Abstract
Mutations in the ATRX protein are associated with the alpha-thalassemia and mental retardation X-linked syndrome (ATR-X). Almost half of the disease-causing mutations occur in its ATRX-Dnmt3-Dnmt3L (ADD) domain. By employing peptide arrays, chromatin pull-down and peptide binding assays, we show specific binding of the ADD domain to H3 histone tail peptides containing H3K9me3. Peptide binding was disrupted by the presence of the H3K4me3 and H3K4me2 modification marks indicating that the ATRX-ADD domain has a combined readout of these two important marks (absence of H3K4me2 and H3K4me3 and presence of H3K9me3). Disease-causing mutations reduced ATRX-ADD binding to H3 tail peptides. ATRX variants, which fail in the H3K9me3 interaction, show a loss of heterochromatic localization in cells, which indicates the chromatin targeting function of the ADD domain of ATRX. Disruption of H3K9me3 binding may be a general pathogenicity pathway of ATRX mutations in the ADD domain which may explain the clustering of disease mutations in this part of the ATRX protein.
INTRODUCTION
ATRX is a large chromatin-associated nuclear protein of ∼280 kDa that belongs to the SNF2 family of chromatin remodeling proteins. It contains two highly conserved domains, namely a plant homeodomain (PHD) zinc finger at the N-terminus and a C-terminal ATPase/helicase domain. Missense mutations in either of these domains lead to the α-thalassemia and mental retardation (ATR-X) syndrome, a hereditary X-linked disease that associates with severe mental retardation, typical dysmorphic features, multiple congenital anomalies and α-thalassemia (1–3). Null mutations of ATRX in mice are embryonically lethal due to a defect in the formation of the extraembryonic trophoblast (4).
ATRX interacts with the transcription cofactor Daxx and the complex shows adenosine-5′-triphosphate-dependent chromatin remodeling activities (5). ATRX is mainly localized in heterochromatin (6) and in promyelocytic leukemia nuclear bodies (5). In the N-terminal region of ATRX, adjacent to the PHD domain, there is a coiled-coil motif region, which is poorly conserved between the human and mouse ATRX (7) and has been reported to interact with murine HP1α and the Su(var)3-9 enhancer of zeste tritorax domain of the polycomb protein EZH2 (an H3K27 histone lysine methyltransferase) (6,8,9). It was also shown that the C-terminal helicase region of ATRX interacts with methyl CpG binding protein 2 (MeCP2), which binds methylated DNA and is mutated in the Rett syndrome. However, it has been shown that MeCP2 is not required for the correct targeting of ATRX, and the N-terminus of ATRX is sufficient for its heterochromatin localization (10).
The PHD finger of ATRX is atypical and shares homology with the PHD domains of Dnmt3a, Dnmt3b and Dnmt3L and, hence, the domain is called an ATRX-Dnmt3-Dnmt3L (ADD) domain (11–13). The ADD domain consists of an N-terminal GATA-like zinc finger, a PHD finger and a C-terminal α-helix. Nearly half of all natural mutations causing the ATRX syndrome occur in the ADD domain suggesting the high functional significance of this domain (3,13). Moreover, mutations in the ADD domain produce a more severe disease phenotype than mutations in the helicase domain of ATRX (14). Some of the disease-causing mutations in the ADD domain affect the folding of the domain and probably destabilize the full-length ATRX protein. Other mutations, however, lie on the surface of the domain and do not disrupt the structure of the domain. These mutations may cause the disease phenotype by disturbing the unknown functions or interaction sites of this domain (13,15).
Despite the significance of the ADD domain in the ATR-X syndrome, the function of this domain is still unknown. It is known that PHD fingers from various nuclear proteins differentially recognize either methylated or unmodified lysine residues in histone tails (15,16). Mutations in PHD fingers of many nuclear proteins are associated with a variety of diseases, including immunodeficiency syndrome, solid and blood cancers and neurological disorders (15). Recently, it has been shown that the ADD domains of Dnmt3a, Dnmt3b and Dnmt3L interact with histone H3 tails that are unmethylated at lysine 4 (H3K4me0) (17–19). In addition, it was already shown that ATRX and Daxx interact with H3.3. This interaction is essential for ATRX localization and Hira-independent localization of H3.3 at telomeres (20–22).
In the present study, we have explored the function of the ADD domain of ATRX by investigating its potential interaction with modified histone tails using peptide arrays. We found that the ADD domain of ATRX interacts with histone H3 tails that are trimethylated at lysine 9 (H3K9me3) and unmethylated at lysine 4. Our results suggest that the ADD domain of ATRX contains an H3K4me0-binding pocket similar to the ADD domains of Dnmt3a and Dnmt3L, but in addition, it also interacts with H3K9me3. We have also studied the influence of some disease-causing mutations in the ADD domain on histone peptide interaction.
RESULTS
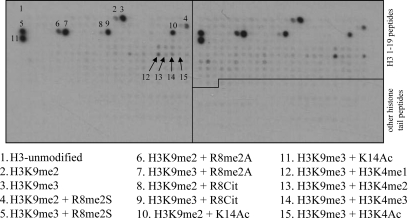
We have applied CelluSpots peptide arrays comprising 384 histone tail peptides, which contain 59 post-translational modifications in different combinations to study the interaction of the purified ADD domain of ATRX with modified histone tails (Fig. 1 and Supplementary Material, Information 1). The ADD domain of ATRX specifically interacted with H3 peptides containing the trimethylation mark at lysine 9 position (Fig. 1). Weaker interaction was observed with H3 peptides that contained H3K9me2, and no binding was seen with H3 peptides that are unmethylated, monomethylated or acetylated at lysine 9 (Fig. 1). No or very weak binding was observed to peptides with H3K4me3 even in the presence of the H3K9me3 modification mark, indicating that the H3K4me3 modification inhibited the binding of the ADD domain to the H3 peptide (Fig. 1). The binding of the ATRX-ADD domain to H3K9me3 peptides was not inhibited by the presence of symmetric or asymmetric dimethylation of R8 or citrullination of this residue (Fig. 1). The ADD domain of ATRX failed to interact with other histone tail peptides viz. H3 (7–26), H3 (16–35), H3 (26–45), H4 (amino acids 1–19), H2A (1–19) and H2B (1–19) irrespective of their modification states (Fig. 1).
Figure 1.
Interaction of the ADD domain with histone tail peptide arrays containing 384 peptides featuring various combinations of known and hypothetical modifications at the H3, H4, H2A and H2B histone tails (Supplementary Material, Information 1). Histone 3 peptides consisting of amino acid residues 1–19 are labeled. The array is set-up in duplicates with two repeats of the same peptides spotted on the right- and on the left-hand side. The modification state of the some peptides is annotated.
Effect of disease-causing mutations on H3 tail binding
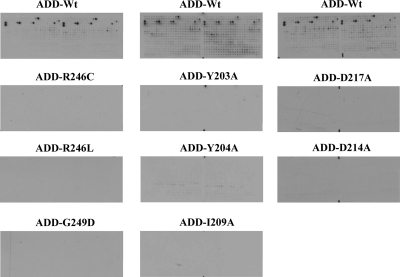
We investigated the influence of disease-causing mutations located in the ADD domain of ATRX on the interaction of the ADD domain with H3K9me3 peptides. For this purpose, we introduced three disease-causing mutations (G249D, R246C and R246L) in the ADD domain. The mutations were selected based on their surface localization, and nuclear magnetic resonance experiments documented their wild-type-like structure (13). The glutathione-S-transferase (GST)-tagged mutant protein domains were overexpressed and purified (Supplementary Material, Fig. S1) and their wild-type-like folding confirmed by circular dichroism (CD) spectroscopy (Supplementary Material, Fig. S2). As reported earlier (13), recombinant mutant protein expression levels were only 30–40% of the wild-type ADD domain, which might contribute to the pathological phenotype. All the three mutant proteins showed a considerable decrease in binding to H3K9me3 peptides, explaining the pathophysiology of these mutations (Fig. 2A and Supplementary Material, Fig. S3).
Figure 2.
Interaction of wild-type and variant ADD domains with histone tail peptide arrays. (A) Effect of disease-causing mutations G249D, R246C and R246L on the interaction of the ADD domain with histone tail peptide arrays. (B) Effect of H3 trimethyl lysine 9 binding pocket mutations Y203A, Y204A and I209A on the interaction of the ADD domain with histone tail peptide arrays. (C) Effect of mutations near H3K4-binding pocket D217A and D214A on the interaction of the ADD domain with histone tail peptide arrays.
Effect of K9me3-binding candidate residues on H3 tail binding
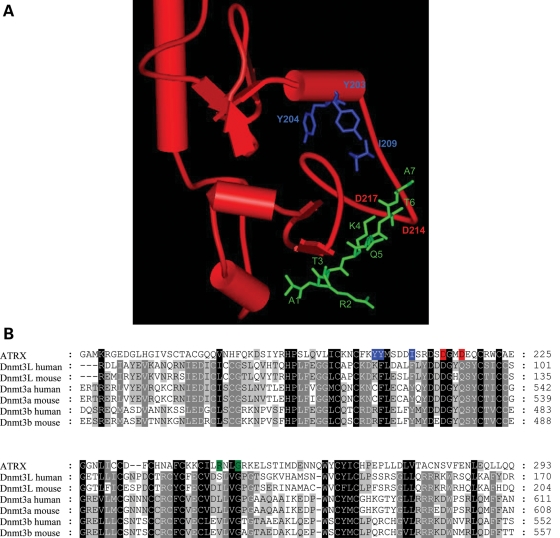
Recently, the structures of the ADD domain of Dnmt3L and Dnmt3a were solved in complex with an H3 tail peptide containing unmethylated lysine at position 4 (17,18). The structures reveal tight interactions with the N-terminal ends of the peptides including K4, but they do not show an interaction of the Dnmt3a or Dnmt3L ADD domains with the K9 position. Based on the observation that binding to trimethylated lysine usually involves hydrophobic aromatic cages (16) and on the amino acid similarity of the ADD domains (Fig. 3), we used the complex of the Dnmt3L-ADD domain with the H3 tail peptide to model the ATRX-ADD domain interaction with histone 3 tail and identified a putative binding pocket for the trimethylated lysine 9 of histone 3 tail formed by Y203, Y204 and I209 (Fig. 3). Trying to disrupt the H3K9me3 interaction of ADD, we individually mutated Y203, Y204 and I209 to alanine. The mutant domain proteins were expressed and purified with similar yields as the wild-type domain (Supplementary Material, Fig. S1) and their wild-type-like folding confirmed by CD spectroscopy (Supplementary Material, Fig. S2). Binding experiments on peptide arrays showed that all the three variants had completely lost their ability to specifically bind to the H3K9me3 peptide (Fig. 2B and Supplementary Material, Fig. S3). Interestingly, the ADD domains of Dnmt3L and Dnmt3a do not read the methylation state of H3K9 (17–19) which may be related to the fact that no aromatic residue is present at the position corresponding to Y203 in Dnmt3L and Dnmt3a, where this Y is replaced by K or C, respectively (Fig. 3B).
Figure 3.
(A) Superposition of the ATRX-ADD domain (red) with the ADD domain of Dnmt3L in complex with H3 1–7 peptide (green). The overlay was generated using Swiss Prot PDB viewer ver. 3.7 and it included 65 Cα atoms with an root mean square deviation (RMSD) of 1.67 Å. The model identified a putative H3K9me3-binding pocket formed by Y203, Y204 and I209 (blue). (B) Sequence alignment of the ADD domains from ATRX, Dnmt3L, Dnmt3a and Dnmt3b. The residues involved in K9me3 and K4me0 binding in ATRX are shaded blue and red, respectively. Disease-associated mutations are shaded green.
Effect of H3K4me0-binding residues on H3 tail binding
In order to probe the H3K4me0-binding pocket of the ADD domain of ATRX, we mutated D217 to alanine based on sequence alignment with ADD domains of Dnmt3L and Dnmt3a (Fig. 3B). The corresponding residues Q93 and Q534 form direct back-bone hydrogen bonds to the K4 amino group in the structures of Dnmt3L and Dnmt3a, respectively. In addition, we exchanged D214, which corresponds to D90 and D531 in Dnmt3L and Dnmt3a, respectively, which form direct side-chain hydrogen bonds to the K4 amino group. The mutation of the corresponding D90 in Dnmt3L led to strong reduction in binding to the H3 tail peptide (17). The mutant proteins were expressed and purified with similar yields as the wild-type domain (Supplementary Material, Fig. S1) and their wild-type-like folding confirmed by CD spectroscopy (Supplementary Material, Fig. S2). As expected, the mutant proteins failed to interact with H3K9me3 peptides (Fig. 2C and Supplementary Material, Fig. S3).
Equilibrium peptide-binding experiments
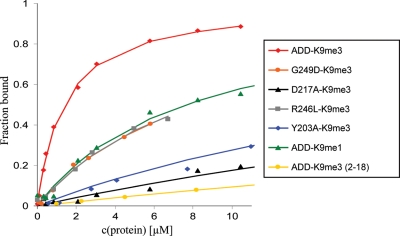
By using fluorescence depolarization and purified H3 tail peptides, we determined the dissociation constant of the ADD domain with the H3K9me3 peptide as 1.38 (±0.37) µm (Fig. 4 and Table 1). Binding to the H3K9me1 peptide and the unmodified H3 tail peptide was 6-fold and >60-fold weaker, respectively. Binding to the H3K4me3/H3K9me3 peptide was >60-fold weaker, indicating that trimethylation of K4 blocks binding of the ADD domain. These results are in good qualitative agreement with the peptide array-binding experiments. We have also examined the binding of the ADD domain of ATRX to an H3K9me3 peptide lacking the first amino acid (H3K9me3, 2–19 amino acids) and observed a very weak binding with a dissociation constant above 80 µm (Fig. 4 and Table 1). It is known that the ADD domains of Dnmt3a and Dnmt3L interact with the N-terminal tail of the H3 peptide and the first amino acid is contacted by hydrogen bonds to the N-terminal amino group (17,18). The Dnmt3a ADD domain failed to interact with H3 peptides acetylated at the N-terminus (18,19), suggesting that the interaction with the end of the H3 tail is similar in all three ADD domains.
Figure 4.
Determination of dissociation constants by equilibrium peptide-binding experiments. The binding constants of the wild-type and variant ADD domains were determined by fluorescence depolarization with H3K9me3 (1–19), H3K9me1 (1–19) or H3K9me3 (2–19) fluorescently labeled peptides.
Table 1.
Dissociation constants of ADD domain or variants as determined by fluorescence depolarization with H3K9me3 (1–19), H3K9me1 (1–19) or H3K9me3 (2–19) fluorescently labeled peptides
| Protein | Peptide | KD value (μm) |
|---|---|---|
| ADD-wt | H3K9me3 | 1.3 ± 0.37 |
| ADD-wt | H3K9me1 | 8.4 ± 1.1 |
| ADD-wt | H3K4me3/H3K9me3 | >80 |
| ADD-wt | H3 (2–19) K9me3 | >80 |
| ADD-wt | H3-unmodified | >80 |
| ADD-G249D | H3K9me3 | 7.0 ± 1.1 |
| ADD-R246L | H3K9me3 | 6.7 ± 0.22 |
| ADD-Y203A | H3K9me3 | 25 ± 0.47 |
| ADD-D217A | H3K9me3 | 44 ± 1.9 |
Error margins represent the standard deviations of independent repeated measurements.
The dissociation constants of representative ADD domain variants were also determined by using fluorescence depolarization experiments. It was found that the disease-causing mutations G249D and R246L reduced the binding affinity of the ADD domain to the H3K9me3 peptide by 5-fold (Fig. 4 and Table 1). The Y203A mutation in the K9me3-binding pocket and the mutation D217A in the K4me0-binding site reduced the binding affinity of the ADD domain with the H3K9me3 peptide by 18-fold and 32-fold, respectively (Fig. 4 and Table 1).
Nucleosome and histone pull-down experiments with the ATRX-ADD domain
The interaction of the ADD domain with the H3K9me3 modification mark was also confirmed by pull-down experiments using native chromatin and histones. The GST pull-down experiments were performed using the GST-tagged ADD domain to pull-down native nucleosomes (mainly mononucleosomes) isolated from HEK293 cells (Supplementary Material, Fig. S4). As predicted by the results of the peptide array-binding experiments and equilibrium peptide-binding experiments, the ADD domain of ATRX interacted with nucleosomes containing the H3K9me3 mark (Fig. 5A and Supplementary Material, Fig. S5A).
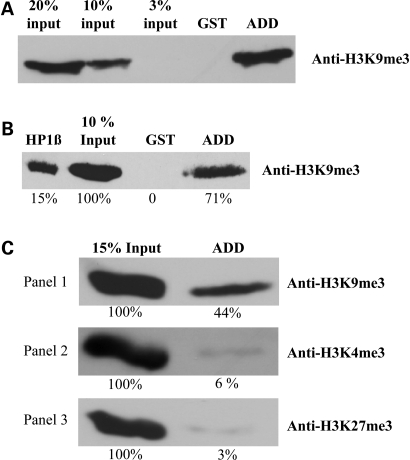
Figure 5.
Interaction analysis of the ADD domain with native oligonucleosomes and native histones, purified from human cells, by using the GST pull-down assay. The bound fractions were separated and immunoblotted with modification-specific antibodies. (A) Interaction of the ADD domain with native oligonucleosomes containing H3K9me3 marks. (B) Interaction of the ADD domain with native histones containing H3K9me3 marks. HP1β was used as a positive control. The band intensities were analyzed quantitatively using Phoretix software and the relative amounts are indicated below each band. (C) Specificity of the H3K9me3 interaction with native histones. For the specificity analysis, the bound fractions were separated and immunoblotted with anti-H3K9me3 antibody, anti-H3K4me3 antibody or anti-H3K27me3 antibody.
It has already been shown that the ADD domain of ATRX could bind DNA homopolymers in vitro (23). Using native histones isolated from HEK293 cells, we observed that the ADD domain of ATRX interacts with histones containing H3K9me3 marks (Fig. 5B and Supplementary Material, Fig. S5B), indicating that the interaction is not DNA mediated. We have analyzed the specificity of the ADD domain interaction with histone tails by probing the bound fraction of the pull down with different modification-specific antibodies. We found that the histone fraction bound by the ADD domain was enriched with the histones containing the H3K9me3 mark, but it did not contain detectable amounts of the H3K4me3 and H3K27me3 marks (Fig. 5C and Supplementary Material, Fig. S5C). Equal protein loading in all pull downs was confirmed by anti-GST staining (Supplementary Material, Fig. S5). We conclude that the ADD domain specifically interacts with histone 3 containing the H3K9me3 modification mark.
Sub-cellular localization of GFP-NT-ATRX and its variants
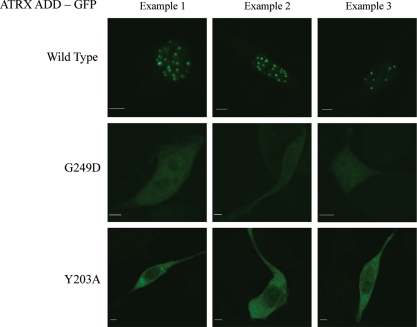
We studied the cellular localization of a GFP-fused N-terminal fragment of ATRX containing the ADD domain. In agreement with the published results (10), we observed that the N-terminal part of ATRX is located in the nucleus, where it formed distinct spots (Fig. 6 and Supplementary Material, Fig. S6), which coincide with 4′,6-diamidino-2-phenylindole (DAPI)-stained pericentromeric heterochromatin spots (Supplementary Material, Fig. S7). We have selected one example each of the disease related (G249D), K9 pocket (Y203A) and K4 interaction (D217A) mutants described previously to investigate whether these mutations, which all disrupt H3 tail binding, interfere with the subnuclear localization of ATRX. Our results show that all ATRX variants lost heterochromatic localization. With G249D, a weak expression and homogenous cellular distribution was observed (Fig. 6). Y203A was expressed at higher levels comparable with the wild-type protein and also showed a homogenous distribution with some depletion in the nucleus. The D217A variant lost its heterochromatic localization, but it showed aggregation in large nuclear and cytoplasmatic spots (Supplementary Material, Fig. S8). While the result obtained with the D217A variant due to its aggregation cannot be interpreted in a straightforward manner, both other variants clearly document that a loss of H3 tail binding of the ADD domain interferes with the heterochromatic localization of the ATRX protein in NIH3T3 cells.
Figure 6.
Localization of the GFP-NT-ATRX to nuclear heterochromatic spots in NIH3T3 cells (see also Supplementary Material, Fig. S6). Spotty nuclear localization is lost in the G249D and Y203A variants. The ATRX spots coincide with DAPI-stained heterochromatic spots (Supplementary Material, Fig. S7). The pictures shown are representative for all cells in the respective experiments (>50 in each case). The scale bars correspond to 5 µm.
DISCUSSION
The ADD domain of the ATRX protein is a hotspot for mutations causing the ATR-X syndrome (3). In spite of this, the molecular function of this domain remained unknown. It had been suggested that it might interact with histone 3 tails (3,13,15) and recently it was shown that ATRX and Daxx interact with H3.3 (20–22). The related ADD domains of Dnmt3a and Dnmt3L interact with H3 tails unmodified at K4 (17–19). In this study, we report that the ADD domain of ATRX indeed interacts with H3 tails. It shares with the related ADD domains that binding is only possible if K4 is unmodified. However, it possesses an additional readout for K9me3, and it actually needs both of these marks for productive binding, indicating a simultaneous readout of these two important chromatin marks. Our data suggest that the interaction of the ATRX, Dnmt3a and Dnmt3L-ADD domains with the N-terminus of the H3 tail, which precludes the binding of peptides with modified K4, is similar among all three proteins. However, the binding to H3K9me3 suggests the presence of an additional aromatic cage-binding pocket for the trimethyl lysine side chain of K9 which is specific for the ATRX-ADD domain. We provide evidence that this pocket is created by Y203, Y204 and I209. We could find these candidate residues on the basis of the superposition of the ATRX-ADD domain with the Dnmt3L-ADD domain, which contains the peptide ligand ordered up to residue 7. The lack of an aromatic residue at the position corresponding to Y203 in Dnmt3a and Dnmt3L may explain why these domains do not show preference for H3K9me3. Simultaneous readout of K4me0 and K9me3 by the ATRX-ADD domain is observed in peptide array binding and with isolated peptides. In addition, we show that disruption of either of the pockets prevents peptide binding. Our observation that disease-causing mutations in the ADD domain reduce the affinity of this interaction, and that this interferes with the heterochromatic localization of the ATRX protein shows how these mutations may lead to the ATR-X syndrome phenotype.
Several lines of previously published evidence strongly support our conclusions. It is established that ATRX is mainly localized on heterochromatin (6), which is one of the major sites of H3K9me3 marks (24,25). Indeed, the co-localization of ATRX and H3K9me3 was demonstrated in pericentric heterochromatin regions in mouse cells (26). The observation that the mutation of lysine 4 in the H3 tail abrogates binding by ATRX (20) also supports our model of a combined readout of K4me0 and K9me3 by the ATRX-ADD domain. The chromatin targeting of ATRX may be further enhanced by its interaction with HP1α (6,8). In addition, ATRX interacts with MeCP2 via its C-terminal domain, which could contribute to heterochromatic targeting, through the interaction of MeCP2 with methylated DNA in post-mitotic neurons where MeCP2 is highly abundant. However, it has been shown that the N-terminal domain of ATRX alone is sufficient for heterochromatin localization, which ruled out the possibility that ATRX heterochromatin localization is based on its interaction with MeCP2 (10). We showed here that the ADD domain of ATRX interacts with H3K9me3, which can directly target the protein to heterochromatin. The association of ATRX and heterochromatin may further be stabilized by its interactions with DNA, HP1α and MeCP2, thereby generating a network of interacting factors and domains. Our results showing that the N-terminal part of ATRX lost its heterochromatic localization when binding of the ADD domain to H3 tails is disrupted suggest that the ADD domain (and its binding to K4me0/K9me3 H3 tails) plays an important role in heterochromatic localization of ATRX.
We have shown that disease-causing mutations G249D, R246C and R246L reduced the interaction of the ADD domain with the H3 tail leading to a loss of heterochromatic localization. These mutations are located on the surface of the ADD domain of ATRX and they do not disrupt the structure of the domain. Our observation suggests that the molecular basis of the ATRX syndrome caused by these mutations lies in the disruption of the chromatin recognition function of the ADD domain. Disruption of H3K9me3 binding may be a general pathogenicity pathway of ATRX mutations, which may explain the clustering of disease mutations in the ADD domain.
EXPERIMENTAL PROCEDURES
Cloning, site-directed mutagenesis, expression and purification
The sequence encoding the human ADD domain of ATRX (residues 163–292 in NP_000480.2) was cloned into pGEX-4T3 vector (GE Healthcare) as a fusion protein with GST using BamHI and XhoI sites. The G249D, R246C, R246L, D217A, D214A, Y203A, Y204A and I209A mutations were introduced by the PCR-megaprimer mutagenesis method as previously described (27). Mutagenesis was confirmed by restriction marker site analysis and DNA sequencing.
For expression, E.coli BL21 (Novagen) carrying the corresponding plasmid were grown in the Luria-Bertani medium at 37°C to OD600 ≈ 0.6, then shifted to 22°C for 20 min and induced overnight with 1 mm isopropyl β-D-thiogalactoside. The collected cells were re-suspended in 20 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.5), 0.5 m KCl, 0.2 mm dithiothreitol (DTT), 1 mm ethylenediaminetetraacetic acid and 10% glycerol and disrupted by sonication. The supernatants were passed through glutathione Sepharose 4B resin (Amersham Biosciences) and washed with the same buffer. The bound proteins were eluted with a similar buffer containing 40 mm glutathione and dialyzed in 20 mm HEPES (pH 7.5), 0.2 m KCl, 0.2 mm DTT, 1 mm EDTA and 10% glycerol for 2 h then overnight in 20 mm HEPES (pH 7.5), 0.2 m KCl, 0.2 mm DTT, 1 mm EDTA and 60% glycerol.
Binding of protein domains to peptide arrays containing modified residues
Peptide arrays are ideal tools to analyze the binding specificity of histone tail antibodies (28) and epigenetic reading domains (29,30). Here, we used a peptide array comprising 384 peptide spots prepared by the CelluSpots method (31) to study the interaction of the purified ADD domain of ATRX with modified histone tails (Fig. 1). The array contains peptides from eight different regions of the N-terminal tails of histones, namely H3 1–19, 7–26, 16–35 and 26–45, H4 1–19 and 11–30, H2A 1–19 and H2B 1–19 (Fig. 1 and Supplementary Material, Information 1), featuring 59 post-translational modifications in many different combinations. Each array presented the same peptides in duplicate for quality control. The results were confirmed by binding of the ADD domain to SPOT peptide arrays originating from an independent peptide synthesis (data not shown).
The CelluSpot arrays were provided by Intavis AG (Köln, Germany). They are now commercially available from Active Motif (Cat. No. 13001). The array was blocked by overnight incubation in the tween 20 containing tris buffered saline (TTBS) buffer (10 mm Tris/HCl pH 8.3, 0.05% Tween-20 and 150 mm NaCl) containing 5% non-fat dried milk at 4°C. The membrane was then washed once with the TTBS buffer, and incubated with 50 nm purified GST-tagged ADD domain of ATRX or mutant domains at room temperature for 1 h in interaction buffer [100 mm KCl, 20 mm HEPES (pH 7.5), 1 mm EDTA, 0.1 mm DTT and 10% glycerol]. The membrane was washed in the TTBS buffer and incubated with goat anti-GST antibody (GE Healthcare #27- 4577-01, at 1:5000 dilution) for 1 h at room temperature. The unbound antibody was washed three times with TTBS and the membrane was incubated with horseradish peroxidase-conjugated anti-Goat antibody (Invitrogen #81-1620 1:12000) in TTBS for 1 h at room temperature. Finally, the membrane was submerged in ECL developing solution (GE Healthcare) and the image was captured on X-ray film. Typical exposure times were 0.5–5 min. The images were analyzed using an in-house program (Array Analyze, available at http://www.activemotif.com/catalog/667.html or from the authors upon request).
Fluorescence depolarization
Purified peptides were purchased from Intavis AG (Köln, Germany). Fluorescence depolarization experiments were carried out at 25°C using a Varian Carry Eclipse fluorescence spectrophotometer. The binding constant of the ADD domain or its variants to the fluorescein isothiocyanate-coupled peptides [H3K9me3 (amino acids 1–19), H3K9me1 (amino acids 1–19) or H3 (2–19) K9me3 (amino acids 2–19)] was determined using 100 nm of peptide incubated with increasing concentrations of the ADD domain in interaction buffer (25 mm Tris pH 8.0, 100 mm KCl, 5 mm MgCl2, 10% glycerol and 0.1% NP40) by fluorescence depolarization (excitation at 494 nm, emission at 524 nm and excitation and emission slits at 5 nm). The data were fitted to a binary-binding equilibrium including a variable baseline (BL) and effect factor (F) to determine the equilibrium-binding constant using the Microsoft Excel Solver module (32).
The binding of the weak binding variants to the H3K9me3 peptide was analyzed using the signal factor observed for wild-type binding to the same peptide. The binding constant of the ADD domain to the unmodified H3 and double-modified H3 K4me3/K9me3 peptides was estimated by competition experiments in which 100 nm of fluorophore-coupled H3K9me3 was incubated with 2.75 μm of the ADD domain in interaction buffer, and their interaction was competed with 5-, 10-, 100-, 200- and 250-fold molar excesses of unlabeled unmodified H3 1–19 peptide. The data were fitted to the ternary-binding equilibrium model in which the labeled und unlabeled peptides compete for binding to the protein. The binding constant to the H3 (2–19) K9me3 peptide was estimated assuming a similar signal factor as observed to H3 (1–19) K9me3.
GST pull-down assay
Native histones were isolated from the HEK293 cell line using the acid extraction method as previously described (33). Native oligonucleosomes were prepared from the HEK293 cell line by partial Micrococcal nuclease (NEB) digestion of nuclei. The oligonucleosomes were isolated as described (34), with the exception that following the Micrococcal nuclease digestion, the nuclei were pelleted and the supernatant containing oligoncleosomes (predominantly mononucleosomes) was used for the GST pull-down assay.
For the GST pull-down with native oligonucleosomes, 20 µl of Glutathione Sepharose 4B resin (Amersham Biosciences) were incubated with 8 µg of the GST-tagged ADD domain of ATRX for 1 h at 4°C in chromatin interaction buffer (50 mm Tris pH 8.0, 100 mm NaCl, 2 mm EDTA and 0.1% Triton X-100). The beads were washed once with interaction buffer and blocked in interaction buffer containing 5% bovine serum albumin (BSA) for 1 h at 4°C. One hundred micrograms of native oligonucleosomes dissolved in interaction buffer were added to the beads and incubated for 3h. Finally, the beads were washed three times with interaction buffer and re-suspended in 2× sodium dodecyl sulfate polyacylamide gel electrophoresis (SDS-PAGE) loading dye. The beads were boiled for 10 min, spun down and the supernatant containing the bound fraction was separated on a 16% SDS–PAGE gel and immunoblotted with anti-H3K9me3 antibody (Abcam, ab8898).
For the GST pull down with native histones, 20 µl of Glutathione Sepharose 4B resin (Amersham Biosciences) was incubated with 8 µg of GST-tagged ADD domain of ATRX or 4 µg of GST-tagged chromo domain of HP1β for 1 h at 4°C in the histone interaction buffer (25 mm Tris pH 8.0, 150 mm KCl, 5 mm MgCl2, 10% glycerol and 0.1% NP40 equivalent). Then the beads were washed once with interaction buffer and blocked with interaction buffer containing 5% BSA for 1 h at 4°C. Twenty micrograms of native histones were added to the beads in interaction buffer and incubated for 3 h in interaction buffer. Finally, the beads were washed three times with wash buffer (25 mm Tris pH 8.0, 400 mm KCl, 5 mm MgCl2, 10% glycerol and 0.1% NP40 equivalent) and re-suspended in 2× SDS–PAGE loading dye. The beads were boiled for 10 min, spun down and the supernatant containing the bound fraction was separated on a 16% SDS–PAGE and immunoblotted with modification-specific histone antibodies: anti-H3K9me3 antibody (Abcam, ab8898), anti-H3K27me3 antibody (Active Motif) and anti-H3K4me3 antibody (Active Motif) or anti-GST antibody (GE Healthcare). GST-tagged HP1β, which is known to bind histones containing H3K9me3 (35), was used as a positive control.
Cell culture and microscopy
An expression construct for the GFP-fused N-terminal to ADD domain part of human ATRX splicing isoform 2 was obtained from Dr Adrian Bird (Edinburgh, UK) (10). The Y203A, D217A and G249D mutants were introduced as described above and sequenced. NIH3T3 cells were grown in Dulbecco's modified eagle's medium with 10% (v/v) fetal calf serum and 2 mm l-glutamine at 37°C in 5% (v/v) CO2. The cells were seeded on cover slips and 1–2 × 105 cells were transfected with the GFP-NT-ATRX constructs in six-well plates using FuGENE 6 (Roche, Basel, Switzerland) and 1 µg of total plasmid DNA per well according to the manufacturer's instructions. Two days after transfection, the cells were fixed with 4% (w/v) paraformaldehyde and embedded with Mowiol (Carl Roth). Confocal images were taken with a Zeiss LSM510 instrument (Carl Zeiss, Jena, Germany, software version 3.0) using a 63× oil immersion objective. The Argon laser line at 514 nm was used to excite GFP fluorescence and a BP530-550 filter was used for image recording. Fluorescence microscopy pictures were taken using an AXIOPLAN2 microscope equipped with AxioCam HRc camera and Achroplan 100× oil immersion objective with filter sets DAPI FT395/LP420 and GFP BP450-490/FT510 (all from Carl Zeiss).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the NIH (DK082678) and DFG (JE 252/6).
Supplementary Material
REFERENCES
- 1.Gibbons R.J., Picketts D.J., Villard L., Higgs D.R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. doi:10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 2.Gibbons R.J., Brueton L., Buckle V.J., Burn J., Clayton-Smith J., Davison B.C., Gardner R.J., Homfray T., Kearney L., Kingston H.M., et al. Clinical and hematologic aspects of the X-linked alpha-thalassemia/mental retardation syndrome (ATR-X) Am. J. Med. Genet. 1995;55:288–299. doi: 10.1002/ajmg.1320550309. doi:10.1002/ajmg.1320550309. [DOI] [PubMed] [Google Scholar]
- 3.Gibbons R.J., Wada T., Fisher C.A., Malik N., Mitson M.J., Steensma D.P., Fryer A., Goudie D.R., Krantz I.D., Traeger-Synodinos J. Mutations in the chromatin-associated protein ATRX. Hum. Mutat. 2008;29:796–802. doi: 10.1002/humu.20734. doi:10.1002/humu.20734. [DOI] [PubMed] [Google Scholar]
- 4.Garrick D., Sharpe J.A., Arkell R., Dobbie L., Smith A.J., Wood W.G., Higgs D.R., Gibbons R.J. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2006;2:e58. doi: 10.1371/journal.pgen.0020058. doi:10.1371/journal.pgen.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue Y., Gibbons R., Yan Z., Yang D., McDowell T.L., Sechi S., Qin J., Zhou S., Higgs D., Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl Acad. Sci. USA. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. doi:10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDowell T.L., Gibbons R.J., Sutherland H., O'Rourke D.M., Bickmore W.A., Pombo A., Turley H., Gatter K., Picketts D.J., Buckle V.J., et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc. Natl Acad. Sci. USA. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. doi:10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picketts D.J., Tastan A.O., Higgs D.R., Gibbons R.J. Comparison of the human and murine ATRX gene identifies highly conserved, functionally important domains. Mamm. Genome. 1998;9:400–403. doi: 10.1007/s003359900781. doi:10.1007/s003359900781. [DOI] [PubMed] [Google Scholar]
- 8.Le Douarin B., Nielsen A.L., Garnier J.M., Ichinose H., Jeanmougin F., Losson R., Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 9.Cardoso C., Timsit S., Villard L., Khrestchatisky M., Fontes M., Colleaux L. Specific interaction between the XNP/ATR-X gene product and the SET domain of the human EZH2 protein. Hum. Mol. Genet. 1998;7:679–684. doi: 10.1093/hmg/7.4.679. doi:10.1093/hmg/7.4.679. [DOI] [PubMed] [Google Scholar]
- 10.Nan X., Hou J., Maclean A., Nasir J., Lafuente M.J., Shu X., Kriaucionis S., Bird A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc. Natl Acad. Sci. USA. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. doi:10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie S., Wang Z., Okano M., Nogami M., Li Y., He W.W., Okumura K., Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. doi:10.1016/S0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 12.Aapola U., Kawasaki K., Scott H.S., Ollila J., Vihinen M., Heino M., Shintani A., Kawasaki K., Minoshima S., Krohn K., et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–298. doi: 10.1006/geno.2000.6168. doi:10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- 13.Argentaro A., Yang J.C., Chapman L., Kowalczyk M.S., Gibbons R.J., Higgs D.R., Neuhaus D., Rhodes D. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc. Natl Acad. Sci. USA. 2007;104:11939–11944. doi: 10.1073/pnas.0704057104. doi:10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Badens C., Lacoste C., Philip N., Martini N., Courrier S., Giuliano F., Verloes A., Munnich A., Leheup B., Burglen L., et al. Mutations in PHD-like domain of the ATRX gene correlate with severe psychomotor impairment and severe urogenital abnormalities in patients with ATRX syndrome. Clin. Genet. 2006;70:57–62. doi: 10.1111/j.1399-0004.2006.00641.x. doi:10.1111/j.1399-0004.2006.00641.x. [DOI] [PubMed] [Google Scholar]
- 15.Baker L.A., Allis C.D., Wang G.G. PHD fingers in human diseases: disorders arising from misinterpreting epigenetic marks. Mutat. Res. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taverna S.D., Li H., Ruthenburg A.J., Allis C.D., Patel D.J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. doi:10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ooi S.K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. doi:10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otani J., Nankumo T., Arita K., Inamoto S., Ariyoshi M., Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. doi:10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Jurkowska R., Soeroes S., Rajavelu A., Dhayalan A., Bock I., Rathert P., Brandt O., Reinhardt R., Fischle W., et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. doi:10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong L.H., McGhie J.D., Sim M., Anderson M.A., Ahn S., Hannan R.D., George A.J., Morgan K.A., Mann J.R., Choo K.H. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. doi:10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg A.D., Banaszynski L.A., Noh K.M., Lewis P.W., Elsaesser S.J., Stadler S., Dewell S., Law M., Guo X., Li X., et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. doi:10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis P.W., Elsaesser S.J., Noh K.M., Stadler S.C., Allis C.D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl Acad. Sci. USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. doi:10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardoso C., Lutz Y., Mignon C., Compe E., Depetris D., Mattei M.G., Fontes M., Colleaux L. ATR-X mutations cause impaired nuclear location and altered DNA binding properties of the XNP/ATR-X protein. J. Med. Genet. 2000;37:746–751. doi: 10.1136/jmg.37.10.746. doi:10.1136/jmg.37.10.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters A.H., Mermoud J.E., O'Carroll D., Pagani M., Schweizer D., Brockdorff N., Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 2002;30:77–80. doi: 10.1038/ng789. doi:10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- 25.Guenatri M., Bailly D., Maison C., Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. doi:10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumann C., Schmidtmann A., Muegge K., De La Fuente R. Association of ATRX with pericentric heterochromatin and the Y chromosome of neonatal mouse spermatogonia. BMC Mol. Biol. 2008;9:29. doi: 10.1186/1471-2199-9-29. doi:10.1186/1471-2199-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeltsch A., Lanio T. Site-directed mutagenesis by polymerase chain reaction. Methods Mol. Biol. 2002;182:85–94. doi: 10.1385/1-59259-194-9:085. [DOI] [PubMed] [Google Scholar]
- 28.Bock I., Dhayalan A., Kudithipudi S., Brandt O., Rathert P., Jeltsch A. Detailed specificity analysis of antibodies binding to modified histone tails with peptide arrays. Epigenetics. 2011;6:263–265. doi: 10.4161/epi.6.2.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao T., Collins R.E., Horton J.R., Zhang X., Zhang R., Dhayalan A., Tamas R., Jeltsch A., Cheng X. The ankyrin repeat domain of Huntingtin interacting protein 14 contains a surface aromatic cage, a potential site for methyl-lysine binding. Proteins. 2009;76:772–777. doi: 10.1002/prot.22452. doi:10.1002/prot.22452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rathert P., Dhayalan A., Murakami M., Zhang X., Tamas R., Jurkowska R., Komatsu Y., Shinkai Y., Cheng X., Jeltsch A. Protein lysine methyltransferase G9a acts on non-histone targets. Nat. Chem. Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. doi:10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winkler D.F., Hilpert K., Brandt O., Hancock R.E. Synthesis of peptide arrays using SPOT-technology and the CelluSpots-method. Methods Mol. Biol. 2009;570:157–174. doi: 10.1007/978-1-60327-394-7_5. doi:10.1007/978-1-60327-394-7_5. [DOI] [PubMed] [Google Scholar]
- 32.Jeltsch A., Hoggett J., Urbanke C. In: Encyclopedia of Molecular Cell Biology and Molecular Medicine. Meyers R.A., editor. Vol. 11. Wiley-VCH; 2005. pp. 391–410. [Google Scholar]
- 33.Shechter D., Dormann H.L., Allis C.D., Hake S.B. Extraction, purification and analysis of histones. Nat. Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. doi:10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 34.Brand M., Rampalli S., Chaturvedi C.P., Dilworth F.J. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat. Protoc. 2008;3:398–409. doi: 10.1038/nprot.2008.8. doi:10.1038/nprot.2008.8. [DOI] [PubMed] [Google Scholar]
- 35.Jacobs S.A., Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. doi:10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.