Abstract
The maintenance of genetic stability depends on the fine-tuned initiation and termination of pathways involved in cell cycle checkpoints and DNA repair. Here, we describe a new pathway that regulates checkpoint kinase 1 (CHK1) activity, a key element controlling both checkpoints and DNA repair. We show that the ubiquitin-specific peptidase 1 (USP1) deubiquitinase participates in the maintenance of both total and phosphorylated levels of CHK1 in response to genotoxic stress. We establish that USP1 depletion stimulates the damage-specific DNA-binding protein 1-dependent degradation of phosphorylated CHK1 in both a monoubiquitinylated Fanconi anaemia, complementation group D2 (FANCD2)-dependent and -independent manner. Our data support the existence of a circuit in which CHK1 activates checkpoints, DNA repair and proliferating cell nuclear antigen and FANCD2 monoubiquitinylation. The latter two events, in turn, switch off activated CHK1 by negative feedback inhibition, which contributes to the downregulation of the DNA damage response. This pathway, which is compromised in the cancer-prone disease Fanconi anaemia (FA), likely contributes to the hypersensitivity of cells from FA patients to DNA damage and to the clinical phenotype of the syndrome; it may also represent a pharmacological target to improve patient care and develop new cancer therapies.
INTRODUCTION
Genetic stability in response to DNA damage and stalled replication forks depends on the relatively well-described interconnected action of cell cycle checkpoints and DNA repair mechanisms (1,2). A crucial role in the maintenance of the genomic stability is played by the signalling pathway initiated by replication protein A (RPA)-coated single-strand DNA stretches that are generated during replication fork stalling or via the resection of double-strand breaks that activate S and G2/M checkpoints through the ataxia telangiectasia and RAD3 related (ATR)-mediated phosphorylation of checkpoint kinase 1 (CHK1). This phosphorylation triggers CHK1 activation and downstream CHK1-mediated events that regulate cell cycle arrest and DNA repair (2,3). In addition to its role in cell cycle regulation, CHK1 is functionally or directly involved in proliferating cell nuclear antigen (PCNA) and Fanconi anemia, complementation group D2 (FANCD2) monoubiquitinylation (4–7) and in the full activation of the homologous recombination (HR) protein RAD51 (8). Consequently, it is likely that CHK1 activity requires a fine-tuned regulation to optimize the cellular response to DNA damage. Indeed, studies in yeast reported that shortening or delaying CHK1 activation results in cellular hypersensitivity to DNA damage despite an opposite effect on mitotic entry (9). Recent studies demonstrated that CHK1 is part of both checkpoint initiation and termination. CHK1 phosphorylation may both participate in CHK1 activation and promote its degradation. The turning off of CHK1 activity can be mediated by the proteasomal degradation of its S345 phosphorylated form following cullin 4A (CUL4A)- or cullin 1 (CUL1)-dependent ubiquitinylation (10,11). Alternatively, CHK1 signalling could be restrained by the targeted proteasomal degradation of the adaptor protein CLASPIN. CLASPIN, which is initially required for optimal CHK1 phosphorylation by ATR, is ubiquitinylated by the E3 ligase complex associating the Skp1-Cul1-F-box proteins to the beta-transducin repeat containing protein (SCFβTrcp), a process impeded by the deubiquitinase ubiquitin specific peptodase 7 (USP7) (12). Indeed, CLASPIN degradation contributes to checkpoint termination, preventing ATR-mediated CHK1 phosphorylation. Consequently, full checkpoint termination is the result of the coordinated action of all these distinct pathways. Because CHK1 expression has been associated with anticancer therapy resistance (13–16), a better understanding of the different mechanisms involved in CHK1 signalling may reveal new predictive biomarkers of tumour responsiveness and has major implications for cancer therapy choices.
Rare genetic diseases, such as Fanconi anaemia (FA) syndrome (17), which are caused by inherited recessive mutations in DNA damage response (DDR) genes, represent fundamental models to approach how cells cope with DNA lesions and replicative stress (18). FA patients present with aplastic anaemia, developmental defects and a predisposition to cancer (19). No fewer than 15 FANC proteins are required for cellular and chromosomal resistance to interstrand DNA crosslinks (ICLs) (19–25). At least eight FANC proteins (FANCA/B/C/E/F/G/L/M) are assembled into the FANCcore complex that catalyses the monoubiquitinylation of FANCD2 (mono-Ub-FANCD2) and FANCI (mono-Ub-FANCI) during S phase or in response to DNA damage, promoting their chromatin localization, assembly in subnuclear foci and interaction with the FANCD2/FANCI-associated nuclease 1 protein (24,26–28). The other FANC proteins, FANCJ/BRIP1, FANCD1/BRCA2 and FANCN/PALB2, act in parallel or downstream of FANCD2, completing the so-called FANC pathway. The FANC pathway participates in the regulation of HR (26–28) and is required for ICL resistance by direct or functional interactions with partners, such as ATR, BReast CAncer predisposition gene/protein 1 (BRCA1), bloom syndrome-mutated protein and the meiotic recombination 11 homolog (MRE11)–RAD50–Nibrin (NBN or NBS1) complex (29–37). Recently, RAD51C mutations have been reported in one family of patients with an FA-like phenotype, supporting the link between FA, HR and ICL repair (38). FA cells undergo a longer G2 phase (39) and an abnormal delay in S and G2 transit following DNA damage (40). The accumulation of FA cells in late S/G2 following treatment with ICL-inducing agents is dependent on CHK1 activity and its ATR-dependent phosphorylation (5). The accumulation of FA cells with a 4N DNA content and CHK1 hyperphosphorylation could be the consequence of a normal checkpoint response in cells with DNA damage hypersensitivity (41). Alternatively, the accumulation of FA cells in late S/G2 could also represent a defect in recovering from the ATR/CHK1 checkpoint in response to ICL-inducing agents. To test this hypothesis and gain insight into the mechanisms involved in CHK1 regulation, we analysed the activation of CHK1 in cells expressing a constitutively ‘activated' FANC pathway by knocking down the deubiquitinase ubiquitin-specific peptidase 1 (USP1), which restrains the monoubiquitinylation of FANCD2/FANCI (42,43). Following this approach, we discovered USP1-regulated and damage-specific DNA-binding protein 1 (DDB1)-dependent mechanisms that are involved in the downregulation of activated CHK1 in human cells in both a FANC pathway-dependent and -independent manner.
RESULTS
USP1 depletion impacts the stability and phosphorylation of CHK1 in both a monoubiquitinylated FANCD2-dependent and -independent manner
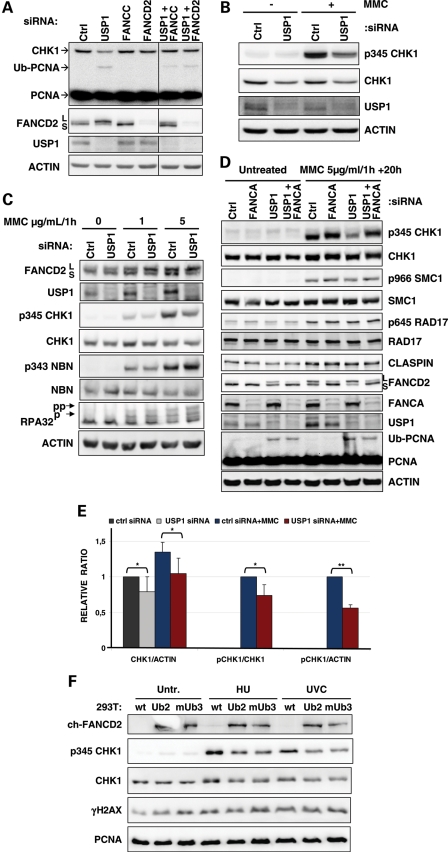
FANC pathway deficiency is associated with DNA damage-induced CHK1 hyperphosphorylation (5), suggesting that FANCD2 monoubiquitinylation could restrain checkpoint signalling. Thus, we hypothesized that a higher constitutional level of mono-Ub-FANCD2 should result in the opposite, i.e. a lower level of phosphorylated CHK1 (p-CHK1). In order to increase FANCD2 monoubiquitinylation and address the involvement of the FANC pathway in CHK1 regulation, we depleted the USP1 deubiquitinase using siRNA in U2OS and HeLa cells. As expected (42,44), USP1 downregulation induced higher levels of both mono-Ub-FANCD2 and mono-Ub-PCNA in the absence of genotoxic stress (Fig. 1A and D). In U2OS cells, USP1 depletion was consistently associated with a clear reduction in the steady-state level of the CHK1 protein (Fig. 1A and B). In HeLa cells, the observed effects of USP1 depletion on the steady-state level of CHK1 were more variable (Fig. 1C and D). Nevertheless, quantitative analysis of seven independent experiments indicated that the CHK1 levels in USP1-deficient HeLa cells were significantly lower than in USP1-proficient HeLa cells (Fig. 1E, P < 0.05). Using U2OS cells transiently transfected with a CHK1–FLAG construct (Supplementary Material, Fig. S1A), we corroborated the observation that the decrease in CHK1 steady-state levels was caused by USP1 downregulation. The CHK1 steady-state decrease was also observed with an unphosphorylatable 2S/A mutant CHK1–FLAG protein (Supplementary Material, Fig. S1A); for this mutant, p-CHK1 is not a prerequisite for its own downregulation. Co-depletion of USP1, together with the FANCcore complex proteins FANCC or FANCD2, rescued the steady-state level of CHK1 (Fig. 1A). Consequently, monoubiquitinylation of FANCD2 could be a major event involved in USP1-dependent regulation of CHK1 steady-state levels.
Figure 1.
USP1 depletion specifically downregulates p-CHK1 and protein levels. (A) Untreated U2OS cells were lysed 72 h after transfection with the indicated siRNAs. The lysates were analysed by western blot using the indicated antibodies. (B) U2OS cells transfected with the indicated siRNA were treated with 2 µg/ml/1h of MMC and lysed 20 h after treatment. The lysates were analysed by western blot using the indicated antibodies. (C) Two days after transfection with the indicated siRNAs, HeLa cells were treated for 1 h with the reported dose of MMC and washed before the addition of pre-warmed complete medium. Cells were lysed 20 h later, and the lysates were analysed by western blot using the indicated antibodies. (D) HeLa cells were treated with 5 µg/ml/1h of MMC 2 days after siRNA transfection, and proteins were extracted 20 h after treatment to examine the activation of several DDR proteins by immunoblotting. (E) Quantification of the ratio of CHK1 to actin, p345CHK1 to CHK1 and p345CHK1 to actin in HeLa cells treated with 5 µg/ml/1h of MMC and lysed 20 h later. Values represent the mean and SD of three (MMC treatment) to seven experiments (untreated). Student's t-test was used: *P < 0.05, **P < 0.01. (F) 293T cells stably expressing a chicken FANCD2-KR-ubiquitin fusion (clone Ub2) or FANCD2-KR-monoubiquitin fusion (clone mUb3) were treated with HU and lysed at the end of the treatment (2 mM for 30 min) or UV (5J) and lysed 1 h after irradiation, and the phosphorylation of CHK1 and H2AX was analysed by immunoblotting.
Subsequently, we exposed the cells to the DNA cross-linking agent mitomycin C (MMC) or to ultra violet C (UVC) to analyse the behaviour of DNA damage-induced p-CHK1 in USP1-depleted cells. USP1 depletion was associated with a consistent and significant reduction in the level of phosphorylated S345-CHK1 in both U2OS and HeLa cells (Fig. 1B–E and Supplementary Material, Fig. S1B). More importantly, the reduced phosphorylation of CHK1 observed in USP1-depleted cells appeared to be highly specific. Indeed, both the DNA damage-induced phosphorylation and the expression of other ATR/ataxia telangiectasia-mutated protein (ATM) targets, including NBN, RPA32, RAD17 and structural maintenance chromosome 1 (SMC1), were not modified by USP1 depletion (Fig. 1C and D). Moreover, depletion of the FANCcore complex component FANCA, which is necessary for FANCD2 monoubiquitinylation, rescued p-CHK1 in USP1-depleted cells exposed to MMC or UVC (Fig. 1D and Supplementary Material, Fig. S1B). Thus, FANC pathway proficiency and FANCD2 monoubiquitinylation may be required for the observed effects of USP1 depletion on p-CHK1.
To directly observe an active role for monoubiquitinylated FANCD2 in CHK1 downregulation, we stably expressed a constitutively monoubiquitinylated chicken form of FANCD2 (45) in HEK293T cells (Fig. 1F). Ectopic expression of mono-Ub-FANCD2 slightly increased p-CHK1 and phosphorylated histone 2 AX (γH2AX) levels, revealing that cells that constitutively express monoubiquitinylated FANCD2 undergo DNA stress and activate a DDR response. Nevertheless, in response to genotoxic stress, cell lines overexpressing monoubiquitinylated FANCD2 showed lower levels of both phosphorylated and unphosphorylated CHK1. In contrast, the increase in the γH2AX level was similar in all cell lines. These observations support the notion that FANCD2 monoubiquitinylation specifically participates in the downregulation of CHK1 expression/activity.
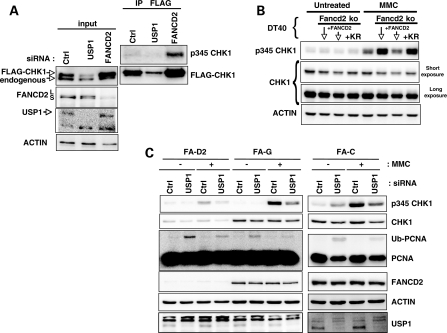
To support the finding that mono-Ub-FANCD2 is a key element controlling CHK1 level/phosphorylation, we analysed the behaviour of a CHK1–FLAG construct stably expressed in MRC5 fibroblasts following deprivation of either USP1 or FANCD2 (Fig. 2A). Again, USP1 depletion was associated with increased FANCD2 monoubiquitinylation and downregulation of CHK1. Notably, in contrast to USP1 depletion, FANCD2 downregulation induced a higher level of CHK1–FLAG, and a fraction of this protein appeared to be phosphorylated (Fig. 2A). Moreover, we analysed CHK1 behaviour following MMC exposure in the chicken DT40 parental cell line, FANCD2 knock-out (KO) DT40 cells, FANCD2 KO DT40 cells ectopically corrected with a wild-type FANCD2 and FANCD2 KO DT40 cells expressing the FANCD2 non-monoubiquitinylable lys (K) 563 arg (R) (KR) form (Fig. 2B). FANCD2-deficient DT40 cells showed a clear hyperphosphorylation of CHK1, which was rescued by the expression of the wild-type FANCD2 gene but not by the expression of the FANCD2-KR form. Notably, in ‘corrected' DT40 cells, the level of CHK1 was also clearly reduced when compared with that of the KO counterpart. Altogether, these observations validate the negative role of the activated FANC pathway in both CHK1 stabilization and phosphorylation.
Figure 2.
USP1 depletion and FANCD2 monoubiquitinylation have implications in CHK1 downregulation. (A) MRC5 fibroblasts stably expressing CHK1–FLAG were transfected with the indicated siRNAs and lysed 72 h later. CHK1–FLAG was then immunoprecipitated, and its phosphorylation was analysed by western blot. (B) p-CHK1 and levels as observed in untreated and MMC-exposed DT40 cells. Fancd2ko indicates the cells carrying knockout Fancd2 alleles; +FANCD2 indicates previous cells (Fancd2ko) transfected with a wt Fancd2 (i.e. Fancd2 corrected); and +KR indicates Fancd2ko cells expressing a Fancd2 KR mutant form. Proteins were isolated 6 h after treatment with 100 ng/ml of MMC and analysed by western blot with the indicated antibodies. (C) FA-D2, FA-G and FA-C human fibroblasts were tranfected with USP1-siRNA. MMC was added 48 h later at 2 µg/ml for 1 h and cells were lysed 24 h later. The lysates were analysed by western blot with the indicated antibodies.
Finally, to ascertain if elements other than mono-Ub-FANCD2 participate in USP1-mediated CHK1 downregulation, we depleted USP1 in FA-C, FA-G and FA-D2 fibroblasts (Fig. 2C). Depletion of USP1 in cells derived from FA patients resulted in decreased CHK1 levels and phosphorylation, suggesting that mono-Ub-FANCD2 is not the only USP1 target involved in CHK1 regulation and that the germline-derived inactivation of the FANC pathway in FA patient-derived cells uncovered or selected other pathways to compensate for their intrinsic deficiency and assure cell survival.
In conclusion, using several approaches and cellular models, USP1 depletion specifically affects the steady-state levels and the phosphorylation of CHK1 without perturbing ATR signalling following both FANC pathway-dependent and -independent mechanisms.
USP1 depletion impacts CHK1 independently of both the cell cycle and DNA repair
Because CHK1 is a cell cycle regulated and DNA damage checkpoint protein, we asked whether (a) the downregulation of CHK1 in USP1-depleted cells is an experimental artefact resulting from modifications of the cell cycle and/or altered DNA repair capabilities caused by USP1 depletion and (b) USP1 depletion modifies the G2 checkpoint in response to DNA damage.
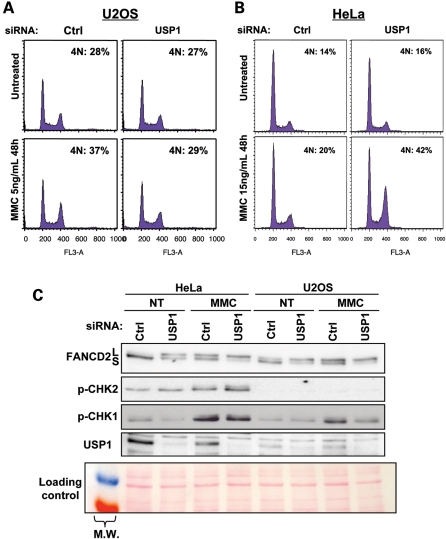
We first examined the cell cycle distribution in USP1-depleted cells. In accordance with published data on Usp1−/− mouse embryonic fibroblasts (MEFs) (46), USP1 depletion had no major effects on the cell cycle progression in unchallenged cells (Fig. 3A and B). Consequently, the lower level of CHK1 in USP1-depleted cells is not merely a consequence of cell accumulation in G1.
Figure 3.
USP1 depletion differentially affects the accumulation in late S/G2 of U2OS and HeLa cells exposed to MMC. Forty-eight hours post-transfection with USP1 siRNA, U2OS (A) and HeLa (B) cells were treated with 15 ng/ml or 5 ng/ml of MMC, respectively, for 48 h before fixation and analysis by flow cytometry. (C) HeLa and U2OS cells were transfected with USP1 siRNAs, treated with 1 µg/ml/1h of MMC and lysed 20 h later. The lysates were analysed by western blot using the indicated antibodies. Red Ponceau staining of the membrane was used as a loading control.
Then we monitored cell cycle recovery in response to MMC in both U2OS and HeLa cells. As reported in Figure 3A, USP1-depleted U2OS cells re-enter the cell cycle more rapidly than USP1-proficient cells, suggesting that USP1 depletion could accelerate recovery from the G2 checkpoint. In contrast, as predicted from studies with Usp1−/− MEFs (46), USP1-depleted HeLa cells showed an increased MMC-induced G2/M content compared with USP1-proficient cells (Fig. 3B). Because U2OS and HeLa cells differ in their p53 status, we repeated experiments in the presence of the p53 inhibitor pifithrin-α, and we observed no difference in the response (data not shown). Consequently, the differences between U2OS and HeLa cells seem to be independent of their p53 status. Western blot analysis of the two downstream cellular checkpoint kinases CHK1 and CHK2 revealed major differences between the HeLa and U2OS cells (Fig. 3C). HeLa cells showed a higher level of phosphorylated CHK1 and CHK2; this result could support the prolonged G2/M checkpoint when CHK1 signalling decreases as a result of USP1 depletion. The observed differences in CHK2 phosphorylation between HeLa and U2OS cells appeared to be linked to higher basal levels of CHK2, which appeared slightly phosphorylated as a consequence of USP1 depletion (Supplementary Material, Fig. S1C). Moreover, data from Usp1−/− MEFs support our observations that p-CHK2 increases significantly in USP1-depleted HeLa cells following MMC (46). Notably, although the authors did not stress this point in their article, the p-CHK1 level is lowered in response to MMC in Usp1−/− MEFs, as we report here. Thus, CHK1 downregulation in USP1-depleted cells is not a consequence of cell cycle modifications and cannot be generally implicated in the exit or recovery from the S or G2/M checkpoints.
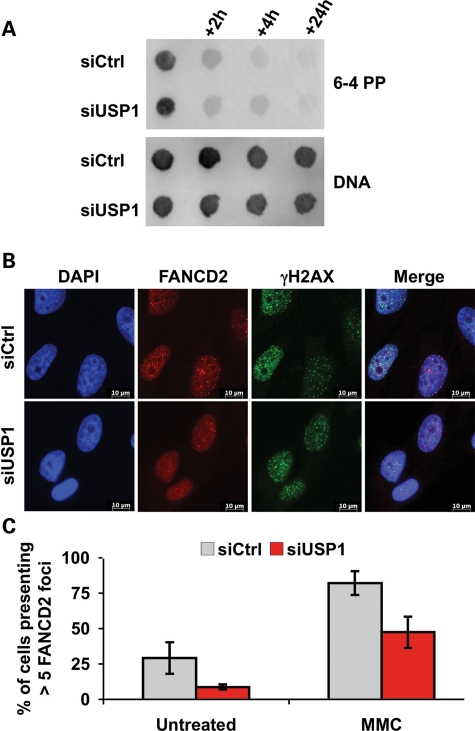
Next, we asked whether CHK1 dephosphorylation and/or degradation in USP1-depleted cells are the consequences of increased DNA repair capabilities. To determine whether USP1 depletion modifies the intrinsic DNA repair capabilities of DNA-damaged cells, we analysed by dot blot the repair kinetics of 6–4 pyrimidine–pyrimidone (6-4 PP) lesions induced by UVC exposure. As presented in Figure 4A, 6-4 PP were eliminated with equal kinetics in both USP1-proficient and -deficient cells, meaning that the DNA repair pathways involved in the elimination of this lesion work normally in the absence of USP1.
Figure 4.
USP1 depletion does not affect the repair of UVC-induced DNA lesions and does not improve FANCD2 foci formation. (A) HeLa cells transfected with Ctrl or USP1 siRNAs were treated with UVC (10 J/m2), and genomic DNA was prepared at the indicated times. Dot blots were performed with antibodies against 6-4 PP. DNA was then stained with an antibody recognizing single-stranded DNA. (B) Immunofluorescence analysis of the γH2AX (green) and FANCD2 (red) foci in Ctrl and USP1-siRNA-transfected cells treated with MMC (200 ng/ml for 24 h). Nuclei were stained with DAPI. Right images represent the merge of the three colours. (C) Quantification of cells with more than five FANCD2 nuclear foci in control- and USP1-siRNA-transfected cells left untreated and/or exposed to MMC (200 ng/ml) and fixed 24 h post-treatment. Data represent the mean of three independent experiments, and at least 100 cells were scored for each experiment.
FANCD2 and PCNA monoubiquitinylation, as well as FANCD2 redistribution in sub-nuclear foci, is considered mandatory for cellular resistance to stalled replication forks. Consequently, the reduction in p-CHK1 and/or stability following DNA damage could be the normal behaviour of cells in which USP1 depletion results in enhanced FANCD2 or PCNA activity as a consequence of higher levels of their monoubiquitinylated forms (42). However, more recent data in avian DT40 cells (47) and mice (46) demonstrated a cellular hypersensitivity to MMC in the absence of USP1. In accordance with these results, we observed a defect in FANCD2 foci formation in USP1-depleted human cells (Fig. 4B and C). Importantly, defective FANCD2 relocalization was not the consequence of an impaired formation of γH2AX foci, which are required for FANCD2 assembling in subnuclear foci (48) (Fig. 4B). Hence, as predicted by data obtained in chicken and mice (46,47), this result suggests that FANCD2 undergoes a finely regulated ubiquitinylation/deubiquitinylation cycle to perform its physiological role and be recruited to and accumulate in nuclear foci. Consequently, this challenges the idea that enhanced DNA repair activity by mono-Ub-FANCD2 is responsible for the weaker CHK1 activation observed in USP1-depleted cells.
The most obvious conclusion of our data (Figs 1 and 4) is that CHK1 dephosphorylation and/or degradation in USP1-depleted cells is not caused by increased DNA repair capabilities in USP1-deficient cells but is directly linked to USP1 downregulation.
USP1 limits DDB1-dependent degradation of CHK1
Multiple pathways and proteins are involved in CHK1 regulation (49). Among these players, the adaptor protein CLASPIN appears to be a master regulator of CHK1 activity (50). We observed that the siRNA-mediated depletion of CLASPIN strongly reduced UVC-induced p-CHK1 but not CHK1 protein levels (Fig. 5A). Compared with CLASPIN depletion, USP1 depletion affected both the CHK1 steady-state level and phosphorylation without discernible effects on CLASPIN phosphorylation or stabilization (Figs 1D and 5A). Interestingly, co-depletion of USP1 and CLASPIN had an additive effect on both UVC-induced p-CHK1 and CHK1 protein levels. Altogether, these results suggest that CLASPIN is not an intermediary player downstream of USP1 in the regulation of CHK1. On the contrary, our data suggest that CLASPIN-dependent and USP1-regulated events work on convergent pathways to turn on and restrain CHK1 activation.
Figure 5.
Phosphorylated CHK1 is degraded in a DDB1-dependent manner in the absence of USP1. (A) U2OS cells were transfected with the indicated siRNAs, treated 72 h later with UVC (10 J/m2) and lysed 4 h later. Proteins were analysed by western blot with the indicated antibodies. (*) indicates a nonspecific band recognized by the anti-p345 CHK1 antibody. CLS and U1 indicate CLASPIN and USP1, respectively. (B) HeLa cells transfected with the indicated siRNAs were treated with MMC (2 µg/ml/1h) or UVC (10 J/m2) and lysed 20 h and 4 h later, respectively. The lysates were analysed by western blot with the indicated antibodies. U1 and D1 indicate USP1 and DDB1, respectively. (C) U2OS cells transfected with Ctrl or USP1 siRNAs were treated with DMSO or the proteasome inhibitor MG132 (10 μm) for 6 h. The cellular extracts were analysed by western blot using the indicated antibodies. (D) Endogenous CHK1 was immunoprecipitated from HEK293T cells that were untreated or treated with HU (5 mm; 4 h), and its association with DDB1 was analysed by western blot. (E) The lysates from HeLa cells transfected with the indicated siRNAs were analysed by immunoblotting to monitor CDT1 levels. DDB1 inhibition stabilized CDT1 while USP1 depletion had no effect. (F) HeLa cells transfected with Ctrl or USP1 siRNAs were treated with MMC (2 µg/ml/1 h) and lysed 20 h later. While USP1 depletion significantly reduced p-CHK1 and protein levels, it did not enhance the degradation of CDT1. (G) A model illustrating the major molecular mechanisms involved in CHK1 regulation and USP1 participation in this phenomenon. The FANCcore complex was voluntarily excluded from the schema to simplify the figure.
Genotoxic stresses may promote the degradation of phosphorylated CHK1 in a Cul1- and/or Cul4A-dependent manner (51). During replication, a fraction of CHK1 is indeed polyubiquitinylated by a CUL4A/DDB1-containing E3 ligase and is subsequently degraded by the proteasome (10). Hence, we directly tested the role of DDB1 in CHK1 stability by silencing it alone or in addition to USP1. Curiously, for unknown reasons, DDB1 depletion reduced USP1 steady-state levels (Fig. 5B). Nevertheless, the reduced level of USP1 in DDB1-depleted cells appeared sufficient to deubiquitinylate FANCD2 and maintain a normal ratio of FANCD2-L to FANCD2-S. Co-depletion of DDB1 with USP1 nearly abrogated the reduced stability and phosphorylation of CHK1 observed following the simple depletion of USP1 in MMC- and UVC-treated cells (Fig. 5B). Moreover, FANCD2 monoubiquitinylation in USP1-depleted cells was not modified by the depletion of DDB1, suggesting that downregulation of CHK1 in USP1-depleted cells requires DDB1 downstream of FANCD2. In agreement with the hypothesis that phosphorylated CHK1 is ubiquitinylated and subsequently degraded in the absence of USP1, treatment with the proteasome inhibitor MG-132 could increase the level of CHK1 in USP1-depleted cells but not in control cells (Fig. 5C). However, because MG-132 could strongly reduce mono-Ub-FANCD2 [(52) and data not shown], the restoration of CHK1 levels could be the consequence of its blocked degradation, the loss of FANCD2 monoubiquitinylation or both. Nevertheless, this result demonstrates that CHK1 downregulation is the consequence of a rapidly reversible mechanism, likely proteasomal degradation. Importantly, the already described interaction between endogenous CHK1 and DDB1 (10) was enhanced in USP1-depleted HEK293T cells (Fig. 5D), which was consistent with the idea that CHK1 degradation is linked to DDB1. Similar results were obtained with exogenous CHK1–FLAG (data not shown). It is important to note that USP1 depletion neither reduced the steady-state level of another known DDB1 target, chromatin licensing and DNA replication factor 1 (CDT1) (53), nor enhanced its DNA damage-induced degradation, suggesting that the DDB1/CUL4A machinery is not simply overactive in USP1-depleted cells (Fig. 5E and F). Altogether, our results suggest that USP1 depletion specifically promotes the degradation of phosphorylated CHK1 in a DDB1-dependent manner.
DISCUSSION
We present evidence that the USP1 deubiquitinase controls a feedback loop that limits CHK1 activity to rescue DNA-damaged cells. A fully proficient DDR cell activates a response to overcome the difficulties imposed by spontaneously arising and genotoxin-induced delayed or stalled DNA replication forks. CHK1 participates in this response by both delaying cell cycle progression and participating in DNA repair. CHK1 is indeed required for the monoubiquitinylation of PCNA and FANCD2, which is involved in the resolution of replicative stress. ATR-mediated phosphorylation of CHK1 may start a sequence of events that simultaneously initiates and limits checkpoints through the targeted degradation of phosphorylated CHK1 (11,51). Similarly, the full activation of both FANCD2 and PCNA, in addition to the resolution of DNA replication difficulties, could also participate in cell recovery from cell cycle checkpoints. We propose that the stimulation of CHK1 degradation by mono-Ub-FANCD2 represents a feedback mechanism that contributes to the recovery of DNA-damaged cells (Fig. 5G). This feedback could participate in the extinction of FANCD2 and RAD51, which is under the control of p-CHK1 (4–6,8), limiting HR events that could be toxic for the cells. In this regard, the compromised HR efficiency of USP1-depleted cells could be the consequence of FANCD2 dysfunction (46), weaker CHK1 activity (8) or both. Our data suggest that DDB1-dependent degradation of phosphorylated CHK1 could be one of the mechanisms explaining these findings.
The regulation of the DDB1/CUL4A-based E3 ligases is a complex process that requires the presence of DDB1 and CUL4-associated factors (DCAFs) for the specific targeting of various substrates of this ubiquitin ligase (54). USP1 depletion specifically affects CHK1. Consequently, it is tempting to speculate that mono-Ub-FANCD2 promotes the association of the yet unidentified CHK1-specific DCAF with DDB1 and/or CHK1, thereby enhancing CHK1 degradation and inactivation.
It is difficult to determine whether CHK1 regulation depends on FANCD2, FANCI or monoubiquitinylated FANCD2/FANCI dimer, and we cannot exclude a role for PCNA or other presently unknown targets of USP1 in the CHK1 control. A PCNA/FANCD2 interaction has been recently identified, and this interaction may be required for proficient FANCD2 monoubiquitinylation (55,56). Thus, PCNA could participate in the control of p-CHK1 via its action on FANCD2. However, without excluding this possibility, the observation that USP1 depletion in FA cells has an effect on phosphorylation/stability of CHK1 indicates that mono-Ub-PCNA could influence CHK1 levels independently from its role in FANCD2 monoubiquitinylation.
We propose that the accumulation of 4N DNA content following ICL-inducing agents in FA cells (57) is a consequence of both defective DNA repair and a failure in recovering from checkpoint arrest through the inhibition of CHK1. Our data predict that FA cells, which cannot monoubiquitinylate FANCD2, display overactive CHK1. Indeed, Gautier and colleagues (58) observed more p-CHK1 at the basal level in FANCA-deficient cells compared with FANC-proficient cells, a phenotype that is expected to result from compromised DDB1-dependent degradation of phosphorylated CHK1. In agreement with this, transient depletion of FANCD2 strongly increased p-CHK1 in untreated cells (Fig. 1E). This CHK1 overactivation may be detrimental for FA patients; recently, an ‘FA-M' patient was identified who did not display classical symptoms of FA and presented with bi-allelic mutations in both FANCA and FANCM (59). The brother of this patient carried only bi-allelic mutations in FANCA and developed ‘typical' FA. It is tempting to speculate that the mild phenotype of the ‘FA-M/FA-A' patient is due to the compensatory effect of the mutations in FANCM. Because FANCM is required for efficient ATR/CHK1 activation independently of the FANC complex (60), we propose that an overactivation of CHK1, triggered by FANCA mutations, is compensated for by FANCM deficiency, leading to normal CHK1 activity and a mild FA phenotype. The same hypothesis could be evoked for all FA patients presenting with a mild clinical phenotype or who develop bone marrow failure later in life, as proposed in a recently published study (61). Thus, bi-allelic FANC mutations could be associated with a partial impairment of CHK1 activation or activity resulting in a better outcome for these patients.
MATERIALS AND METHODS
Cell culture and drugs
HeLa cells, U2OS, HEK293T, FA cells (PD20, PD331 and PD352) and wild-type SV40-immortalised MRC5 fibroblasts were routinely grown in DMEM (Gibco) supplemented with 10% fetal calf serum (Dutcher), 2 mm glutamine, 1 mm sodium pyruvate, 100 U/ml penicillin and 100 µg/ml streptomycin (all from Gibco) at 37°C in 5% CO2. MRC5 stably expressing CHK1–FLAG cells were selected and maintained in G418-containing medium (400 µg/ml). HEK293T cells were transfected with the chicken FANCD2 constructs previously described (45), and clones were selected using Zeocin. DT40 cells were grown in the same medium described above but supplemented with 1% chicken serum and 50 µm β2-mercaptoethanol (45). Genotoxic treatments were performed as previously described (5).
Transfections
Single siRNAs were purchased from Eurogentec. The following sequences were used: Ctrl (CGUCGACGGAAUACUUCGATT), FANCA (GGGUCAAGAGGGAAAAAUATT), and USP1a (CGACAGCUAUGGAUUAUUUTT) and USP1b (GAGAAGGACUUUCUGAAUUTT) used as a mix. Luciferase (used as a control in a few experiments), FANCD2 and CLASPIN siRNAs were previously described (5). DDB1 and FANCC smartpool siRNAs were obtained from Dharmacon. Transfection of siRNA (usually at 100 nm) was performed with Oligofectamine (Invitrogen), and experiments were carried out 48–72 h later. In the case of double transfection, the same total amount of siRNA was used by adding control siRNA to the single siRNA of interest. The CHK1 construct was transfected in U2OS cells with Lipofectamine LTX (Invitrogen) according to the manufacturer's instructions. In a few experiments, siRNA transfections were performed with Interferin (Polyplus Transfection) at a concentration of 20 nm.
Cell extracts and antibodies
Cells were washed in ice-cold PBS and disrupted in lysis buffer [50 mm Tris pH 7.9, 150 mm NaCl, 1 mm EDTA and 0.5% NP-40, supplemented with protease and phosphatase inhibitors (Roche)]. After 20 min of incubation on ice, lysates were combined with 4X Laemmli buffer containing β2-mercaptoethanol, sonicated, denaturated by boiling and separated by SDS–PAGE. The antibodies used were directed against FANCD2, RAD17, CHK1, actin, NBS1 and PCNA (Santa Cruz Biotechnology); FANCD2, RPA32, SMC1, p966SMC1 and vinculin (Abcam); p645RAD17, p345CHK1 and p343NBS1 (Cell Signalling); p68CHK2 and RPA32 (Calbiochem); CLASPIN, FANCI, DDB1 and CDT1 (Bethyl); and γH2AX (Upstate). Antibodies that had been produced and kindly provided by the Fanconi Anemia Research Fund were also used against USP1 (D3598) and FANCA (D5300 and D6512). Signals were detected by the Gene-Gnome apparatus, and their intensities were quantified using the Genetools (Syngene Bio Imaging) software or Image J.
Immunofluorescence
Cells grown on glass cover slips were fixed in 4% paraformaldehyde for 15 min before permeabilization in 0.5% Triton for 5 min. After blocking in 5% BSA, cells were stained with antibodies against FANCD2 (Abcam ab2187; 1/1000) and γH2AX (Upstate; 1/2000) for 1 h in IF buffer (0.5% Tween 20, 12% BSA, 0.036% NaN3 in PBS). Primary antibody detection was achieved by incubation with donkey anti-rabbit Alexafluor 594 and donkey anti-mouse Alexafluor 488 secondary antibodies (Molecular Probes; 1/1000) for 1 h at room temperature. Slides were mounted in Vectashield containing DAPI (Vector Laboratories) and examined at a magnification of ×100 using a fluorescence microscope (Olympus). Only nuclei showing more than five discrete FANCD2 foci were considered as FANCD2 foci-positive.
Co-immunoprecipitation
HEK293T cells were seeded in 100 mm Petri dishes and transfected with the indicated siRNAs. Three days later, cells were treated for 4 h with HU (5 mm) and were disrupted in lysis buffer. Lysates were clarified with protein G/agarose and normal mouse IgG (Santa Cruz Biotechnology). Immunoprecipitation was performed on 1–2 mg of cell lysates using 2 µg of monoclonal anti-CHK1 (G4) antibody captured with protein G/agarose. Immunoprecipitates were washed four times in lysis buffer before eluting the beads in Laemmli buffer. To avoid cross-reactions with the IgG heavy chains, a secondary antibody that does not recognize denaturated IgG (TrueBlot™; eBioscience) was used to detect CHK1. Anti-FLAG beads (Sigma) were also used to immunoprecipitate exogenous FLAG-tagged CHK1.
Repair of UV-induced DNA lesions
Genomic DNA was extracted using the NucleoSpin Tissue kit (Macherey-Nagel) before denaturation for 5 min at 95°C. Dot blotting on Nylon membranes was performed with 200 ng of DNA for detection of 6-4 PP. Membranes were incubated in 0.4N NaOH for 20 min at room temperature before blocking in 5% non-fat dried milk in PBS-T. An antibody recognizing 6-4 PP was used at 1/2000 (Cosmo Bio), and anti-single-stranded DNA was used as a loading control (Chemicon).
SUPPLEMENTARY MATERIAL
FUNDING
J.H.G. was supported by fellowships from the Ministère de l'Enseignement supérieur et de la Recherche and the Association pour la Recherche sur le Cancer (ARC). This work was supported by grants from the Ligue Nationale contre le Cancer (Equipe Labellisée 2006 and 2009), Electricité de France (EDF, 2009) and Agence Nationale pour la Recherche (ANR, Genopat 2008) to F.R. Funding to pay the Open Access publication charges for this article was provided by Ligue Nationale contre le Cancer.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all members of F. Rosselli's lab. We are grateful to K.K. Khanna for providing the CHK1 construct and V. Pennaneach for the U2OS cells. We are indebted to P. Kannouche and her lab members for helpful discussions, reagents and technical help.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Harper J.W., Elledge S.J. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. doi:10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. doi:10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cimprich K.A., Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. doi:10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collis S.J., Barber L.J., Clark A.J., Martin J.S., Ward J.D., Boulton S.J. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat. Cell Biol. 2007;9:391–401. doi: 10.1038/ncb1555. doi:10.1038/ncb1555. [DOI] [PubMed] [Google Scholar]
- 5.Guervilly J.H., Mace-Aime G., Rosselli F. Loss of CHK1 function impedes DNA damage-induced FANCD2 monoubiquitination but normalizes the abnormal G2 arrest in Fanconi anemia. Hum. Mol. Genet. 2008;17:679–689. doi: 10.1093/hmg/ddm340. doi:10.1093/hmg/ddm340. [DOI] [PubMed] [Google Scholar]
- 6.Wang X., Kennedy R.D., Ray K., Stuckert P., Ellenberger T., D'Andrea A.D. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol. Cell Biol. 2007;27:3098–3108. doi: 10.1128/MCB.02357-06. doi:10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X.H., Shiotani B., Classon M., Zou L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008;22:1147–1152. doi: 10.1101/gad.1632808. doi:10.1101/gad.1632808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorensen C.S., Hansen L.T., Dziegielewski J., Syljuasen R.G., Lundin C., Bartek J., Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. doi:10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 9.den Elzen N.R., O'Connell M.J. Recovery from DNA damage checkpoint arrest by PP1-mediated inhibition of Chk1. EMBO J. 2004;23:908–918. doi: 10.1038/sj.emboj.7600105. doi:10.1038/sj.emboj.7600105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung-Pineda V., Huh J., Piwnica-Worms H. DDB1 targets Chk1 to the Cul4 E3 ligase complex in normal cycling cells and in cells experiencing replication stress. Cancer Res. 2009;69:2630–2637. doi: 10.1158/0008-5472.CAN-08-3382. doi:10.1158/0008-5472.CAN-08-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y.W., Brognard J., Coughlin C., You Z., Dolled-Filhart M., Aslanian A., Manning G., Abraham R.T., Hunter T. The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol. Cell. 2009;35:442–453. doi: 10.1016/j.molcel.2009.06.030. doi:10.1016/j.molcel.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faustrup H., Bekker-Jensen S., Bartek J., Lukas J., Mailand N. USP7 counteracts SCFbetaTrCP- but not APCCdh1-mediated proteolysis of Claspin. J. Cell Biol. 2009;184:13–19. doi: 10.1083/jcb.200807137. doi:10.1083/jcb.200807137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. doi:10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 14.Cavelier C., Didier C., Prade N., Mansat-De Mas V., Manenti S., Recher C., Demur C., Ducommun B. Constitutive activation of the DNA damage signaling pathway in acute myeloid leukemia with complex karyotype: potential importance for checkpoint targeting therapy. Cancer Res. 2009;69:8652–8661. doi: 10.1158/0008-5472.CAN-09-0939. doi:10.1158/0008-5472.CAN-09-0939. [DOI] [PubMed] [Google Scholar]
- 15.Didier C., Cavelier C., Quaranta M., Galcera M.O., Demur C., Laurent G., Manenti S., Ducommun B. G2/M checkpoint stringency is a key parameter in the sensitivity of AML cells to genotoxic stress. Oncogene. 2008;27:3811–3820. doi: 10.1038/sj.onc.1211041. doi:10.1038/sj.onc.1211041. [DOI] [PubMed] [Google Scholar]
- 16.Hu B., Zhou X.Y., Wang X., Zeng Z.C., Iliakis G., Wang Y. The radioresistance to killing of A1–5 cells derives from activation of the Chk1 pathway. J. Biol. Chem. 2001;276:17693–17698. doi: 10.1074/jbc.M009340200. doi:10.1074/jbc.M009340200. [DOI] [PubMed] [Google Scholar]
- 17.Lobitz S., Velleuer E. Guido Fanconi (1892–1979): a jack of all trades. Nat. Rev. Cancer. 2006;6:893–898. doi: 10.1038/nrc2009. doi:10.1038/nrc2009. [DOI] [PubMed] [Google Scholar]
- 18.Kerzendorfer C., O'Driscoll M. Human DNA damage response and repair deficiency syndromes: linking genomic instability and cell cycle checkpoint proficiency. DNA Repair (Amst.) 2009;8:1139–1152. doi: 10.1016/j.dnarep.2009.04.018. doi:10.1016/j.dnarep.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. doi:10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 20.Crossan G.P., van der Weyden L., Rosado I.V., Langevin F., Gaillard P.H., McIntyre R.E., Gallagher F., Kettunen M.I., Lewis D.Y., Brindle K., et al. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat. Genet. 2011;43:147–152. doi: 10.1038/ng.752. doi:10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kee Y., D'Andrea A.D. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. doi:10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y., Lach F.P., Desetty R., Hanenberg H., Auerbach A.D., Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat. Genet. 2011;43:142–146. doi: 10.1038/ng.750. doi:10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy-Lahad E. Fanconi anemia and breast cancer susceptibility meet again. Nat. Genet. 2010;42:368–369. doi: 10.1038/ng0510-368. doi:10.1038/ng0510-368. [DOI] [PubMed] [Google Scholar]
- 24.O'Donnell L., Durocher D. DNA repair has a new FAN1 club. Mol. Cell. 2010;39:167–169. doi: 10.1016/j.molcel.2010.07.010. doi:10.1016/j.molcel.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Stoepker C., Hain K., Schuster B., Hilhorst-Hofstee Y., Rooimans M.A., Steltenpool J., Oostra A.B., Eirich K., Korthof E.T., Nieuwint A.W., et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat. Genet. 2011;43:138–141. doi: 10.1038/ng.751. doi:10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 26.Adamo A., Collis S.J., Adelman C.A., Silva N., Horejsi Z., Ward J.D., Martinez-Perez E., Boulton S.J., La Volpe A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol. Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. doi:10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 27.Dray E., Etchin J., Wiese C., Saro D., Williams G.J., Hammel M., Yu X., Galkin V.E., Liu D., Tsai M.S., et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 2010;17:1255–1259. doi: 10.1038/nsmb.1916. doi:10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pace P., Mosedale G., Hodskinson M.R., Rosado I.V., Sivasubramaniam M., Patel K.J. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. doi:10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 29.Naim V., Rosselli F. The FANC pathway and mitosis: a replication legacy. Cell Cycle. 2009;8:2907–2911. doi: 10.4161/cc.8.18.9538. doi:10.4161/cc.8.18.9538. [DOI] [PubMed] [Google Scholar]
- 30.Pichierri P., Averbeck D., Rosselli F. DNA cross-link-dependent RAD50/MRE11/NBS1 subnuclear assembly requires the Fanconi anemia C protein. Hum. Mol. Genet. 2002;11:2531–2546. doi: 10.1093/hmg/11.21.2531. doi:10.1093/hmg/11.21.2531. [DOI] [PubMed] [Google Scholar]
- 31.Pichierri P., Rosselli F. Fanconi anemia proteins and the s phase checkpoint. Cell Cycle. 2004;3:698–700. [PubMed] [Google Scholar]
- 32.Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M.S., Timmers C., Hejna J., Grompe M., D'Andrea A.D. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. doi:10.1016/S1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 33.Hirano S., Yamamoto K., Ishiai M., Yamazoe M., Seki M., Matsushita N., Ohzeki M., Yamashita Y.M., Arakawa H., Buerstedde J.M., et al. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 2005;24:418–427. doi: 10.1038/sj.emboj.7600534. doi:10.1038/sj.emboj.7600534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naim V., Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Biol. 2009;11:761–768. doi: 10.1038/ncb1883. doi:10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- 35.Nakanishi K., Yang Y.G., Pierce A.J., Taniguchi T., Digweed M., D'Andrea A.D., Wang Z.Q., Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl Acad. Sci. USA. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. doi:10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pichierri P., Franchitto A., Rosselli F. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J. 2004;23:3154–3163. doi: 10.1038/sj.emboj.7600277. doi:10.1038/sj.emboj.7600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andreassen P.R., D'Andrea A.D., Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. doi:10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaz F., Hanenberg H., Schuster B., Barker K., Wiek C., Erven V., Neveling K., Endt D., Kesterton I., Autore F., et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 2010;42:406–409. doi: 10.1038/ng.570. doi:10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 39.Dutrillaux B., Aurias A., Dutrillaux A.M., Buriot D., Prieur M. The cell cycle of lymphocytes in Fanconi anemia. Hum. Genet. 1982;62:327–332. doi: 10.1007/BF00304549. doi:10.1007/BF00304549. [DOI] [PubMed] [Google Scholar]
- 40.Kaiser T.N., Lojewski A., Dougherty C., Juergens L., Sahar E., Latt S.A. Flow cytometric characterization of the response of Fanconi's anemia cells to mitomycin C treatment. Cytometry. 1982;2:291–297. doi: 10.1002/cyto.990020505. doi:10.1002/cyto.990020505. [DOI] [PubMed] [Google Scholar]
- 41.Heinrich M.C., Hoatlin M.E., Zigler A.J., Silvey K.V., Bakke A.C., Keeble W.W., Zhi Y., Reifsteck C.A., Grompe M., Brown M.G., et al. DNA cross-linker-induced G2/M arrest in group C Fanconi anemia lymphoblasts reflects normal checkpoint function. Blood. 1998;91:275–287. [PubMed] [Google Scholar]
- 42.Nijman S.M., Huang T.T., Dirac A.M., Brummelkamp T.R., Kerkhoven R.M., D'Andrea A.D., Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. doi:10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R., 3rd, Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D'Andrea A.D., et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. doi:10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang T.T., Nijman S.M., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W., Gygi S.P., Ploegh H.L., Bernards R., D'Andrea A.D. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 45.Matsushita N., Kitao H., Ishiai M., Nagashima N., Hirano S., Okawa K., Ohta T., Yu D.S., McHugh P.J., Hickson I.D., et al. A FancD2-monoubiquitin fusion reveals hidden functions of Fanconi anemia core complex in DNA repair. Mol. Cell. 2005;19:841–847. doi: 10.1016/j.molcel.2005.08.018. doi:10.1016/j.molcel.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 46.Kim J.M., Parmar K., Huang M., Weinstock D.M., Ruit C.A., Kutok J.L., D'Andrea A.D. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev. Cell. 2009;16:314–320. doi: 10.1016/j.devcel.2009.01.001. doi:10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oestergaard V.H., Langevin F., Kuiken H.J., Pace P., Niedzwiedz W., Simpson L.J., Ohzeki M., Takata M., Sale J.E., Patel K.J. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol. Cell. 2007;28:798–809. doi: 10.1016/j.molcel.2007.09.020. doi:10.1016/j.molcel.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bogliolo M., Lyakhovich A., Callen E., Castella M., Cappelli E., Ramirez M.J., Creus A., Marcos R., Kalb R., Neveling K., et al. Histone H2AX and Fanconi anemia FANCD2 function in the same pathway to maintain chromosome stability. EMBO J. 2007;26:1340–1351. doi: 10.1038/sj.emboj.7601574. doi:10.1038/sj.emboj.7601574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iliakis G. New partners for Chk1. Cell Cycle. 2010;9:2059–2062. doi:10.4161/cc.9.11.11858. [PubMed] [Google Scholar]
- 50.Liu S., Bekker-Jensen S., Mailand N., Lukas C., Bartek J., Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol. Cell Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. doi:10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y.W., Otterness D.M., Chiang G.G., Xie W., Liu Y.C., Mercurio F., Abraham R.T. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol. Cell. 2005;19:607–618. doi: 10.1016/j.molcel.2005.07.019. doi:10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 52.Jacquemont C., Taniguchi T. Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Res. 2007;67:7395–7405. doi: 10.1158/0008-5472.CAN-07-1015. doi:10.1158/0008-5472.CAN-07-1015. [DOI] [PubMed] [Google Scholar]
- 53.Hu J., McCall C.M., Ohta T., Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. doi:10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- 54.Lee J., Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell. 2007;26:775–780. doi: 10.1016/j.molcel.2007.06.001. doi:10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 55.Howlett N.G., Harney J.A., Rego M.A., Kolling F.W.T., Glover T.W. Functional interaction between the Fanconi Anemia D2 protein and proliferating cell nuclear antigen (PCNA) via a conserved putative PCNA interaction motif. J. Biol. Chem. 2009;284:28935–28942. doi: 10.1074/jbc.M109.016352. doi:10.1074/jbc.M109.016352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song I.Y., Palle K., Gurkar A., Tateishi S., Kupfer G.M., Vaziri C. Rad18-mediated translesion synthesis of bulky DNA adducts is coupled to activation of the Fanconi anemia DNA repair pathway. J. Biol. Chem. 2010;285:31525–31536. doi: 10.1074/jbc.M110.138206. doi:10.1074/jbc.M110.138206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akkari Y.M., Bateman R.L., Reifsteck C.A., D'Andrea A.D., Olson S.B., Grompe M. The 4N cell cycle delay in Fanconi anemia reflects growth arrest in late S phase. Mol. Genet. Metab. 2001;74:403–412. doi: 10.1006/mgme.2001.3259. doi:10.1006/mgme.2001.3259. [DOI] [PubMed] [Google Scholar]
- 58.Ben-Yehoyada M., Wang L.C., Kozekov I.D., Rizzo C.J., Gottesman M.E., Gautier J. Checkpoint signaling from a single DNA interstrand crosslink. Mol. Cell. 2009;35:704–715. doi: 10.1016/j.molcel.2009.08.014. doi:10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh T.R., Bakker S.T., Agarwal S., Jansen M., Grassman E., Godthelp B.C., Ali A.M., Du C.H., Rooimans M.A., Fan Q., et al. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood. 2009;114:174–180. doi: 10.1182/blood-2009-02-207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Collis S.J., Ciccia A., Deans A.J., Horejsi Z., Martin J.S., Maslen S.L., Skehel J.M., Elledge S.J., West S.C., Boulton S.J. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol. Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. doi:10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 61.Ceccaldi R., Briot D., Larghero J., Vasquez N., Dubois d'Enghien C., Chamousset D., Noguera M.E., Waisfisz Q., Hermine O., Pondarre C., et al. Spontaneous abrogation of the G DNA damage checkpoint has clinical benefits but promotes leukemogenesis in Fanconi anemia patients. J. Clin. Invest. 2011;121:184–194. doi: 10.1172/JCI43836. doi:10.1172/JCI43836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.