Abstract
The extended twin kinship design allows the simultaneous testing of additive and nonadditive genetic, shared and individual-specific environmental factors, as well as sex differences in the expression of genes and environment in the presence of assortative mating and combined genetic and cultural transmission (Eaves et al., 1999). It also handles the contribution of these sources of variance to the (co)variation of multiple phenotypes. Keller et al. (2008) extended this comprehensive model for family resemblance to allow or a flexible specification of assortment and vertical transmission. As such, it provides a general framework which can easily be reduced to fit subsets of data such as twin-parent data, children-of-twins data, etc. A flexible Mx specification of this model that allows handling of these various designs is presented in detail and applied to data from the Virginia 30,000. Data on height, body mass index, smoking status, church attendance, and political affiliation were obtained from twins and their families. Results indicate that biases in the estimation of variance components depend both on the types of relative available for analysis, and on the underlying genetic and environmental architecture of the phenotype of interest.
Keywords: extended twin kinship design, Mx
Introduction
Genetic epidemiological models are used in research to delineate the role of genes and environment to individual differences. Typically, models are a simplification of the reality which allows us to test whether the observed data are consistent with the model. Galton (1875) recognized that comparing the similarity of identical and fraternal twins provided insight in the relative importance of nature and nurture. This design, referred to as the classical twin study, has been used extensively to quantify the role of genetic and environmental factors, both shared between family members and unique to each individual, for a range of phenotypes. It has been extended in a variety of ways to allow for sex or other covariate differences, multiple phenotypes, or measures across development, to name a few. However, the twin design remains limited in the number of sources of variance that can be estimated as it relies in information gained from the twin correlations and variance of the trait. Extending it by including other relatives that have different degrees of genetic and environmental relatedness allows the identification of additional sources of variance.
Eaves et al. (1999) developed an extended twinship model that allows the simultaneous estimation of additive and non-additive genetic and shared and unique environmental influences, in the presence of assortative mating and sex differences in these sources of variance. This model requires data collected from twin pairs, their parents, siblings, spouses and children. Data from 88 sex-specific relationships are used to estimate the combined effects of genetic and cultural transmission, and the resulting genotype by environment covariance. This specification modeled phenotypic assortative mating and phenotypic cultural transmission and was implemented in Mx (Neale et al., 1994; Neale et al., 2006) for use with raw data and applied to a range of phenotypes (Kirk et al., 1999; Lake et al., 2000; Maes et al., 1999). Keller et al (2009) further extended the model by allowing assortment and vertical transmission to be specified more flexibly, allowing testing of, for example, social homogamy (SH) versus primary phenotypic assortment (PA). This ‘Cascade’ model is described in detail within Keller et al. (this issue) which includes the complete algebra for each of the relationships and was translated into Mx as such. Furthermore, assumptions and biases are discussed, as well as the power to detect each of the sources of variance (Medland et al. 2009). We have also implemented the cascade model in Mx building on the previous multivariate extended twin (ET) version which uses building blocks to generate the expectations for the relationships (Maes et al. 1999) and appears to be more efficient. We also added various features to facilitate the fitting of models to various combinations of relatives.
In this paper, we describe the main features of the flexible Mx specification for extended twin kinship designs. We also apply the cascade model and reduced models to kinship data on the Virginia 30,000 on five phenotypes with varying degrees of genetic and environmental architecture. We hope to show that accuracy of results and potential biases - when extended twin kinship data are not available - vary according to the underlying architecture of the phenotype.
Materials and Methods
The Virginia 30,000
The Virginia 30,000 sample contains data from 14,763 twins, ascertained from two sources (Eaves et al., 1999; Truett et al., 1994). Public birth records and other public records in the Commonwealth of Virginia were used to obtain current address information for twins born in Virginia between 1915 and 1971, with questionnaires mailed to twins who had returned at least one questionnaire in previous surveys. A second group of twins was identified through their response to a letter published in the newsletter of the American Association of Retired Persons (AARP, 9476 individuals). Twins participating in the study were mailed a 16 page ‘Health and Lifestyles’ questionnaire, and were asked to supply the names and addresses of their spouses, siblings, parents and children for the follow-up study of relatives of twins. Completed questionnaires were obtained from 69.8% of twins invited to participate in the study, which was carried out between 1986 and 1989.
The original twin questionnaire was modified slightly to provide two additional forms, one appropriate for the parents of twins and another for the spouses, children and siblings of twins. Modifications affected only those aspects of the questionnaire related to twinning, in order to obtain self-report data. The response rate from relatives (44.7%) was much lower than that from the twins. Of the complete sample of 28,521 individuals (from 5670 extended kinships) with valid data, 59.7% were female, with 50% of respondents under 50 years of age.
Zygosity determination
Zygosity of twins was determined on the basis of responses to standard questions about similarity and the degree to which others confused them. This method has been shown to give at least 95% agreement with diagnosis based on extensive blood typing (Eaves et al., 1989).
Measures
In all questionnaires mailed to twins and their relatives, self-report data on height and weight were obtained. Body mass index (BMI) was calculated and BMI data were logtransformed to reduce skewness. An ordinal church attendance was derived from a single item which asked respondents to indicate the number corresponding to the frequency at which they attend church services. The 6 possible response values were: ‘never’, ‘rarely’, ‘a few times a year’, ‘once or twice a month’, ‘once a week’ and ‘more than once a week’ (Maes et al., 1999). Several questions were asked regarding the frequency, quantity and age of onset of the respondents’ lifetime smoking habits, from which a dichotomous variable reflecting whether they had ever smoked or not (Maes et al., 2004). Political affiliation was based on two items from a larger set of social attitudes (Eaves et al., 1999) and reflects one position of the Democratic-Republican dimension.
Statistical methods
Raw continuous data were used for height and body mass index. Smoking status, church attendance and political affiliation were analyzed as raw ordinal measures with respectively two, six and five categories.
Structural modeling of the data was undertaken using methods described in Keller et al. (2009) and based on Eaves et al. (1999) and Truett et al. (1994), which assess the contributions of additive and dominant genetic effects in the presence of effects such as vertical cultural inheritance, phenotypic assortative mating or social homogamy, shared twin and sibling environments and within-family environment. Phenotypic assortment occurs when mate selection is based at least partly on the trait being studied, and is evidenced by a correlation between the observed phenotypes of spouses. Such a correlation may also result from shared social background which can be modeled alternatively. Vertical cultural inheritance is the transmission of non-genetic information from parent to child, and refers to the environmental effects the parents create for their children based on their phenotype. The models of assortment and cultural transmission tested here represent some of the possible mechanisms for family resemblance (Cloninger et al., 1979; Fulker, 1988; Heath & Eaves, 1985). Between-family environmental effects make family members relatively more similar, whereas sibling environments are those environmental factors shared between all types of offspring. A special twin environment is an additional correlation between the environment of twins (in addition to the sibling environment) which makes both MZ and DZ twins more alike than ordinary siblings even in the absence of genetic effects (Neale & Cardon, 1992). While all these sources of common environment contribute to variation among individuals regardless of relationship, they differ in their effect on the covariation between types of relatives. The contribution of genetic and environmental factors may be depend on both magnitude and nature upon an individual’s sex.
A FORTRAN program ‘Famfit’ was originally written by Lindon Eaves to fit an extended twin kinship model to correlations of twins and their first degree and collateral relatives, including parents, siblings, spouses and children. A mathematically equivalent version of the model was implemented in Mx (Maes et al., 1999) to (i) fit models directly to the raw data to obtain maximum likelihood estimates of the model parameters with appropriate confidence intervals (Neale & Miller, 1997) and handling missing data (Little and Rubin, 1987), (ii) analyze multiple variables simultaneously using the rules of multivariate path analysis (Vogler, 1985), and (iii) make it easier to develop and modify as necessary for other pedigree structures and other models of familial resemblance. To accommodate alternative specifications of assortment, major changes were required to the Mx specifications which led to a more concise script. In the new version, we also added in various data handling options which have greatly increased the flexibility of the code which can now be used to analyze data on any combination of relatives including twins, parents, siblings, spouses and children of twins. Thus basically any type of twin design, from the classical twin design to the nuclear twin family design to ET and cascade, can be fit using the same script. We hope that as such it will become a starting point for further developments and improvements. To help with this goal, we will describe here how the program is constructed.
The principles behind the Mx version, which is available on http://www.vcu.edu/mx, are simple. The full model is broken up into a number of building blocks which are precalculated in the top part of the program. These also include a set of constraints which are necessary to uniquely identify all the model parameters. The expectations of each of the existing relationships including twins and their first degree and collateral relatives can then be formed by combining the building blocks in the appropriate way, each of which is done in a series of calculation groups. Further calculation groups are specified to combine the various relationships in order to construct the expected matrices for relatives for all five types of twin pair (MZM, DZM, MZF, DZF, DZO). The data groups then provide the observed data as well as these expected covariance matrices in terms of the precalculated expectations. Finally, calculation groups are added to print the various parameter estimates and to derive components of variance. The full model allows for a complete treatment of sex differences, both in the magnitude and the kind of the effects. Thus both the building blocks and the expectations for the relationships have to be specified for the four combinations (male-male, female-female, male-female and female-male).
The Mx script starts with a number of ‘#define’ statements which control various parts of the job to be run. They are set up in such a way that to apply the model to different sets of data, only a number of parameters have to be changed at the top of the script while the main part of the code remains unchanged. The choices to be made up front include (i) ordinal or continuous data, (ii) confidence intervals or not, (iii) extensive or essential output, (iv) individual likelihood statistics or not, (v) save matrix of expected correlations, (vi) sex differences or not, (vii) full ET design or sub design with limited set of relatives, (viii) dominance versus shared environment in submodels that do not allow both to be simultaneously estimated, (ix) phenotypic assortment or social homogamy. #define’d variables are also use to provide filenames for the observed data, for saving various outputs, for details regarding the variable(s) being analyzed and thresholds given ordinal data, specifications for the variable means, start values and boundaries for the parameters, and the number of variables to be analyzed. Additional variables are used to control which design is being fitted to the data. Finally, each of the 64 groups are referred to by names also declared with ‘#define’ statements to make it easier to insert or delete groups without extensive renumbering.
Calculation groups are used to declare matrices for additive genetic (both common to both sexes and male-specific) and cultural transmission latent factors. These groups also calculate the covariance between an individual’s genotype and his phenotype, including paths through a correlated set of genes and through genotype-environment covariance resulting from the combined presence of genetic and cultural transmission. This g-e covariance is one of the building blocks that are generated for each combination by sex. An assortment path between spouses is specified and additional parameters for additive genetic factors which allow the specification of assortment through the phenotype versus social homogamy. The two sets of genetic paths are set equal to test for phenotypic assortment, or the second set of paths is set to zero for social homogamy. Now all the parameters are declared to compute the covariance between the genotypes of siblings (either MZ or DZ twins/siblings), which may include effects due to assortment. These are then combined with GE covariance paths and the covariance between the cultural transmission latent factors of siblings as building blocks (ABC) for sibling, avuncular and cousin relationships in each of the zygosities.
Matrices are also declared for the non-additive genetic latent factors as well as shared sibling, twin and unique environmental factors and correlations between these factors across sex. These factors together with the additive genetic ones (and associated GE covariance) form the phenotypic variances which are set up as constrained parameters. Corresponding paths are set up to control which sources of variance contribute to assortment. The combination of all sources of variance and their counterparts to control assortment then allow the calculation of the covariance between a person’s actual phenotype P and the phenotype on which assortment P~ is based. Finally parameters for cultural transmission and their covariances need to be declared in matrices.
Constraints to ensure equilibrium of genetic, environmental and GE covariances over consecutive generations are then set up. Three constraints are needed for the genetic latent factors, one for the common set of genes, one for the male-specific genes and one for the covariance between the common and male-specific genetic factors. There are also three constraints for the residual environmental covariance between male, female and opposite sex pairs. The covariances between genetic and environmental factors are also sex-specific and require four constraints.
Additional groups are used to create larger building blocks to be used in acrossgeneration relationships. The covariances between the parental phenotype and the additive genetic and cultural transmission latent factors of the children are precalculated as are the covariances of these factors across generations. These blocks involve both direct genetic and cultural transmission paths from parent to offspring. Similarly, blocks for covariances due to genetic and cultural transmission that involve assortment are constructed and combined to generate (grand)parent-offspring, avuncular and cousin relationships.
The expectations for each of the 88 sex-specific relationships in the extended twin kinship design are then specified. In addition to expected covariances between the actual phenotypes of the relatives involved, we also calculate expected covariances between the actual phenotype (P) of one relative with the ‘mating phenotype’ (P~) of the other relative, referred to as PP~ covariances, or between the mating phenotypes of both relatives (P~P~ covariances) which are used as part of covariances between relatives further apart. First the twin covariances for the five zygosities are generated, followed by PP~ and P~P~ covariances. Second are the sibling and PP~ covariances. The expectations for the correlations between twins use the blocks for ABC covariances across siblings, latent factors representing genetic dominance, non-parental shared environment and special twin environment and the correlations between these factors in males and females in opposite sex twins. The sibling expectations are similar to those for twins except for the special twin environment contribution.
The third group of first degree relationships consists of parent-offspring relatives. The parent-offspring correlations are made of building blocks between direct and indirect (through assortment) paths from the parental phenotype to latent ABC factors of the children and the matrices defining the links between these latent factors and phenotypes. The same building blocks multiplied by additional blocks connecting ABC factors across generations are used to compute expected grandparent-grandchildren correlations. The Famfit program did not include expectations for these relationships as the number of observed pairs of these relationships was relatively small the VA 30,000 sample. However, when fitting to the raw data, all possible relationships have to be explicitly specified. Given the assumption that the correlation between the twins and their parents is identical to the correlation between the twins and their children, the grandparent-grandchild correlations can be computed by combining the expected parent-offspring correlations in the appropriate way.
Next the expected covariances for avuncular relationships through MZ twins, DZ twins and siblings are computed. The matrix algebra for each of these correlations consists of seven matrices: i) paths from the phenotype of an uncle/aunt to his/her latent ABC factors, multiplied with ii) paths from the latent ABC factors to the genetic latent AB factors of a niece/nephew, and iii) a twin or sibling correlation from an uncle/aunt to his/her cotwin, multiplied with iv) cultural transmission path, and v) a twin or sibling PP~ correlation from an uncle/aunt to the mating phenotype of his/her cotwin, multiplied with vi) genetic and cultural transmission paths through assortment, all of which are multiplied finally by vii) a matrix of paths from the latent factors in the child to his/her phenotype. In addition to the regular avuncular covariances, we also specify PP~ covariances for such relationships through twins, which are used in the cousin covariances. The cousin relationships which may exist through MZ twins or DZ twins are specified next. These are also built up by combining the various building blocks in the appropriate fashion, in a similar way as the avuncular relationships with a few extra matrices.
Next a number of calculation groups are used to combine the various individual expected correlations into larger units which can then be combined to produce a table with all the expected correlations. More importantly, they are organized in such a way that they can be put together to generate the expected covariance matrices for the extended kinships by zygosity. Separate groups are used to organize the twin, sibling, parent-offspring, grandparent-grandchild, avuncular and cousin correlations. Following this are groups that calculate the relationships through marriage including first degree relatives and their spouse, spouses through twins and nieces/nephews and the spouse of their uncle/aunt.
Finally all the building blocks that do not vary according to the zygosity of the twin pairs, for example, the covariance between brothers and sisters, are organized in one group. These are then combined with matrices specific to each zygosity to generate the expected covariance matrices for each of the five types. An extra group is used to set up matrices to be used across the data groups to handle regression of covariates. It also generates matrices to produce the relevant subsets of the extended kinship expected covariance matrices when fitting one of the subdesigns. The data groups read the observed raw data for all the relatives. In addition to specifying the model for the covariances between relatives, the data groups also contain models for the means. The latter can include constraints across birth order, zygosity, generation and sex, or be estimated freely. The order of the relatives in the expected mean and covariance statements needs to match those in the observed data files. An additional calculation group summarizes the expected means and covariance matrices for all five zygosity groups. Note that various groups include start values and boundary statements to limit the range of values for the parameter estimates, and options statements for the output.
To obtain relevant information from the output in a organized fashion, several calculation groups are used that create tables. The first one of these generates a table of expected covariances for the 88 relationships by sex combination. Following groups summarize parameter estimates and calculate derived parameters and compute unstandardized and standardized variance components separately for males and females. Other groups report the function values for each of the data groups and list the results of the constraints groups to make it easy to check that all the constraints are satisfied. Statements are included to calculate confidence intervals around parameters of interest. Given the number of parameters in the full model and the size of the observed dataset, it is wise to restrict the number of requested confidence intervals until after evaluation of the model. The final group calls up all the computed tables to print. Also a number of optimization options and options for saving output files are specified in this group. If no sex differences are requested, a set of parameters will be equated, or dropped to specific values. If instead of the full extended kinship model a sub design is fitted, several parameters may have to be dropped from the model to ensure identification of the remaining parameters. Finally, if several subdesigns are being analyzed with the same dataset, a loop function can be used to generate the appropriate output.
Results
Descriptive statistics of selected phenotypes
Descriptive statistics for all the variables are listed in Table 1. These include means and variances for the continuous variables (height and body mass index) and response frequencies for the ordinal variables (smoking status, church attendance and political affiliation). We purposefully selected phenotypes with varying degrees of genetic and environmental architecture to evaluate potential biases in parameter estimates when data are only available on few types of relative. Thus we re-analyzed the same twin kinship data sets by pretending we did not have the full family data available. First, we fitted the full cascade model using all relatives (twins, parents, siblings, spouses & children). The sub designs include: (i) the classical twin study (CTD) which only uses MZ and DZ twin pairs, (ii) twins and parents, (iii) twins, parents and sibs, (iv) twins, parents, sibs and spouses, (v) twins and sibs, (vi) twins and spouses, (vii) twins and children, also known as the children of twins (COT) design, and (viii) twins, spouses and children. For designs including spousal pairs, we tested phenotypic assortment and social homogamy alternatively.
Table 1.
Descriptive Statistics for Five Analyzed Phenotypes
(sample sizes, means and standard deviations for continuous variables: height and body mass index; sample sizes and percentages per category for categorical variables: smoking status, church attendance and political affiliation)
| Height | Body mass index | Smoking status | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Mean | SDev | N | Mean | SDev | N | % 0 | % 1 | |
| Male twin | 5270 | 17.81 | 0.71 | 5243 | 32.14 | 1.37 | 5207 | 36.78 | 63.22 |
| Female twin | 9342 | 16.3 | 0.65 | 9271 | 31.55 | 1.75 | 9102 | 56.04 | 43.96 |
| Father | 841 | 17.84 | 0.68 | 837 | 32.6 | 1.29 | 888 | 20.83 | 79.17 |
| Mother | 1350 | 16.35 | 0.64 | 1336 | 32.22 | 1.79 | 1379 | 52.94 | 47.06 |
| Brother | 1177 | 17.94 | 0.67 | 1177 | 32.33 | 1.29 | 1199 | 35.11 | 64.89 |
| Sister | 1774 | 16.43 | 0.64 | 1753 | 31.77 | 1.82 | 1788 | 55.43 | 44.57 |
| Husband | 2465 | 17.85 | 0.68 | 2454 | 32.48 | 1.34 | 2486 | 30.17 | 69.83 |
| Wife | 1817 | 16.35 | 0.64 | 1796 | 31.75 | 1.69 | 1832 | 56.17 | 43.83 |
| Son | 1820 | 18.07 | 0.69 | 1818 | 32.22 | 1.38 | 1816 | 50.06 | 49.94 |
| Daughter | 2739 | 16.57 | 0.64 | 2718 | 31.43 | 1.85 | 2735 | 54.63 | 45.37 |
| Church attendance | Political affiliation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % 0 | % 1 | % 2 | % 3 | % 4 | % 5 | N | % 0 | % 1 | % 2 | % 3 | % 4 | |
| Male twin | 5233 | 14 | 25 | 11 | 19 | 23 | 8.2 | 5135 | 13 | 7.1 | 42 | 12 | 25 |
| Female twin | 9256 | 17 | 32 | 11 | 16 | 17 | 6.8 | 8939 | 13 | 10 | 44 | 13 | 20 |
| Father | 840 | 14 | 30 | 11 | 17 | 21 | 6.3 | 859 | 15 | 6.3 | 40 | 14 | 26 |
| Mother | 1332 | 17 | 37 | 11 | 16 | 15 | 4.4 | 1361 | 14 | 8 | 45 | 11.2 | 22 |
| Brother | 1211 | 12 | 25 | 11.2 | 19 | 24 | 9.2 | 1188 | 9.4 | 5.7 | 46 | 14 | 25 |
| Sister | 1808 | 17 | 30 | 11.6 | 19 | 16 | 6.2 | 1770 | 11 | 8.2 | 50 | 11.2 | 20 |
| Husband | 2455 | 14 | 29 | 11 | 16 | 21 | 8.4 | 2419 | 14 | 7.2 | 39 | 11.2 | 29 |
| Wife | 1844 | 19 | 33 | 11.7 | 15 | 16 | 5.6 | 1807 | 14 | 7.7 | 42 | 13 | 24 |
| Son | 1817 | 8.4 | 19 | 11.2 | 21 | 25 | 15 | 1822 | 8.7 | 8.7 | 47 | 13 | 22 |
| Daughter | 2732 | 11.6 | 26 | 11.4 | 19 | 21 | 11 | 2711 | 11.1 | 10 | 51 | 13 | 14 |
Maximum likelihood estimation from individual observations
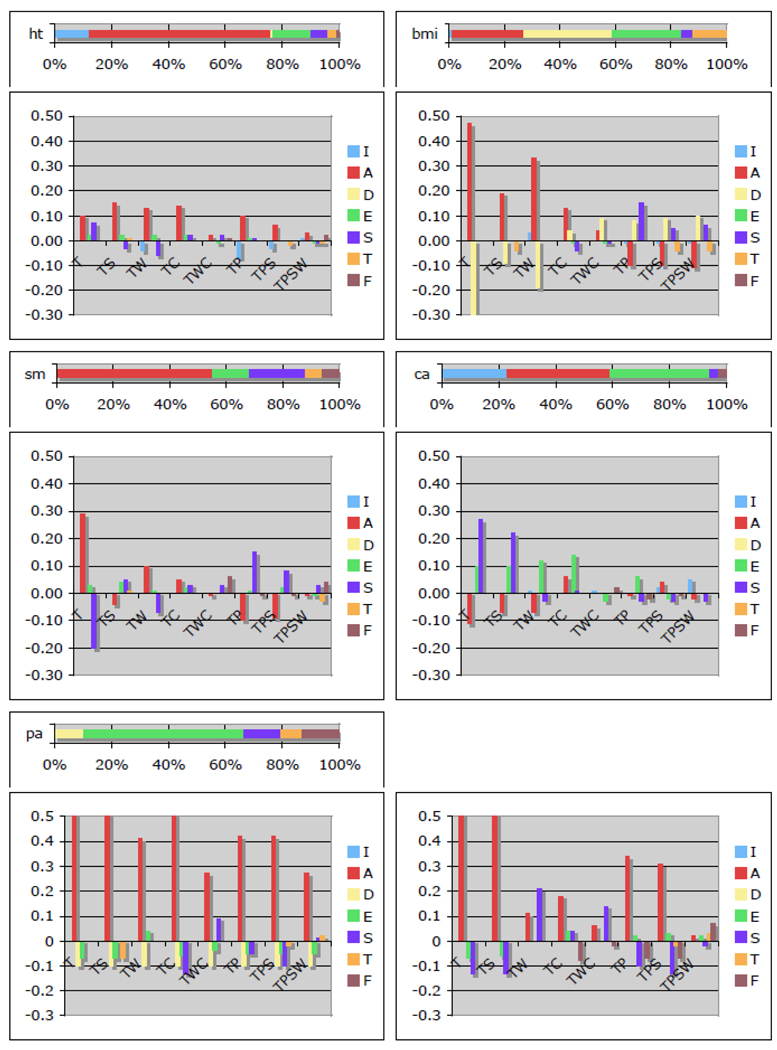
Raw data maximum likelihood methods were used to obtain unbiased estimates of all parameters under the full cascade model and the eight sub-designs. The major sources of variation in the full cascade model are: additive genetic (A), non-additive genetic (D), unique environmental (E), common sibling environmental (C), and twin environmental factors (T), cultural transmission (F) and genotype-environment covariance (GE). Note that GE will be included when both genetic and cultural transmission are present. The 95% confidence intervals could be obtained from Mx using the method of Neale and Miller (1997). We report results from the full model, which may include sources of variance that are not significantly different from zero, to avoid biases of parameters when dropping others from the model. However, when fitting sub-designs, parameters that are not identified by the design have to be dropped. For CTD [referred to as T in figure 1], only ACE (or ADE) factors were estimated. The addition of parents to the twin design [TP] allows the estimation of assortment (I) and F. Adding siblings [TPS] provides an estimate of T. With data on twins, parents, siblings and spouses, [TPSW] either an ACETFI or and ACETDI can be fitted. Augmenting the twin design with additional siblings [TS] provides information to estimate ACET or ADET. Twins and spouses [TW] add an estimate of I onto the traditional ACE or ADE models. Children of twins [TC] allow estimation of ACEF or ACED. The addition of the spouses of twins [TWC] gives a handle on ACEFI or ACEDI. Appendix 1 presents likelihood statistics for each of the models. Estimates of the variance components under different models are presented in Appendix 2. Discrepancies between any of the sub-designs and the full cascade model are presented in Figure 1. As these models allow for sex differences, we have opted to present results for males only as those for females are similar.
Figure 1.
Discrepancies in estimates of variance components compared to those of the cascade model (top graph) by type of relative included in the design and source of variance.
[ht: height, bmi: body mass index, sm: smoking status, ca: church attendance, pa: political affiliation; relative types and their combinations: T: twins, S: sibings, W: spouses, C: children, P: parents sources of variance: I: genetic variance due to assortment, A: additive genetic variance, D: dominance, E: unique environmental variance, S: sibing environmental variance, T: twin environmental variance, F: cultural transmission]
Height
When the full cascade model was fitted to data on height, the majority of the variance was accounted for by additive genetic factors (63%) with an additional proportion (.12) resulting from genetic variance increased through assortment. The remainder of the variance was split between genetic dominance (01%), unique environment (13%), sibling environment (6%), twin environment (3%) and cultural transmission (1%). Phenotypic assortment was a better explanation for the spousal correlation than social homogamy. Fitting any of the reduced models to a subset of the relatives and comparing it the full phenotypic assortment model resulted in an overestimation of A between 2 and 15% and an underestimation (0–7%) of the genetic variance through assortment. Biases in the proportion of variance accounted for by environmental sources was limited (−7 to 7%) with slightly bigger discrepancies for GE covariance in designs that include parent-offspring relationships.
Body Mass Index
Not one source of variance accounted for more than a third of the variance when fitting the full model to log-transformed BMI. As the spousal correlation for BMI was low, models including phenotypic assortment or social homogamy fitted almost equally well. Genetic factors accounted for 58% of the variance of which the largest part is due to dominance. Of the environmental contributions, 25% were unique, 4% shared and 12% twin environment. Cultural transmission was estimated at zero in the full model. In all the sub-designs, dominance and cultural transmission cannot be estimated simultaneously Models without dominance tended to overestimate the contribution of additive genetic factors (by 19–49%) and slightly underestimate most environmental contributions (mostly between −8 to 0%) except for cultural transmission which is overestimated between 3–7%. Models without cultural transmission appeared to overestimate dominance (4–10%) and underestimate additive genetic effects (~-10%) with biases in sibling environment going both ways.
Smoking Status
An ordinal measure was used to represent lifetime tobacco use. For smoking status spousal correlations were significant and the social homogamy fitted consistently better than the phenotypic assortment one. The majority of the variance was accounted for by additive genetic factors (~55%). Twenty percent of the variance was due to shared sibling environment, an additional 6% to twin environment and cultural transmission each, with the remaining 13% to unique environment. Biases observed in fitting models to reduced data varied as a function of the relatives included. When the design included parent-offspring pairs, additive genetic factors were typically underestimated (up to 10%), sibling environment was mostly overestimated (up to 15%), cultural transmission slightly underestimated. When only twins and possibly their spouses were available, the reverse was true meaning that genetic factors were biased upward and sibling environmental factors downward.
Church Attendance
The frequency of attending church was measured on an ordinal scale with 6 categories. Spousal correlations were highly significant, and appeared to be best represented by phenotypic assortment. About 60% of the variance in church attendance could be ascribed to additive genetic contributions of which almost half resulted from the consequences of assortment. The second major source of variance was the unique environment (35%). About three percent of the variance was accounted for by sibling environment and cultural transmission each. The bias in the estimates of variance components from fitting models to subsets of the data were generally small (mostly less than 5% in either direction), except for fitting data to twins only. Genetic factors are underestimated in CTD (−11%) and sibling environment is overestimated (27%).
Political Affiliation
An ordinal measure based on two items was created to reflect political affiliation. Social homogamy explained the observed data slightly better than the phenotypic assortment. Unique environmental factors explained the majority of the variance (55%) following by sibling environment (13%), cultural transmission (13%), genetic dominance (10%) and twin environment (7%). Given none of the sub-designs considered here allow for the simultaneous estimation of dominance and cultural transmission as well as ACE, we fit two series of sub-models, the first without cultural transmission and the second without dominance. Fitting sub-designs without cultural transmission, the additive genetic variance component was biased upwards up to 50%, while all other sources of variance were underestimated. Fitting models to fewer relatives that did not include dominance resulted in overestimation of additive genetic factors, especially in designs with twins (and siblings), and underestimation to a lesser degree of sibling environment, and in some models of cultural transmission. These biases appear substantial, but may be a function of the model for assortment. While the only genetic source of variance under the social homogamy model was dominance explaining 10% of the variance, the results of fitting the phenotypic assortment model suggested a total genetic component of 45% without dominance. Evaluating alternative models of assortment may thus prove important in understanding individual differences.
Discussion
We analyzed five variables with known varying genetic and environmental architecture to illustrate the impact of assumptions and resulting biases when analyzing genetic models to different combinations of relatives. Obviously when fewer relatives types were available for analysis, as in e.g. data on CTD versus additional relatives, discrepancies between estimates of the variance components and the true underlying architecture were greater. If variation in the phenotype of interest was primarily explained by additive genetic and unique environmental factors (as in height or church attendance), biases were relatively minor (less than 10%). Significant assortment that was not taken into account, when no spousal pairs were available, resulted in biases in the estimation of the additive genetic and shared environmental contributions, which vary according to the relative magnitude of all sources of variance. The mechanism of assortment, here we evaluated phenotypic assortment versus social homogamy, also appeared to have an impact on the estimates of both genetic and environmental factors.
When both additive and non-additive genetic contributions were substantial as well as shared environmental factors (sibling and/or twin, as for e.g. body mass index), designs without parent-offspring pairs appeared to overestimate additive genetic factors and underestimated non-additive contributions significantly. On the other hand, designs with parent-offspring pairs underestimated additive genetic sources and overestimated both dominance and shared environment to some extent. Thus sources of variance that are confounded in CTD, as they might have opposite effects to the DZ pairs relative to MZ pairs, require additional relatives for unbiased estimation of genetic and environmental contributions, as noted in Keller & Coventry (2005).
For phenotypes where besides additive genetic and sibling environmental factors, twin environment and/or cultural transmission contribute variance, biases varied according to whether or not the designs include parent-offspring pairs. Fitting CTD resulted in additive genetic sources being biased upward and sibling environment downward, while the opposite occurred when fitting TP designs. Note that there were subtle differences according to which relatives were included. Also note that most of the biases would be within the estimated confidence intervals, which suggests that rather than reporting the point estimates from the best fitting most parsimonious model with any given data set, it may be preferable to report estimates with confidence intervals (even those that include zero) from models that include all sources of variance that can be estimated with the available data. Furthermore, we compare sub-designs here to the full cascade model and refer to discrepancies as biases. The cascade model itself, however, is still a model and may be biased compared to the real world (Keller et al., 2009).
Designs that do not allow simultaneously estimation of additive and non-additive genetic factors and various sources of environmental factors may be particularly prone to biased estimates when the majority of variance is explained by environmental factors. Any design including data on MZ and DZ twins will have typically more power to detect additive genetic sources than shared environmental sources of variance. However, it is important to note that all sources of variance were in some instances overestimated and in other underestimated depending on both the types of relatives included and the true underlying genetic and environmental architecture of the phenotype.
Furthermore, given the complexity of the model and the large number of estimated parameters, caution is needed in the interpretation of any results. Even with a large sample as the Virginia 30,000, information may be limited to estimate some parameters, especially those which are highly correlated or only identified by one or few relationships. We have shown here that even with limited relatives, some parameters can be estimated with limited bias. Others may be more biased or not identified, depending on which relatives are available. Unfortunately, there is not an absolute picture of which parameters are more or less biased, as this depends heavily on the underlying genetic and environmental architecture of the trait. Although we believe that in theory the full cascade model is identified, any particular dataset may not have enough information to identify particular parameters. Also, relatively few samples are available with the full extended kinship data analyzed here. However, increasingly more twin studies include data on other relatives (primarily parents and siblings).
Acknowledgements
This research has been supported by grants AG04954, GM30250, GM32732, AA06781, AA07728, AA07535 and MH40828 from NIH, grant 941177 from the NH&MRC, a gift from R.J.R. Nabisco and grants from the JM Templeton Foundation. The authors would also like to thank the twins and their families for their participation in this project. The first author is supported by grants MH020030, CA093423, DA016977, MH068521, DA018673, VTSF, CA085739, DA022989 and DA024413.
References
- Cloninger CR, Rice J, Reich T. Multifactorial inheritance with cultural transmission and assortative mating. II. A general model of combined polygenic and cultural inheritance. American Journal of Human Genetics. 1979;31:176–198. [PMC free article] [PubMed] [Google Scholar]
- Eaves LJ, Eysenck HJ, Martin NG. Genes, Culture and Personality: An empirical approach. London: Oxford University Press; 1989. [Google Scholar]
- Eaves LJ, Heath AC, Martin NG, Neale MC, Meyer JM, Silberg JL, Corey LA, Truett K, Walters E. Comparing the biological and cultural inheritance of stature and conservatism in the kinships of monozygotic and dizygotic twins. In: Cloninger CR, editor. Proceedings of 1994 APPA Conference; Washington: American Psychiatric Press; 1999. pp. 269–308. [Google Scholar]
- Fulker DW. Genetic and cultural transmission in human behavior. In: Weir SB, Eisen EJ, Goodman MM, Namkoong G, editors. Proceedings of the Second International Conference on Quantitative Genetics. 1988. pp. 318–340. [Google Scholar]
- Galton F. History of Twins, in Inquiries into Human Faculty and its Development. 1875:155–173. [Google Scholar]
- Heath AC, Eaves LJ. Resolving the effects of phenotype and social background on mate selection. Behavior Genetics. 1985;15:15–30. doi: 10.1007/BF01071929. [DOI] [PubMed] [Google Scholar]
- Keller MC, Coventry WL. Quantifying and addressing parameter indeterminancy in the classical twin design. Twin Research and Human Genetics. 2005;8:201–213. doi: 10.1375/1832427054253068. [DOI] [PubMed] [Google Scholar]
- Keller MC, Medland SE, Duncan LE, Hatemi PK, Neale MC, Maes HHM, Eaves LJ. Modeling Extended Twin Family Data I: Description of the Cascade model. Twin Research. 2009 doi: 10.1375/twin.12.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk KM, Maes HH, Neale MC, Heath AC, Martin NG, Eaves LJ. Frequency of church attendance in Australia and Virginia: models of family resemblance. Twin Research. 1999;2:99–107. doi: 10.1375/136905299320565960. [DOI] [PubMed] [Google Scholar]
- Lake RIE, Eaves LJ, Maes HHM, Heath AC, Martin NG. Further evidence against the environmental transmission of individual differences in neuroticism from a collaborative study of 45,850 twins and relatives on two continents. Behavior Genetics. 2000;30:223–233. doi: 10.1023/a:1001918408984. [DOI] [PubMed] [Google Scholar]
- Little RJA, Rubin DB. Statistical analysis with missing data. New York: John Wiley and Sons; 1987. [Google Scholar]
- Maes HH, Neale MC, Martin NG, Heath AC, Eaves LJ. Religious attendance and frequency of alcohol use: same genes or same environments: a bivariate extended tiwn kinship model. Twin Research. 1999;2:169–179. doi: 10.1375/136905299320566031. [DOI] [PubMed] [Google Scholar]
- Maes HH, Sullivan PF, Bulik CM, Neale MC, Prescott CA, Eaves LJ, Kendler KS. A twin study of genetic and environmental influences on tobacco initiation, regular tobacco use and nicotine dependence. Psychological Medicine. 2004;34:1–11. doi: 10.1017/s0033291704002405. [DOI] [PubMed] [Google Scholar]
- Medland SE, Keller MC. Modeling Extended Twin Family Data II: Power associated with different family structures. Twin Research. 2009 doi: 10.1375/twin.12.1.19. [DOI] [PubMed] [Google Scholar]
- Neale MC, Boker SM, Xie G, Maes HH. Mx: Statistical Modelling (6th ed.) Richmond, VA: Department of Psychiatry; 2006. [Google Scholar]
- Neale MC, Cardon LR. Methodology for genetic studies of twins and families. Dortrecht: Kluwer Academic Publishers; 1992. [Google Scholar]
- Neale MC, Miller MB. The use of likelihood-based confidence intervals in genetic models. Behavior Genetics. 1997;27:113–120. doi: 10.1023/a:1025681223921. [DOI] [PubMed] [Google Scholar]
- Neale MC, Walters EE, Eaves LJ, Maes HH, Kendler KS. Multivariate genetic analysis of twin-family data on fears: Mx models. Behavior Genetics. 1994;24:119–139. doi: 10.1007/BF01067816. [DOI] [PubMed] [Google Scholar]
- Truett KR, Eaves LJ, Walters EE, Heath AC, Hewitt JK, Meyer JM, Silberg J, Neale MC, Martin NG, Kendler KS. A model system for analysis of family resemblance in extended kinships of twins. Behavior Genetics. 1994;24:35–49. doi: 10.1007/BF01067927. [DOI] [PubMed] [Google Scholar]
- Vogler GP. Multivariate path analysis of familial resemblance. Genetic Epidemiology. 1985;2:35–53. doi: 10.1002/gepi.1370020105. [DOI] [PubMed] [Google Scholar]