiPS cells retain an inactive X-chromosome (38). Furthermore, individual female hiPS cell lines exhibit a nonrandom pattern of XCI as they reflect the XCI status of the single fibroblast from which they were derived (38). This is advantageous for the study of heterozygous X-linked diseases, such as RTT, as it allows the generation of hiPS cell lines that express either the WT or mutant form of the protein depending on the pattern of XCI, ultimately allowing the generation of isogenic control (expressing WT MECP2) and experimental (expressing mutant MECP2) hiPS cell lines. Therefore, we sought to determine whether RTT-hiPS cells retain an inactive X-chromosome, and if so, whether it exhibits a nonrandom pattern of XCI.
To investigate the XCI status of Δ3–4-hiPS cells, we probed for the expression of XIST RNA, a key molecule transcribed from the inactive X-chromosome during the initiation of XCI (48), by RNA fluorescence in situ hybridization (RNA-FISH). We observed a single XIST RNA signal in 67–100% of Δ3–4-hiPS cell colonies (Fig. 3A). We note that of the 73% Δ3–4-hiPS #6 colonies where a single XIST RNA signal was detectable, 38% of the colonies (categorized as mix) were below the threshold for a positive colony (>90% of cells per colony). We further assessed the XCI status of Δ3–4-hiPS cells by immunocytochemistry for histone H3 lysine 27 trimethylation (H3K27me3), a repressive chromatin mark which accumulates on the inactive X-chromosome during the initiation of XCI (49). Similar to XIST RNA, we observed a single H3K27me3 signal in 55–100% of the Δ3–4-hiPS cell colonies (Fig. 3B). Similar results were observed in the T158M-hiPS cells as they expressed a single XIST RNA and H3K27me3 signal in 60–77% and 56–75% of the hiPS cell colonies, respectively (Supplementary Material, Fig. S5).
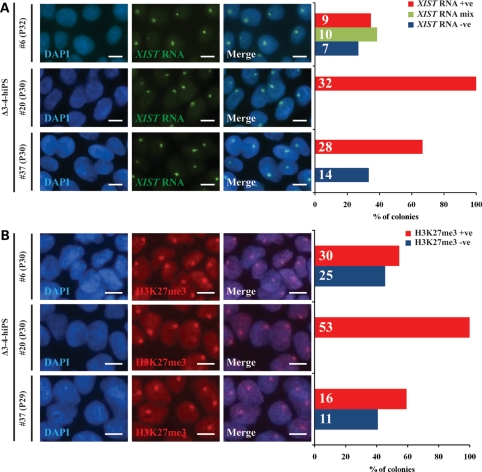
Figure 3.
Female Δ3–4-hiPS cells retain an inactive X-chromosome. Δ3–4-hiPS cells were assessed by RNA-FISH and immunocytochemistry for XIST RNA (A) and H3K27me3 (B), respectively. Graph depicts the percentage of colonies with a single XIST RNA (A) and H3K27me3 (B) signal. Positive (+ve) colonies indicate that >90% of the cells observed within the colony were positive for the signal. Examples of positive colonies are shown on the left panels. Negative (−ve) colonies indicate that no cells within the colony were positive for the signal. Mix colonies indicate colonies where a single XIST RNA signal was detectable, but were below the threshold for a positive colony. The detection of these XCI marks suggests that Δ3–4-hiPS cells retain an inactive X-chromosome, at least in a large fraction of cells. The number of colonies analysed is indicated within each bar. Passage number of analysis is indicated in brackets. P, passage. Scale bars, 10 µm.
The accumulation of a single XIST RNA and H3K27me3 signal, combined with a skewed XCI pattern (see below), in the RTT-hiPS cells suggests they retain an inactive X-chromosome. Intriguingly, in all but one RTT-hiPS cell line, variability within colonies was not observed. The lack of a single XIST RNA and H3K27me3 signal in some of the RTT-hiPS cells could be interpreted as: (i) the loss of an X-chromosome (i.e. 45, XO), (ii) the reactivation of the inactive X-chromosome, or (iii) an inactive X-chromosome that has lost XIST RNA and H3K27me3 but remains transcriptionally suppressed. We exclude the possibility for the loss of an X-chromosome as Δ3–4-hiPS #37 possess two X-chromosomes as observed by X-centromere DNA-FISH (Supplementary Material, Fig. S6), consistent with a normal female karyotype (Fig. 2G). We also exclude the possibility that RTT-hiPS cells carry two active X-chromosomes since neuronal derivates of RTT-hiPS cells do not exhibit random XCI (see below). Therefore, we favour the final possibility as it has been previously demonstrated that female hiPS and hES cells can be subject to the loss of XIST RNA and other repressive chromatin marks (including H3K27me3) during in vitro culture (38,50,51). However, the inactive X-chromosome in the female hiPS and hES cells is maintained in an inactive state where X-linked genes (i.e. MECP2) retain a monoallelic expression pattern (38,50,51). To that end, we confirmed the retention of an inactive X-chromosome in RTT-hiPS cells by allele-specific expression analysis of MECP2 (see below).
To investigate the pattern of XCI in Δ3–4-hiPS cells, we used the androgen receptor (AR) assay which has been used previously for investigating XCI patterns in RTT-patient brains (24). The AR assay detects the heterozygous trinucleotide repeat polymorphism in the first exon of the X-linked AR gene by PCR to distinguish between the paternal and maternal X-chromosome. Genomic DNA is digested with methylation-sensitive enzymes prior to PCR to allow detection of the (methylated) inactive X-chromosome. The AR assay revealed that Δ3–4-fibroblasts exhibited a random pattern of XCI (69:31) as shown by the detection of two different-sized amplicons of the AR gene after digestion with methylation-sensitive enzymes (Fig. 4A, Supplementary Material, Table S1). On the other hand, all Δ3–4-hiPS cell lines exhibited an extreme XCI skewing pattern (96:4 to 99:1). Furthermore, Δ3–4-hiPS #6 and #37 skewed towards the same parental X-chromosome being inactivated, while Δ3–4-hiPS #20 skewed towards the alternative parental X-chromosome being inactivated. Male BJ-fibroblasts were used as a positive control for the complete digestion of the (unmethylated) active X-chromosome resulting in the lack of an AR signal (Supplementary Material, Fig. S7).
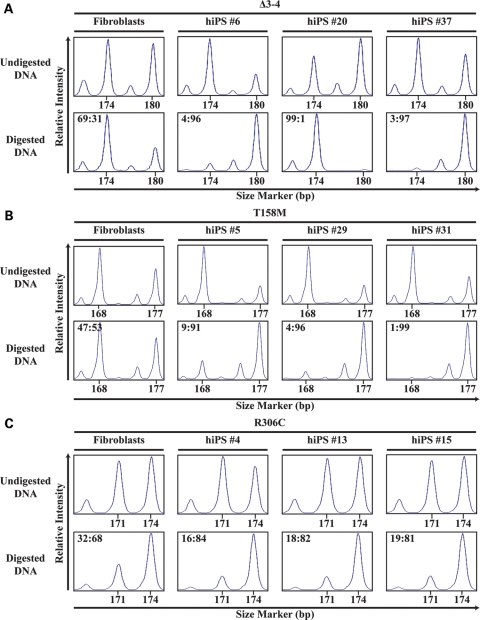
Figure 4.
XCI is nonrandom in female RTT-hiPS cells. RTT-fibroblasts and -hiPS cells were assessed for XCI patterns via the AR assay. Δ3–4- (A), T158M- (B), and R306C- (C) fibroblasts exhibited a random XCI pattern as shown by the presence of two different-sized amplicons, 174 and 180 bp, 168 and 177 bp, and 171 and 174 bp, respectively, after digestion with methylation-sensitive enzymes. Δ3–4- (A), T158M- (B), and R306C- (C) hiPS cells exhibited an extreme XCI skewing pattern as shown by the preferential detection of a single-sized peak after digestion with methylation-sensitive enzymes. The corrected XCI ratio (see Materials and Methods) is indicated on the top left of the digested graph.
When the AR assay was performed on T158M-hiPS cells, all T158M-hiPS cell lines showed an extreme XCI skewing pattern (91:9 to 99:1) towards the same parental X-chromosome being inactivated, while T158M-fibroblasts exhibited a random pattern of XCI (53:47) (Fig. 4B, Supplementary Material, Table S1). We also performed the AR assay on a previously published RTT-hiPS cell line with the point mutation p.R306C (30,46), as well as two additional hiPS cell lines from the same patient. Similar to T158M-hiPS cells, all R306C-hiPS cell lines showed a highly skewed pattern of XCI (84:16 to 81:19) towards the same parental X-chromosome being inactivated, while R306C-fibroblasts exhibited a random pattern of XCI (68:32) (Fig. 4C, Supplementary Material, Table S1). The relatively lesser degree of skewing observed in R306C-hiPS cells is potentially attributed to the smaller AR amplicon (171 bp) including the stutter peak of the larger AR amplicon (174 bp) and hence overestimates the former (see Materials and Methods). The extreme XCI skewing pattern observed in the RTT-hiPS cells is similar to previous findings that female hiPS cells exhibit a nonrandom XCI pattern (38). Collectively, these data suggest that RTT-hiPS cells retain an inactive X-chromosome in a nonrandom pattern. Interestingly, Δ3–4-hiPS cell lines appear to have alternative parental X-chromosomes inactivated, but the T158M- and R306C-hiPS cell lines examined here appear to have the same parental X-chromosome inactivated.
MECP2 expression follows the pattern of XCI in RTT-hiPS cells and their neuronal derivatives
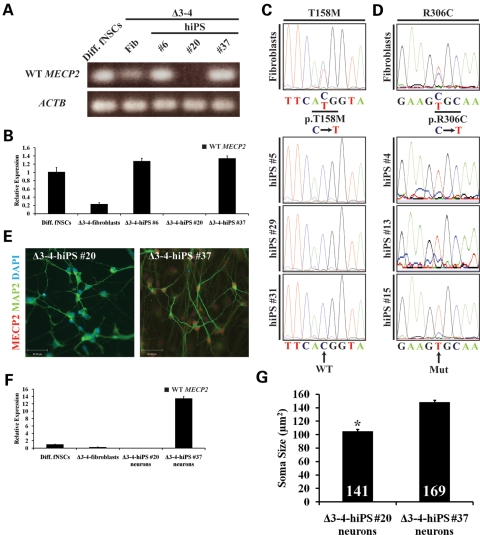
The importance of the XCI pattern with respect to RTT is the expression pattern of the WT or mutant MECP2 in neurons. In addition, to further assess whether RTT-hiPS cells retain an inactive X-chromosome despite the loss of XIST RNA and H3K27me3 in some cells and whether the extreme XCI skewing pattern represents a nonrandom XCI pattern, we performed allele-specific expression analysis of MECP2. By taking advantage of the MECP2 expression in hiPS cells, we performed reverse transcriptase PCR (RT–PCR) and quantitative (q)RT–PCR in Δ3–4-hiPS cells using primers that specifically detect the WT MECP2 transcript and not the mutant Δ3–4 MECP2 transcript. RT–PCR and qRT–PCR detected the expression of WT MECP2 transcripts exclusively in Δ3–4-hiPS #6 and #37 but not in Δ3–4-hiPS #20, suggesting that Δ3–4-hiPS #20 expresses the mutant Δ3–4 MECP2 transcript (Fig. 5A and B). This is in agreement with the AR assay data that Δ3–4-hiPS #6 and #37 have the same parental X-chromosome inactivated, while Δ3–4-hiPS #20 has the alternative parental X-chromosome inactivated. It is worth noting that when qRT–PCR was performed using primers upstream of the Δ3–4 MECP2 mutation (and hence detect both the WT and mutant Δ3–4 MECP2 transcripts), transcripts were detected from Δ3–4-hiPS #20 (Supplementary Material, Fig. S8a). This suggests that mutant Δ3–4 MECP2 transcripts are expressed in Δ3–4-hiPS #20, at least upstream of the Δ3–4 MECP2 mutation.
Figure 5.
MECP2 expression follows the pattern of XCI in RTT-hiPS cells and their neuronal derivatives. RT–PCR (A) and qRT–PCR (B) of Δ3–4-hiPS cells detected WT MECP2 transcripts in Δ3–4-hiPS #6 and #37 but not in Δ3–4-hiPS #20, indicating Δ3–4-hiPS #20 expresses the Δ3–4 mutant MECP2 transcript. Diff. fNSCs, differentiated fetal neural stem cells; Fib, fibroblasts. Data are expressed as mean ± SEM (B). Sequencing of T158M- (C) and R306C- (D) fibroblasts' cDNA revealed that the fibroblasts express a mixture of WT and mutant MECP2 transcripts as indicated by the ‘C' and ‘T' nucleotide peaks, respectively. Sequencing of T158M- (C) and R306C- (D) hiPS cells revealed that they only express the WT and mutant MECP2 transcripts, respectively. Mut, Mutant. Δ3–4-hiPS #20 and #37 were differentiated into neurons for 9–10 weeks. WT MECP2 expression was assessed by immunocytochemistry (E) and qRT–PCR (F) and was detected in Δ3–4-hiPS #37-derived neurons but not in Δ3–4-hiPS #20-derived neurons. Scale bars, 39 µm (E). Diff. fNSCs, differentiated fetal neural stem cells; Fib, fibroblasts. Data are expressed as mean ± SEM (F). (G) Δ3–4-hiPS #20 and #37 were differentiated into neurons for 9 weeks. Δ3–4-hiPS #20-derived neurons exhibited significant reduction in soma size compared with Δ3–4-hiPS #37-derived neurons (*, P < 0.0001, Student's t-test). Total number of neurons (n) analysed per hiPS cell line from two independent biological replicates is indicated at the bottom of each bar. Data are expressed as mean ± SEM.
To determine the pattern of MECP2 expression in T158M- and R306C-hiPS cells, we performed sequencing of the cDNA to detect either the expression of the WT nucleotide (C) or the mutant nucleotide (T) of the p.T158M and p.R306C mutations, respectively. Sequencing revealed that T158M- and R306C-fibroblasts have a heterogeneous pattern of XCI and expressed both the WT and mutant transcripts of MECP2 consistent with a random XCI status (Fig. 5C and D). On the other hand, all T158M-hiPS cell lines expressed the WT MECP2 transcript (Fig. 5C), while all R306C-hiPS cell lines expressed the mutant MECP2 transcript (Fig. 5D). This is in agreement with the AR assay that all T158M- and R306C-hiPS cell lines had the same parental X-chromosome inactivated. Therefore, the published R306C-hiPS cells do in fact express the mutant MECP2 transcript, but the T158M-hiPS cells appear to express the WT MECP2 transcript. Collectively, the nonrandom monoallelic expression pattern of the MECP2 transcript in RTT-hiPS cells demonstrate that RTT-hiPS cells retain an inactive X-chromosome in a nonrandom pattern despite the loss of XIST RNA and H3K27me3 in some cells.
Finally, we sought to determine whether the pattern of WT and mutant-specific expression of MECP2 seen in the different Δ3–4-hiPS cell lines is maintained upon differentiation. Since RTT is primarily a neurodevelopmental disorder, we performed directed differentiation of Δ3–4-hiPS #20 and #37 into the neuronal lineage using previously published protocols designed for hES cells (52). We chose to focus on Δ3–4-hiPS #20 and #37 because they have alternative parental X-chromosomes inactivated. Furthermore, the nature of the predicted null mutation (compared with missense mutations in p.T158M and p.R306C) allows for direct visualization of the WT MECP2 but not the mutant Δ3–4 MECP2 at the protein level via immunocytochemistry using an antibody raised against the C-terminus of MECP2. Δ3–4-hiPS cells were able to differentiate into abundant MAP2-positive neurons (Fig. 5E). Co-labelling for MECP2 detected WT MECP2 protein expression exclusively in the nuclei of Δ3–4-hiPS #37-derived neurons but not in Δ3–4-hiPS #20-derived neurons (Fig. 5E). This was further confirmed by qRT–PCR as WT MECP2 transcripts were detected exclusively in Δ3–4-hiPS #37-derived neurons but not in Δ3–4-hiPS #20-derived neurons (Fig. 5F). This is in agreement with the RT–PCR and qRT–PCR results for WT MECP2 transcripts in the Δ3–4-hiPS cells and suggests that the Δ3–4 MECP2 mutation results in the complete absence of the full-length MECP2 protein and is functionally a null mutation. Similarly, qRT–PCR using primers upstream of the Δ3–4 MECP2 mutation detected transcripts from the Δ3–4-hiPS #20-derived neurons, suggesting the mutant Δ3–4 MECP2 transcript is expressed in these neurons, at least upstream of the Δ3–4 MECP2 mutation (Supplementary Material, Fig. S8b). Finally, we performed the AR assay on Δ3–4-hiPS #20- and #37-derived neurons and observed an extreme XCI skewing pattern similar to the parental hiPS cell lines (Supplementary Material, Fig. S9 and Table S1), excluding the possibility that XIST RNA- and H3K27me3-negative Δ3–4-hiPS cells carry two active X-chromosomes which would have otherwise resulted in random XCI. From these data, we conclude that the nonrandom monoallelic expression of MECP2 in Δ3–4-hiPS cells is maintained upon neuronal differentiation and that Δ3–4-hiPS #37 is an isogenic control of the mutant Δ3–4-hiPS #20.
To demonstrate the utility of the isogenic control and mutant Δ3–4-hiPS cells, we performed phenotyping of soma size in Δ3–4-hiPS cell-derived neurons. We observed that mutant Δ3–4-hiPS #20-derived neurons exhibited a significant reduction in soma size compared with isogenic control Δ3–4-hiPS #37-derived neurons (Fig. 5G), consistent with previous findings in Mecp2−/y mice and postmortem tissues from RTT patients (20,53,54). From these data, we conclude that MECP2 expression is monoallelic in a nonrandom pattern from a single active X-chromosome in RTT-hiPS cells and their neuronal derivatives, and that isogenic Δ3–4-hiPS cell lines have been generated that express either the WT or mutant MECP2 after directed differentiation into neuronal lineages. Finally, we provide a proof-of-principle experiment to demonstrate the utility of the isogenic Δ3–4-hiPS cell-derived neurons for elucidating the pathogenesis of RTT in vitro.
DISCUSSION
In this study, we established isogenic RTT-hiPS cell lines that express either the WT or mutant allele of MECP2. We mapped a functionally null mutation in MECP2 for a classic RTT patient that consisted of a pair of deletions, one of which is also associated with an insertion, that removes exon 3 and the 5′ end of exon 4. This mutation ultimately removes the entire MBD and TRD from the MECP2 coding region. The pair of deletions seem to be caused by two different mechanisms with the larger deletion potentially caused by Alu recombination-mediated deletions and is associated with an insertion, while the smaller deletion is potentially caused by microhomology-mediated processes (42–45). We established Δ3–4-hiPS cell lines from this patient and from fibroblasts carrying another RTT-associated p.T158M point mutation in MECP2. Δ3–4- and T158M-hiPS cells were shown to be pluripotent and fully reprogrammed. Consistent with recent findings (38), female RTT-hiPS cells retain an inactive X-chromosome as suggested by the expression of a single XIST RNA and H3K27me3 signal in the RTT-hiPS cell colonies. Furthermore, the pattern of XCI is nonrandom as suggested by the extreme XCI skewing pattern detected by the AR assay. Most interestingly, we obtained Δ3–4-hiPS cell lines that have alternative parental X-chromosomes inactivated with Δ3–4-hiPS cell lines #6 and #37 inactivating one of the parental X-chromosomes, while Δ3–4-hiPS #20 inactivated the alternative parental X-chromosome. This XCI pattern raised the prospect of isolating a pair of isogenic control and experimental Δ3–4-hiPS cell lines. Indeed, Δ3–4-hiPS #6 and #37, but not Δ3–4-hiPS #20, specifically expressed WT MECP2 transcripts. Furthermore, when directed differentiation was performed towards the neuronal lineage on Δ3–4-hiPS #20 and #37, the primary cell type affected in RTT (20), WT MECP2 transcripts and protein were detected only in neuronal derivates of Δ3–4-hiPS #37 but not of Δ3–4-hiPS #20. This indicates that the Δ3–4 MECP2 mutation results in the complete absence of the full-length MECP2 protein and is functionally a null mutation. From these data, we conclude that Δ3–4-hiPS #37 is an isogenic control for the mutant Δ3–4-hiPS #20.
In contrast to Δ3–4-hiPS cells, we were not able to isolate T158M- and R306C-hiPS cell lines that had alternative parental X-chromosomes inactivated. This is unlikely to be due to differences in reprogramming efficiencies of fibroblasts expressing WT or mutant MECP2, as both alleles of MECP2 expression were obtained, where all the T158M-hiPS cell lines expressed the WT MECP2 transcript while all R306C-hiPS cell lines expressed the mutant MECP2 transcript. It is also unlikely to be due to a skewed XCI pattern in the parental fibroblasts, as they had a random XCI pattern and expressed both the WT and mutant MECP2 transcripts. Therefore, we propose that the distribution of MECP2 allele expression in T158M- and R306C-hiPS cell lines was due to chance, and it is crucial to screen a larger cohort of hiPS cell lines for heterozygous X-linked disorders compared with autosomal disorders to ensure the generation of isogenic control and experimental hiPS cell lines. We recommend screening for hiPS cell lines carrying alternative parental X-chromosomes inactivated using the AR assay followed by confirmation of the WT or mutant RNA or protein expression directly in hiPS cells or the differentiated cell type of interest prior to performing extensive pluripotency characterization.
While our work was being completed, Muotri and colleagues (55) published elegant work demonstrating the utility of RTT-hiPS cells for understanding RTT. Interestingly, however, and distinct from our present study, their described RTT-hiPS cells possessed two active X-chromosomes with a random pattern of XCI upon differentiation into neuronal derivatives and hence their phenotyping relied on comparisons to control hiPS cells generated from unrelated healthy individuals. The generation of female RTT-hiPS cells retaining an inactive X-chromosome with a nonrandom XCI pattern in the present study is consistent with the female hiPS cells described by Plath and colleagues (38), but not with the female RTT-hiPS cells described by Muotri and colleagues (55). This discrepancy may be attributed to differences in reprogramming methods and culturing of hiPS cells which remain to be determined.
The generation of isogenic RTT-hiPS cell lines is advantageous for several reasons. For disease phenotyping, appropriate healthy control hiPS cells are essential and isogenic cells from the same patient eliminate the diversity of genetic backgrounds that exist between individuals. Previous studies of disease phenotyping in patient-specific hiPS cells have used unaffected parents (32), unrelated healthy individuals (33–37,55) and hES cells (33) as controls. Although these studies report disease-associated phenotypes in affected hiPS cells, confounding effects due to differences in genetic background and modifier genes cannot be entirely excluded. Furthermore, isogenic Δ3–4-hiPS cell lines may respond to directed differentiation cues in a more uniform manner compared with hiPS and hES cell lines generated from different individuals (56,57). For these reasons, we hypothesize that future downstream applications with Δ3–4-hiPS cell lines will be able to identify phenotypes that are specific to the MECP2 mutation. Finally, it would be of interest to mix the mutant and isogenic control Δ3–4-hiPS cells in different relative proportions prior to neuronal differentiation to recapitulate the mosaic mutant and WT MECP2 expression pattern that exists in RTT girls for downstream applications.
The Δ3–4 MECP2 mutation mapped in this study presents several advantages over the common missense and nonsense point mutations for studying RTT in vitro (11). The complete absence of the full-length MECP2 protein with the two functional domains, MBD and TRD, in this mutation will likely lead to more pronounced phenotypes for downstream applications compared with hypomorphic missense p.T158M and p.R306C alleles and nonsense Mecp2308 alleles that retain partial function (21,40,41). Indeed, it has been reported that RTT patients with large deletions in MECP2 present with a higher severity score than non-deleted patients (13). The Δ3–4 MECP2 mutation is also more amenable to rescue experiments using transgenes as there is no residual functional full-length MECP2. In contrast, rescue of the p.T158M and p.R306C point mutations will require targeted correction of the expressing allele, or transgenes that simultaneously knockdown the hypomorphic mutant MECP2 and express WT MECP2 at normal levels.
Finally, the generation of Δ3–4-hiPS derived-neurons allow the study of MECP2 function in human neurons and for disease phenotyping which has traditionally been difficult due to lack of human brain tissues from RTT patients for research purposes. To this end, we provide proof-of-principle evidence to demonstrate the utility of the isogenic RTT-hiPS cells as mutant Δ3–4-hiPS #20-derived neurons exhibited a significant reduction in soma size compared with isogenic control Δ3–4-hiPS #37-derived neurons, consistent with previous findings suggesting neuronal maturation is affected in RTT (20,53,54). With the identification of RTT-associated phenotypes in Δ3–4-hiPS cell-derived neurons, it may be possible to use these neurons for drug screens. This is of particular interest as drug targets will act on pathways that compensate for the complete loss of functional MECP2 in the Δ3–4 MECP2 mutant. This is in contrast to p.T158M and p.R306C point mutations where drug targets may act on increasing the hypomorphic MECP2 function or reversing the effect of specific mutations, such as conformation changes of the protein.
In conclusion, to the best of our knowledge, this is the first report where a large complex genetic mutation has been mapped, patient fibroblasts successfully reprogrammed and a disease-specific phenotype observed in cell types differentiated from experimental hiPS cell lines compared with those from isogenic control hiPS cell lines generated from the same patient. We confirm recent findings that female hiPS cells retain an inactive X-chromosome in a nonrandom XCI pattern and can be exploited to generate isogenic control and experimental hiPS cells from heterozygous X-linked diseases (38). The isogenic Δ3–4-hiPS cells reported in the present study will have important implications for determining the pathogenesis of RTT and the role of MECP2 in human neurons.
MATERIALS AND METHODS
MECP2 genotyping
To map the Δ3–4 MECP2 mutation, the region of the deletion was first determined via qPCR using primers (Supplementary Material, Table S2), some of which were previously described (12), spanning the MECP2 locus to determine copy number variations. Genomic DNA was isolated using phenol/chloroform extraction. qPCR was performed with 50 ng of DNA using SYBR Green PCR Master Mix on a 7900 HT Fast Real Time PCR System (all from Applied Biosystems) as per manufacturer's instructions. All reactions were done in triplicates with the average used for subsequent analysis. Standard curves were generated for each primer set and product specificity was assessed using melting curve analysis. The MECP2 amplicon of interest and the FOXP2 reference amplicon (Supplementary Material, Table S2) were amplified for the Δ3–4-fibroblasts and -hiPS cells, normal male BJ-fibroblasts were used as a reference for one copy and normal female IMR90-fibroblasts were used as a positive control for two copies. The copy number of MECP2 was normalized to that of FOXP2 to normalize for differences in DNA input. The delta–delta Ct method was used to determine copy number. The data are expressed as mean ± standard error of the mean (SEM). To map the precise breakpoints of the Δ3–4 MECP2 mutation, the WT MECP2 allele was amplified using KR6-Fwd and KR16-Rev primers, and the Δ3–4 MECP2 allele was amplified using AC7-Fwd and KR16-Rev primers (Supplementary Material, Table S2). The Δ3–4 MECP2 mutant amplicon was gel purified using NucleoSpin Extract II (Macherey-Nagel) as per manufacturer's instructions and sequenced [The Centre for Applied Genomics (TCAG), The Hospital for Sick Children, Canada] using the same primers for amplification. Breakpoints and insertions were determined by aligning with the MECP2 genomic DNA sequence (NCBI Reference Sequence: NG_007107.1). AluSx elements were found using RepeatMasker Open-3.0 (http://www.repeatmasker.org). The precise position of the g.67072_67200del was determined by identifying the 3bp homology upstream of the breakpoint as the breakpoint occurs 3′ to the microhomology (45). Sequencing of the p.T158M mutation was performed by amplifying DNA with RTT primers (Supplementary Material, Table S2). The amplicon was gel purified and sequenced using the same primers for amplification.
Cell culture
Δ3–4-fibroblasts were generated and expanded from a skin punch biopsy of a classic RTT patient at The Hospital for Sick Children, Canada, under the approval of the SickKids Research Ethics Board. T158M (GM17880)- and R306C (GM11270)- fibroblasts were acquired from the Coriell Cell Repository. Human male BJ-fibroblasts (CRL-2522) and female IMR90- fibroblasts (CCL-186) were obtained from American Type Culture Collection (ATCC). Fibroblasts from Coriell and ATCC were maintained in Dulbecco's modified Eagle medium and minimum Essential medium, respectively, supplemented with 10% fetal bovine serum and penicillin/streptomycin (100×) (all from Invitrogen). hiPS cell generation from fibroblasts and culture was performed as previously described (30,46). CA1 and H9 hES cells were obtained from A. Nagy (Mount Sinai Hospital, Toronto, Canada) and The WiCell Research Institute (Wisconsin, USA), respectively, and cultured under the approval of the Canadian Institutes of Health Research Stem Cell Oversight Committee. Culture conditions for hES cells are identical to those for hiPS cells (30,46).
Immunocytochemistry
Cells were rinsed with 2× phosphate-buffered saline (1×) (PBS) (Invitrogen) washes, fixed with 4% formaldehyde (EMD Biosciences) diluted in PBS for 10 min at room temperature (RT), rinsed with 3× PBS washes for 5 min at RT, permeabilized with 0.1% Nonidet P-40 (Sigma) diluted in PBS for 10 min at RT and rinsed with 3× PBST [PBS + 0.1% Tween-20 (Sigma)] washes for 10 min at RT. Cells were blocked in 10% serum of host species [Normal Goat Serum (Cedarlane), Normal Donkey Serum (Millipore)] of secondary antibody and 1% bovine serum albumin (Sigma) diluted in PBST (block solution) overnight at 4°C. Cells were then incubated with primary antibodies (Supplementary Material, Table S3) diluted in block solution overnight at 4°C. Cells were rinsed with 3× PBST washes for 10min at RT and incubated with appropriate Alexa Fluor secondary antibodies (Invitrogen) diluted (1:500) in block solution for 1 h at RT, followed by 3× PBST washes for 10 min at RT. Nuclei were stained with 0.5 µg ml−1 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma) diluted in PBS for 10 min at RT followed by 2× PBS washes. Neurons grown on cover slips (Bellco) were processed in an identical manner except that it was mounted on slides with 0.5 µg ml−1 DAPI diluted in fluorescent mounting medium (Dako). Images were captured using a Leica DMI4000B microscope equipped with Leica DFC340FX camera and Leica Application Suite software or Zeiss Axiovert 200M microscope equipped with a Hamamatsu C9100-13 EMCCD camera and Improvision Volocity software. Soma size analysis of neurons was performed using Improvision Volocity software on 40× images from 50 randomly selected fields from 26 cover slips over two independent biological replicates of neuronal differentiation per hiPS cell line and blinded to the observer.
RNA isolation and PCR analysis
Total RNA was isolated using TRIzol Reagent (Invitrogen) as per manufacturer's instructions. Complementary DNA (cDNA) was generated from 1 µg of DNase I (Invitrogen)-treated total RNA using SuperScript II (Invitrogen) as per manufacturer's instructions. RT–PCR and qRT–PCR were performed using specific primer sequences (Supplementary Material, Table S2). qRT–PCR was performed as described for qPCR in MECP2 genotyping. The housekeeping gene, ACTB (BETA-ACTIN), was used to normalize for differences in cDNA input. For exogenous reprogramming factors, IMR90-fibroblasts freshly infected with OCT4, SOX2, KLF4, c-MYC and mRFP1 retroviral vectors (IMR90-4F + RFP) were used as a positive control; for endogenous pluripotency loci, H9 hES cells were used as a positive control. Sequencing of p.T158M and p.R306C from cDNA was performed by amplifying cDNA with MBD-Fwd and MECP2 Ex4-Rev primers or MECP2 Ex4-Fwd and RTT-Rev primers, respectively (Supplementary Material, Table S2). Amplicons were purified using a QIAquick PCR Purification Kit (Qiagen) as per manufacturer's instructions and sequenced using the same primers for amplification. Differentiated fetal neural stem cells which differentiate into 10–20% neurons (astrocytes make up the remaining percentage) were used as a positive control for RT–PCR and qRT–PCR for MECP2 (kind gift from P. Dirks). RT–PCR and qRT–PCR for WT MECP2 were performed using WT MECP2 and MECP2 Ex4 primers, respectively (Supplementary Material, Table S2). fetal neural stem cells (fNSCs) were isolated from tissues obtained under the approval of the SickKids Research Ethics Board.
In vitro and in vivo differentiation
For in vitro differentiation, hiPS cells were detached (Collagenase IV, Invitrogen) and grown in suspension on low attachment surfaces (Corning) in hiPS cell medium (30,46) without basic fibroblast growth factor for 8 days to form embryoid bodies with medium changes every other day. Embryoid bodies were adhered on to plastic surfaces and allowed to further differentiate for 8 days with medium changes every other day. Cells were then analysed via immunocytochemistry for markers representative of the three germ layers (Supplementary Material, Table S3). For in vivo differentiation, one 10 cm dish of hiPS cells were detached and resuspended in a mixture of Knockout DMEM (Invitrogen), Matrigel (BD Biosciences), Collagen (STEMCELL Technologies) (ratio 2:1:2) and 10 µm ROCK Inhibitor (Sigma) and injected intramuscularly into immunodeficient mice. Tumours were harvested 9–12 weeks after injection. Fixed tumours were embedded in paraffin, sectioned and stained with haematoxylin and eosin for pathological analysis. hES cells and mouse embryonic fibroblasts were used as positive and negative controls, respectively (data not shown). All procedures using animals were approved by the SickKids Animal Care Committee under the auspices of The Canadian Council on Animal Care, and conducted with the approval of the Canadian Institutes of Health Research Stem Cell Oversight Committee.
Karyotyping and DNA fingerprinting
Standard G-banding chromosome analysis with a 400–500 banding resolution was performed at TCAG. DNA fingerprinting was performed using the GenomeLab Human STR Primer Set and analysed by the GenomeLab GeXP Genetic Analysis System (all from Beckman Coulter) as per manufacturer's instructions.
RNA-FISH and DNA-FISH
XIST RNA-FISH and/or X-centromere DNA-FISH was performed using probes and protocols as previously described (58,59). Probes were directly labelled using Nick Translation and Ares Alexa Fluor DNA labelling kits (Invitrogen). Slides were viewed on Nikon TE2000-U microscope with a Roper Scientific CCD camera and NIS elements software package. At least 12 colonies were scored for each line. Results within a colony are based on scoring cells from three independent fields of vision at 60× magnification (>75 cells scored per colony). XIST RNA-positive colonies had a large accumulation of XIST RNA in >90% of the cells in each observed region. Colonies that were scored as XIST RNA-negative had no observable XIST RNA. Occasionally, colonies exhibit a mixture of XIST RNA-positive and -negative cells. For DNA-FISH, the same criteria were used to classify colonies as diploid or aneuploid for the X chromosome.
AR assay
To analyse XCI patterns, 200 ng of DNA was digested for 2 h at 37°C with methylation-sensitive enzymes HpaII and HhaI (Invitrogen) simultaneously to obtain the (methylated) inactive X-chromosome only. To differentiate between the two parental X-chromosomes, 20 ng of digested and undigested DNA was amplified with primers (Supplementary Material, Table S2) designed against the polymorphic trinucleotide (CAG) repeat in the first exon of the AR gene for 32 cycles. The 5′ end of the forward primer is labelled with FAM fluorescein (Invitrogen). PCR products were analysed at TCAG. In brief, PCR products were separated on an ABI3100 Genetic Analyzer with 500 LIZ size standard and analysed by Peak Scanner software (all from Applied Biosystems). The XCI ratio was calculated as previously described (55). In brief, a correction factor was calculated using peak areas of the undigested samples to normalize for preferential amplification of one of the two AR alleles. The XCI ratio was then calculated using the corrected peak area in the digested samples. Male BJ-fibroblasts were used as a positive control for complete digestion of the (unmethylated) active X-chromosome. For R306C-fibroblasts and -hiPS cells, the 171 bp allele will include the stutter peak of the 174 bp allele. Therefore, the signal of the 171 bp allele will be an overestimate. Stutter peak is defined as a 3 bp smaller peak due to polymerase slippage over trinucleotide repeat sequences.
Neuronal differentiation
Neuronal differentiation was performed as previously described with slight modifications (52). In brief, hiPS cells were detached and resuspended in hiPS cell medium (30,46) without basic fibroblast growth factor for 3 days with medium changes every day. Cellular aggregates were then cultured for 3 days in neuronal medium, with medium changes every other day, consisting of Dulbecco's modified Eagle medium: nutrient mixture F-12 (1×), N2 (100×), MEM non-essential amino acids (100×) (all from Invitrogen), and 2 µg ml−1 heparin (Sigma). Suspended cellular aggregates were then adhered on to a plastic surface coated with 20 µg ml−1 laminin (Roche) for 11 days in neuronal medium to generate neuroepithelial cells with medium changes every other day. Neuroepithelial cells were manually detached and cultured as neuroepithelial clusters for 4 days in neuronal medium with medium changes every other day. For neuronal differentiation, neuroepithelial clusters were adhered on to 0.1 mg ml−1 poly-l-ornithine (Sigma) and 20 µg ml−1 laminin-coated cover slips in neuronal differentiation medium consisting of neurobasal, N2 (100×), MEM non-essential amino acid (100×), B27 without vitamin A (50×), penicillin/streptomycin (100×) (all from Invitrogen), brain-derived neurotrophic factor, glial cell line-derived neurotrophic factor, insulin-like growth factor-1 (all from Peprotech at 10 ng ml−1), 1 µm N6,2′-O-dibutyryladenosine 3′,5′-cyclic monophosphate sodium salt, 200 ng ml−1 ascorbic acid (all from Sigma), and 1 µg ml−1 laminin. Neuronal differentiation medium was changed every other day as neurons differentiate from the neuroepithelial clusters and mature over the next seven weeks (52,60).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Canadian Institute for Health Research (MOP102649, IG194505, RMF92090 to J.E.); the Ontario Ministry for Research and Innovation to J.E.; the Beta Sigma Phi International Endowment Fund to J.E.; the National Institutes of Health (HD056452 to L.C.); the Autism Training Research Program from the McGill University to D.G. and the Natural Sciences and Engineering Research Council of Canada—Canada Graduate Scholarship Masters Award and Postgraduate Scholarship Doctoral Award to A.Y.L.C. Funding to pay the Open Access publication charges for this article was provided by Canadian Institute for Health Research.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank S.J. White and B. Minassian for patient interactions and S. Meyn for performing the skin biopsy, T. Thompson for expanding patient fibroblasts, J. Christodoulou for primer sequences, the ESC Facility (The Hospital for Sick Children, Toronto, Canada) for reagents, the Imaging Facility (The Hospital for Sick Children, Toronto, Canada), P. Dirks for fNSC and sharing of reagents, R. Ching for preparing slides for RNA and DNA-FISH experiments, J. Rossant for sharing of reagents, and all members of the Ellis laboratory for helpful discussions. TCAG performed sequencing, karyotyping, and AR assays. H.T. Chao, A.D. Ebert, X.J. Li, S. Russell, and J.P. Weick provided technical advice.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Chahrour M., Zoghbi H.Y. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. doi:10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Hagberg B., Aicardi J., Dias K., Ramos O. A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett's syndrome: report of 35 cases. Ann. Neurol. 1983;14:471–479. doi: 10.1002/ana.410140412. doi:10.1002/ana.410140412. [DOI] [PubMed] [Google Scholar]
- 3.Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. doi:10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 4.Nan X., Meehan R.R., Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. doi:10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nan X., Ng H.H., Johnson C.A., Laherty C.D., Turner B.M., Eisenman R.N., Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. doi:10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 6.Nan X., Campoy F.J., Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. doi:10.1016/S0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 7.Chahrour M., Jung S.Y., Shaw C., Zhou X., Wong S.T., Qin J., Zoghbi H.Y. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. doi:10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Shachar S., Chahrour M., Thaller C., Shaw C.A., Zoghbi H.Y. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum. Mol. Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. doi:10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trappe R., Laccone F., Cobilanschi J., Meins M., Huppke P., Hanefeld F., Engel W. MECP2 mutations in sporadic cases of Rett syndrome are almost exclusively of paternal origin. Am. J. Hum. Genet. 2001;68:1093–1101. doi: 10.1086/320109. doi:10.1086/320109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan M., Lee S.S., Zhang X., Houwink-Manville I., Song H.R., Amir R.E., Budden S., Naidu S., Pereira J.L., Lo I.F., et al. Rett syndrome and beyond: recurrent spontaneous and familial MECP2 mutations at CpG hotspots. Am. J. Hum. Genet. 1999;65:1520–1529. doi: 10.1086/302690. doi:10.1086/302690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Percy A.K., Lane J.B., Childers J., Skinner S., Annese F., Barrish J., Caeg E., Glaze D.G., MacLeod P. Rett syndrome: North American database. J. Child Neurol. 2007;22:1338–1341. doi: 10.1177/0883073807308715. doi:10.1177/0883073807308715. [DOI] [PubMed] [Google Scholar]
- 12.Hardwick S.A., Reuter K., Williamson S.L., Vasudevan V., Donald J., Slater K., Bennetts B., Bebbington A., Leonard H., Williams S.R., et al. Delineation of large deletions of the MECP2 gene in Rett syndrome patients, including a familial case with a male proband. Eur. J. Hum. Genet. 2007;15:1218–1229. doi: 10.1038/sj.ejhg.5201911. doi:10.1038/sj.ejhg.5201911. [DOI] [PubMed] [Google Scholar]
- 13.Scala E., Longo I., Ottimo F., Speciale C., Sampieri K., Katzaki E., Artuso R., Mencarelli M.A., D'Ambrogio T., Vonella G., et al. MECP2 deletions and genotype-phenotype correlation in Rett syndrome. Am. J. Med. Genet. A. 2007;143A:2775–2784. doi: 10.1002/ajmg.a.32002. doi:10.1002/ajmg.a.32002. [DOI] [PubMed] [Google Scholar]
- 14.Archer H.L., Whatley S.D., Evans J.C., Ravine D., Huppke P., Kerr A., Bunyan D., Kerr B., Sweeney E., Davies S.J., et al. Gross rearrangements of the MECP2 gene are found in both classical and atypical Rett syndrome patients. J. Med. Genet. 2006;43:451–456. doi: 10.1136/jmg.2005.033464. doi:10.1136/jmg.2005.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravn K., Nielsen J.B., Skjeldal O.H., Kerr A., Hulten M., Schwartz M. Large genomic rearrangements in MECP2. Hum. Mutat. 2005;25:324. doi: 10.1002/humu.9320. doi:10.1002/humu.9320. [DOI] [PubMed] [Google Scholar]
- 16.Amos-Landgraf J.M., Cottle A., Plenge R.M., Friez M., Schwartz C.E., Longshore J., Willard H.F. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am. J. Hum. Genet. 2006;79:493–499. doi: 10.1086/507565. doi:10.1086/507565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Archer H., Evans J., Leonard H., Colvin L., Ravine D., Christodoulou J., Williamson S., Charman T., Bailey M.E., Sampson J., et al. Correlation between clinical severity in patients with Rett syndrome with a p.R168X or p.T158M MECP2 mutation, and the direction and degree of skewing of X-chromosome inactivation. J. Med. Genet. 2007;44:148–152. doi: 10.1136/jmg.2006.045260. doi:10.1136/jmg.2006.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armstrong D.D. Neuropathology of Rett syndrome. J. Child Neurol. 2005;20:747–753. doi: 10.1177/08830738050200090901. [DOI] [PubMed] [Google Scholar]
- 19.Guy J., Hendrich B., Holmes M., Martin J.E., Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 2001;27:322–326. doi: 10.1038/85899. doi:10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 20.Chen R.Z., Akbarian S., Tudor M., Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 2001;27:327–331. doi: 10.1038/85906. doi:10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 21.Shahbazian M., Young J., Yuva-Paylor L., Spencer C., Antalffy B., Noebels J., Armstrong D., Paylor R., Zoghbi H. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. doi:10.1016/S0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 22.Schule B., Armstrong D.D., Vogel H., Oviedo A., Francke U. Severe congenital encephalopathy caused by MECP2 null mutations in males: central hypoxia and reduced neuronal dendritic structure. Clin. Genet. 2008;74:116–126. doi: 10.1111/j.1399-0004.2008.01005.x. doi:10.1111/j.1399-0004.2008.01005.x. [DOI] [PubMed] [Google Scholar]
- 23.Amir R.E., Van den Veyver I.B., Schultz R., Malicki D.M., Tran C.Q., Dahle E.J., Philippi A., Timar L., Percy A.K., Motil K.J., et al. Influence of mutation type and X chromosome inactivation on Rett syndrome phenotypes. Ann. Neurol. 2000;47:670–679. doi:10.1002/1531-8249(200005)47:5<670::AID-ANA20>3.0.CO;2-F. [PubMed] [Google Scholar]
- 24.Shahbazian M.D., Sun Y., Zoghbi H.Y. Balanced X chromosome inactivation patterns in the Rett syndrome brain. Am. J. Med. Genet. 2002;111:164–168. doi: 10.1002/ajmg.10557. doi:10.1002/ajmg.10557. [DOI] [PubMed] [Google Scholar]
- 25.Young J.I., Zoghbi H.Y. X-chromosome inactivation patterns are unbalanced and affect the phenotypic outcome in a mouse model of Rett syndrome. Am. J. Hum. Genet. 2004;74:511–520. doi: 10.1086/382228. doi:10.1086/382228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. doi:10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. doi:10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 28.Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. doi:10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 29.Dimos J.T., Rodolfa K.T., Niakan K.K., Weisenthal L.M., Mitsumoto H., Chung W., Croft G.F., Saphier G., Leibel R., Goland R., et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. doi:10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 30.Hotta A., Cheung A.Y., Farra N., Vijayaragavan K., Seguin C.A., Draper J.S., Pasceri P., Maksakova I.A., Mager D.L., Rossant J., et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat. Methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. doi:10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- 31.Park I.H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M.W., Cowan C., Hochedlinger K., Daley G.Q. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. doi:10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebert A.D., Yu J., Rose F.F., Jr, Mattis V.B., Lorson C.L., Thomson J.A., Svendsen C.N. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. doi:10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carvajal-Vergara X., Sevilla A., D'Souza S.L., Ang Y.S., Schaniel C., Lee D.F., Yang L., Kaplan A.D., Adler E.D., Rozov R., et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. doi:10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee G., Papapetrou E.P., Kim H., Chambers S.M., Tomishima M.J., Fasano C.A., Ganat Y.M., Menon J., Shimizu F., Viale A., et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. doi:10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moretti A., Bellin M., Welling A., Jung C.B., Lam J.T., Bott-Flugel L., Dorn T., Goedel A., Hohnke C., Hofmann F., et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. doi:10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J., Lian Q., Zhu G., Zhou F., Sui L., Tan C., Mutalif R.A., Navasankari R., Zhang Y., Tse H.F., et al. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8:31–45. doi: 10.1016/j.stem.2010.12.002. doi:10.1016/j.stem.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Itzhaki I., Maizels L., Huber I., Zwi-Dantsis L., Caspi O., Winterstern A., Feldman O., Gepstein A., Arbel G., Hammerman H., et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011 doi: 10.1038/nature09747. 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 38.Tchieu J., Kuoy E., Chin M.H., Trinh H., Patterson M., Sherman S.P., Aimiuwu O., Lindgren A., Hakimian S., Zack J.A., et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. doi:10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. doi:10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 40.Ballestar E., Yusufzai T.M., Wolffe A.P. Effects of Rett syndrome mutations of the methyl-CpG binding domain of the transcriptional repressor MeCP2 on selectivity for association with methylated DNA. Biochemistry. 2000;39:7100–7106. doi: 10.1021/bi0001271. doi:10.1021/bi0001271. [DOI] [PubMed] [Google Scholar]
- 41.Yusufzai T.M., Wolffe A.P. Functional consequences of Rett syndrome mutations on human MeCP2. Nucleic Acids Res. 2000;28:4172–4179. doi: 10.1093/nar/28.21.4172. doi:10.1093/nar/28.21.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sen S.K., Han K., Wang J., Lee J., Wang H., Callinan P.A., Dyer M., Cordaux R., Liang P., Batzer M.A. Human genomic deletions mediated by recombination between Alu elements. Am. J. Hum. Genet. 2006;79:41–53. doi: 10.1086/504600. doi:10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Smith A.J., Walters R.G., Coin L.J., Steinfeld I., Yakhini Z., Sladek R., Froguel P., Blakemore A.I. Small deletion variants have stable breakpoints commonly associated with alu elements. PLoS ONE. 2008;3:e3104. doi: 10.1371/journal.pone.0003104. doi:10.1371/journal.pone.0003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kidd J.M., Graves T., Newman T.L., Fulton R., Hayden H.S., Malig M., Kallicki J., Kaul R., Wilson R.K., Eichler E.E. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. doi:10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conrad D.F., Bird C., Blackburne B., Lindsay S., Mamanova L., Lee C., Turner D.J., Hurles M.E. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat. Genet. 2010;42:385–391. doi: 10.1038/ng.564. doi:10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hotta A., Cheung A.Y., Farra N., Garcha K., Chang W.Y., Pasceri P., Stanford W.L., Ellis J. EOS lentiviral vector selection system for human induced pluripotent stem cells. Nat. Protoc. 2009;4:1828–1844. doi: 10.1038/nprot.2009.201. doi:10.1038/nprot.2009.201. [DOI] [PubMed] [Google Scholar]
- 47.Chan E.M., Ratanasirintrawoot S., Park I.H., Manos P.D., Loh Y.H., Huo H., Miller J.D., Hartung O., Rho J., Ince T.A., et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. doi:10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 48.Marahrens Y., Panning B., Dausman J., Strauss W., Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. doi:10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 49.Plath K., Fang J., Mlynarczyk-Evans S.K., Cao R., Worringer K.A., Wang H., de la Cruz C.C., Otte A.P., Panning B., Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. doi:10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 50.Shen Y., Matsuno Y., Fouse S.D., Rao N., Root S., Xu R., Pellegrini M., Riggs A.D., Fan G. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc. Natl Acad. Sci. USA. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. doi:10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silva S.S., Rowntree R.K., Mekhoubad S., Lee J.T. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc. Natl Acad. Sci. USA. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. doi:10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X.J., Zhang X., Johnson M.A., Wang Z.B., Lavaute T., Zhang S.C. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136:4055–4063. doi: 10.1242/dev.036624. doi:10.1242/dev.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bauman M.L., Kemper T.L., Arin D.M. Pervasive neuroanatomic abnormalities of the brain in three cases of Rett's syndrome. Neurology. 1995;45:1581–1586. doi: 10.1212/wnl.45.8.1581. [DOI] [PubMed] [Google Scholar]
- 54.Kishi N., Macklis J.D. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol. Cell Neurosci. 2004;f27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Marchetto M.C., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. doi:10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu B.Y., Weick J.P., Yu J., Ma L.X., Zhang X.Q., Thomson J.A., Zhang S.C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl Acad. Sci. USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. doi:10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osafune K., Caron L., Borowiak M., Martinez R.J., Fitz-Gerald C.S., Sato Y., Cowan C.A., Chien K.R., Melton D.A. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. doi:10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 58.Chadwick B.P., Willard H.F. A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J. Cell Biol. 2001;152:375–384. doi: 10.1083/jcb.152.2.375. doi:10.1083/jcb.152.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li N., Carrel L. Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc. Natl Acad. Sci. USA. 2008;105:17055–17060. doi: 10.1073/pnas.0807765105. doi:10.1073/pnas.0807765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson M.A., Weick J.P., Pearce R.A., Zhang S.C. Functional neural development from human embryonic stem cells: accelerated synaptic activity via astrocyte coculture. J. Neurosci. 2007;27:3069–3077. doi: 10.1523/JNEUROSCI.4562-06.2007. doi:10.1523/JNEUROSCI.4562-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.