Abstract
Gene duplication is mainly recognized by its primary role in the origin of new genes and functions. However, the idea that gene duplication can be a central player in resolving sexual genetic conflicts, through its potential to generate sex-biased and sex-specifically expressed genes, has been overlooked almost entirely. We review recent data and theory that support gene duplication as a theoretically predicted and experimentally supported means of resolving intralocus sexual antagonism. We believe this role is likely the consequence of sexual conflict for housekeeping genes that are required in both males and females, and that are expressed in sexually dimorphic tissues, i.e., where sexually antagonistic selection is exerted. We think that these genes cannot evolve tissue-specific expression unless they duplicate.
Intralocus sexual antagonism drives genome evolution
In species with two sexes, a single genome encodes for two different organisms, males and females and a large portion of the genes are expressed in both sexes [1]. However, given the ecological, developmental, morphological, physiological, and reproductive differences between sexes, males and females are under distinct selective regimes [2, 3], and hence, the genome that can make well-fitted females is often the one that makes unfitted males [4]. This last observation reveals the existence of intralocus sexually antagonistic variation (i.e., the existence of alleles of genes that are being selected in opposite directions in males and in females). This kind of variation is capable of driving fast genomic changes [5-8]. Notably, intralocus sexually antagonistic variation might be even more prevalent in genomes with heteromorphic sex chromosomes [5, 9, 10] and it should keep shaping genomes in different ways in those genomes. For instance, as we argue below, recent data suggest that sex-specific duplicated genes might often be selected in those genomes to resolve intralocus sexually antagonistic conflicts. We present a detailed model of how this might occur and propose ways to explicitly test the model.
Testis- and sperm-specific gene duplicates: the data
A compilation of examples reveals that a non-random set of genes are being duplicated recurrently some of the time, are evolving testis-specific expression by means of duplication into a new genomic location, and are often evolving under recurrent positive selection or becoming specialized. We argue that these data support the idea that intralocus sexual antagonism is being resolved through gene duplication and are consistent with testis being one of the most sexually antagonistic tissues.
A first very compelling example is the observation that 83% of the nuclearly encoded mitochondrial genes that relocated exhibit testis-specific expression, a pattern that is not shared by the respective parental genes [11]. Significantly, most of these duplicated genes are X-to-autosome or autosome-to-autosome copies and encode for proteins with energy-production functions, while nuclear genes encoding for other mitochondrial functions (e.g., transcription, translation, and biosynthesis) remain in the genome mostly as single, broadly expressed, copies [11]. Gallach et al. [11] suggested that, because sperm have a short life span and will not transfer their mitochondria to the next generation [12], natural selection might favor males which produce large amounts of sperm, or fast sperm, despite the high mutation rate that might be associated with high-energy production [13]. Therefore, while it could be beneficial to decrease the mutation rate in other tissues (i.e., soma and ovary) by preventing the formation of reactive molecules, in the case of sperm, there might be a higher benefit obtained from producing a lot of energy for fertilization, despite the mutations associated with this. Note that this situation would generate a conflict among tissues ("tissue antagonism"; Box 1), under which a certain allelic variant beneficial to a particular tissue(s) (e.g., testis) would increase the fitness of some individuals (males carrying this allele), despite the detrimental effects in other tissues (somatic tissues and ovaries). In agreement with this idea, Gallach et al. [11] concluded that mitochondria with high energy-producing rates could be essential for male competition, but detrimental in females, leading to intralocus sexually antagonistic selection [2]. In addition, Gallach et al. proposed that the observed testis-specific paralogs lead to the resolution of this intralocus sexual antagonism [11]. Some of these paralogs (CG18418 and CG6255) have been evolving under positive selection [14] revealing that selection for a different function of these genes in testis is still been exerted.
Box 1. Intralocus sexual antagonism resolved through gene duplication: a model with strong positive selection at every step.
Here, we introduce a model that depicts a gene that begins to segregate for sexually antagonistic alleles as a consequence of the antagonistic effects of the new allele on different tissues. The resolution of this conflict trough time by gene duplication is outlined.
Gene A is initially monomorphic (only allele A1 is present; Figure 1) and bestows the same fitness to every cell type in both sexes (Time 0, rows A and B). A mutation in the protein-coding region of the gene creates a new allele (allele A2) encoding for a protein variant that increases fitness in testis, with a cost in the soma and the ovary (Time 1, row A). Because natural selection acts at the individual 's level, if the cost in the soma is not very high, A2 might still be a better allele for males than A1, but it will come with a cost for females (Time 1, row B). This new allele causes a conflict between tissues, as well as a conflict between sexes. We do not depict the exact effects of the alleles, nor do we depict their chromosomal location or the dominance of the alleles (see Box 2). We only illustrate that, in general, the effect of A2 depends on the sex and that A2 is a better allele for males and comes with a cost for females. Once the antagonistic allele begins segregating (A2), selection will favor its duplication and relocation (i.e., testis-specific expression and differentiation). Situations analogous to this have been modeled before for cases of heterozygote advantage [15, 16], and it has been emphasized that allelic divergence is expected to promote gene duplication [15]. In addition, the feasibility of this model increases when we consider that the rate at which new gene duplications are introduced every generation is now known to be very high, as revealed by new genomic data [17]. Whenever allele A2 duplicates creating a new gene (gene B) and evolves testis-specific expression (Time 2, row A), selection will favor the individuals (both, males and females) bearing the combination of A1B (Time 2, row B). This situation will lead to the fixation of the allele A1 (gene A) and the duplicated gene B, resolving the sexually antagonistic conflict (Time 3, rows A and B). The outcome will be the presence of the old housekeeping gene (gene A) and a new tissue-specific gene (gene B) as it has been observed in the presented data.
Figure 1 of Box 1.
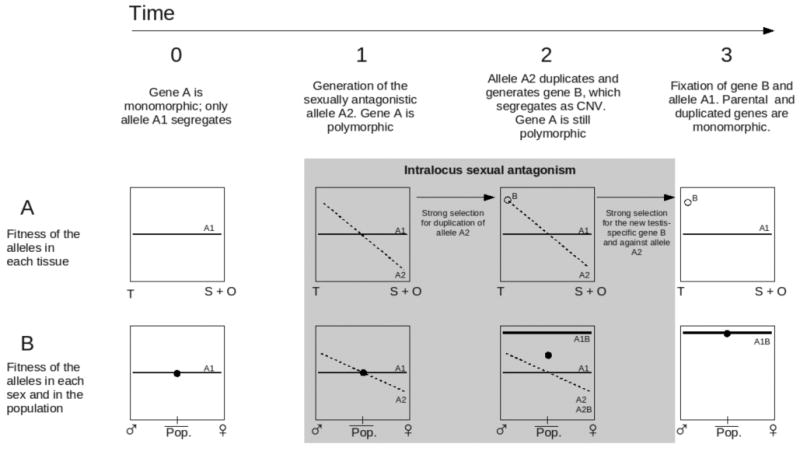
The process of intralocus sexual antagonism resolved through gene duplication is shown. The columns reflect four consecutive events in time while the rows reflect the fitness of thealleles in the tissues (row A) and in the sexes and population (row B). T: fitness of the alleles in testis. S + O: fitness of the alleles in somatic tissues of both sexes and in the ovaries. Pop.: population. Black dot: fitness of the population as a rough estimate of the average fitness between both sexes. White dot: fitness of the duplicated gene B in testis. Note that no line can be represented for the fitness of gene B since gene B is not expressed in somatic tissues or the ovary. Before fixation, gene B segregates as a copy number variant (CNV). Framed in gray, we depict the time gene A is segregating the antagonistic allele.
Box 2. Intralocus sexual antagonism resolved by gene duplication: supporting data for the model, and predictions.
Our duplicative model proposes that intralocus sexual antagonism can be resolved by gene duplication (Box 1). The conditions under which the antagonistic allele will increase in frequency and for how long it will segregate in the population have been studied in detail by other authors [10, 49] and depend on the chromosomal location, the fitness effects and the dominance of the segregating alleles. Importantly, Fry [49] recently discussed previous literature and the conditions for the maintenance of polymorphism under intralocus sexual antagonism and concluded that under equal dominance between sexes, conditions for maintenance of polymorphism are more restrictive for autosomes, in agreement with Rice's [10] conclusions. However, if dominance is sex dependent, partial dominance of A2 (Box 1) in males and of A1 (Box 1) in females can maintain polymorphism for a broad set of selection coefficients for autosomes. In conclusion, these studies showed that sexual antagonism could maintain intralocus variation both in the X chromosome and in autosomes (Box 3).
The predictions of this model are several and can be tested: [1] sexual antagonistic variation should map to housekeeping genes even if they already have produced a sex-specific gene (Box 3), and [2] the study of the function of the duplicated (e.g., testis-specific) genes should reveal a new or specialized function. Interestingly, in organisms with ZW sex determining systems, the heterogametic sex (i.e., females ZW) does not have dosage compensation [50] but still has MSCI [51]. If, as we propose, testis is the most sexually antagonistic tissue, we should find a high number of relocations, particularly (although not exclusively) Z-to-autosome relocations of male-biased and male-specific duplicated genes. Note that an excess of an out-of-the-X pattern of female-biased genes would give support to the MSCI. Therefore, analyzing the genome of ZW organisms may be particularly useful to test not only our model, but also the MSCI model.
Similarly, in mammals, glycolytic enzymes are also known to have testis-specific paralogs (Pgk2 retrogene, Gapdhs, Ldhc, and two, Aldoart1 and Aldoart2, Aldolase retrogenes) [18]. It has been suggested that unique characteristics of the enzymes encoded by these paralogs might be required to localize along the sperm tail [19] to increase the stability of the enzyme until fertilization and/or for sperm metabolism [18]. Interestingly, while purifying selection was inferred acting on Aldoart1 and Aldoart2 protein active sites, positive selection and convergent amino acid substitutions in both enzymes were detected at many other sites [20]. It is known that it is mainly glycolytic enzymes distributed along the longest segment of the flagellum of mammalian sperm, rather than mitochondria metabolism, that contribute most of the ATP needed for sperm motility [18]. We again conjecture that the need for sperm-specific functions likely leads to intralocus sexual antagonism and that gene duplication might allow the resolution of this sexual conflict. Interestingly, Pgk2 has been recurrently retroduplicated [21], arguing in favor of the strength of these selective pressures. Pgk2 is an X-to-autosome copy, but these duplicated glycolytic enzymes are both X-to-autosome and also autosome-to-autosome copies.
Another interesting example involves the Drosophila proteasome, a protein complex involved in protein degradation. In Drosophila melanogaster, 36% (i.e., 12 of 33) of the genes encoding for the 26S proteasome subunit have been observed to undergo multiple relocations (DNA- and RNA-mediated) and further independent evolution of testis-specific expression [22]. Again, these duplicated genes are not only X-to-autosome, but also copies in other directions. The authors of this report argue that a fraction of these genes are probably specialized proteasome proteins for sperm individualization [22].
Recent studies show that two nuclear transport proteins (i.e., Dntf-2 and Ran [23]) have been recurrently retroduplicated to produce testis-biased genes that have evolved under recurrent positive selection [24]. Male germline conflicts related to sexual selection, segregation distortion, and/or parasite-related conflicts [24-26] have been proposed to explain the recurrent fixation, positive selection and losses of some of these genes in some lineages [24]. This example reveals a potential additional source for sexual antagonism: hence, for genes involved in male germline conflicts (possibly Dntf-2 and/or Ran), natural selection might favor different alleles for males and females. Gene duplication and sex-specific expression of the male beneficial allele would again resolve this sexual antagonism ([24] and Box 1).
Interestingly, relocation (both DNA- and RNA-mediated) seems to be repeatedly involved in the above examples and in acquiring testis-specific expression [11, 27-29]. How relocation favors the evolution of this tissue-specific pattern of expression is a question that has been partially answered [29-32]. Insertional biases and a correlation between being expressed in testis and proximity to testis-expressed genes have been proposed. However, although there are testis-expression domains in Drosophila [33] that are probably controlled through different modifications in the structure of the chromatin in testis and somatic tissues [34], it is still unclear how much the chromatin context or bidirectional regulatory regions [32] contribute to these effects. In some instances, the evolution of testis-specific regulatory regions might need to be invoked [30]. In addition, acquiring tissue-specific expression would reduce the pleiotropic constraints over the sex-biased gene [35] and would release the gene from genome-wide sequence homogenizing forces like gene conversion [36], facilitating the evolution of new functions of the relocated sex-specific genes.
We argue that the plethora of testis-specific copies of housekeeping genes that have specialized or are under recurrent positive selection (including duplicates of the TATA-binding protein associated factors, or TAFs, or of telomere capping proteins) [28, 37-42] might have frequently originated by gene duplication to resolve intralocus sexual conflicts. In agreement with our model, data indicate that male-biased genes originate more frequently via gene duplication than non-biased or female-biased genes [37, 41, 43, 44], suggesting that gene duplication is particularly involved in the evolution of new male-biased genes. Interestingly, a big fraction of those male-biased gene duplications are testis-specific duplicates of broadly expressed genes [45].
We propose that the intralocus sexual antagonism often begins in the parental gene via male selection for an allele (i.e., a sexually antagonistic allele) that performs better in testis, but worse in other tissues (i.e., male and female soma and ovaries) and culminates with the fixation of a relocated specialized male-specific gene (Boxes 1 and 2). We do not think that meiotic sex chromosome inactivation (MSCI; [46]), subfunctionalization [47] or selection for higher expression given that males carry only one X chromosome and need to dosage compensate [48] can completely explain the data introduced above. Hence, the MSCI hypothesis (which states that autosomal duplicated genes will be favored if they express in male germline because they will provide an active copy of the gene inactivated by MSCI [27]) cannot explain why there are so many autosome-to-autosome duplications. Also, subfunctionalization does not explain why the partition of expression is always testis vs. all other tissues or even the enrichment and recurrent duplications for certain functions. The need for dosage compensation (DC) cannot explain autosome to autosome relocation patterns or just average level of expression of the parental X-borne genes [11]. Finally, selection for higher expression alone does not explain why duplicated genes often seem to be under recurrent positive selection or to become specialized.
Sexual antagonism and sex-biased expression
The resolution of intralocus sexual antagonism is believed to occur through the evolution of sex-biased expression [10]. In his classic paper, Rice demonstrated that given the hemizygosity in males and longer X chromosome residency in females than in males (2/3 vs. 1/3) both sexes could bring to high frequency new sexually antagonistic alleles despite the harm on the other sex [10]. Males could increase the frequency of sexually antagonistic alleles that are recessive and beneficial to males while females could similarly increase the frequency of sexually antagonistic alleles that are dominant and beneficial to females. This chromosome would, therefore, harbor most intralocus sexually antagonistic variation. Rice also suggested that this intralocus sexual battle would be resolved through the evolution of modifiers of expression that would lower the expression of the particular gene in the harmed sex (i.e., through the origin of a sexually dimorphic trait; Box 3) [10]. However, this idea is currently a topic of debate for several reasons: first, intralocus sexual conflict does not seem to be resolved, but rather to persist [52]; second, most sex-biased expression, particularly male-biased expression, does not appear to evolve by decreasing expression level in one of the sexes [48, 53]; and third, the chromosomes that contain most of the sexually antagonistic variation (e.g., the X chromosome; [9, 10, 54]) would be predicted to accumulate sex-biased genes, and this is not always the case [55-57]. In addition, it has been overlooked that many sexually antagonistic genes might be needed by both sexes, and losing the expression of the gene in one sex is not feasible.
Box 3. Comparison of the resolution of intralocus sexual antagonism through evolution of modifiers of expression and through gene duplication.
Widely expressed (i.e., housekeeping) genes (black lines; Figure 1) performing basic cellular tasks can be located on the X chromosome or on autosomes. At some point, an antagonistic allelic variant of a housekeeping gene will appear, so that this gene will segregate an allele with the original protein function (black dot) and another one with the new antagonistic protein function (blue dot for male-beneficial/female-detrimental allele, and pink dot for female-beneficial/male-detrimental allele). Theory predicts that a large amount of antagonistic alleles will increase in frequency over time and remain polymorphic in the population when located on the X chromosome, while this is much more improbable when located on the autosomes (see Box 2 for details). Under the classic model, the antagonism will be resolved through the evolution of modifiers of expression that will favor sex-biased, and optimally, sex-specific expression (blue and pink lines, for male- and female-biased genes, respectively). Under the duplicative model, the antagonistic allele will duplicate into another chromosomal position, favoring sex-specific expression of the duplicated allele and the fixation of both genes in the population, leading to the resolution of the sexual antagonism (see Box 1 and Figure 1). For this model, two additional features are depicted: 1) sexually antagonistic variation on autosomal genes may also be resolved through the duplication of the sexually antagonistic allele and acquisition of sex-biased expression; and 2) because parental genes keep their function and broad expression pattern, a new antagonistic allele (sky blue dot) may appear, explaining therefore the persistence of the intralocus sexual antagonism, and recurrent duplication can be found (sky blue line).
Figure 1 Box 3.
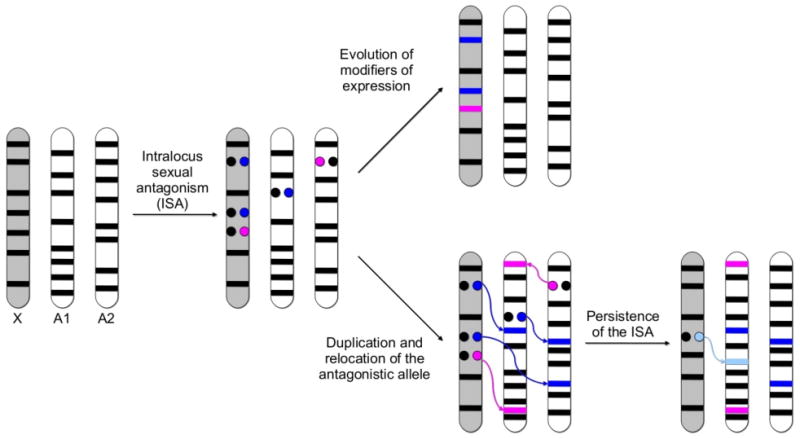
Illustration of two modes of resolution of intralocus sexual antagonism: evolution of modifiers of expression (upper side) and gene duplication (lower side). For simplicity, only the haploid set of chromosomes representing one X chromosome (gray bar) and two autosomes (white bar) are shown.
While Rice's model might apply to genes that can lose or decrease expression in one sex (i.e., sex-specific “dispensable” genes), the conflict might instead be resolved as we suggest above through the duplication of the antagonistic allele in other instances (see Box 3 for differences between the classic model and our model). Other models that include gene duplication have been suggested to resolve sexual antagonism but all of them involve the creation of a male and a female gene [2, 3, 58, 59]. In one of them [59], gene duplication has been even proposed to explain MSCI, but does not account for the continuous duplication of genes after the heteromorphic sex chromosomes and MSCI have evolved or for duplications from autosome to autosome. Our hypothesis predicts an excess of X-to-autosome duplications for sexually antagonistic genes, but in addition, because many housekeeping genes are located on autosomes, we also predict autosome-to-autosome duplications of sexually antagonistic genes (Box 2 and 3). In consequence, our model may explain many gene duplication cases for which MSCI, or level of expression limitations (i.e., DC) are not satisfactory. The proposed model might be the best explanation for gene duplication patterns, particularly when other hypotheses, such as MSCI [46], subfunctionalization [47] or DC [48] cannot satisfactorily explain the observed data. We do not think that our hypothesis replaces, but instead, might complement other hypotheses.
In the following, we briefly introduce why we think that tissue sexual identity (i.e., the fact that some sex determination cascades are triggered only in particular tissues) and sexual antagonism are intimately related biological phenomena.
Tissue dimorphism, sex determination and sexually antagonistic conflict
Data from Drosophila suggest that sex-biased expression is mostly due to shifts in particular tissues (largely testis-specific genes [41]), instead of shifts in expression that affect the whole body. Why do only some tissues contribute to sex-biased expression? In Drosophila, the ratio of X chromosomes to autosomes determines the absence or presence of the Sex lethal (Sxl) protein. In the absence of Sxl protein, the dosage compensation system is activated. In parallel, Sxl also triggers male- or female-specific pathways ending in sex-specific isoforms of the doublesex (dsx) and fruitless (fru) proteins that confer sexual identity to male and female cells [60, 61]. Interestingly, however, not all tissues express dsx or fru, so that, although most cells “know” how many X chromosomes they have, not all of them can translate the X chromosome number into sexual identity. Robinett et al. [61] recently showed that dsx is expressed in almost all of the cells of only a few tissues (basically, genitalia, brain and central nervous system, and fat body), but in only a few or no cells in the other tissues. In other words, only these tissues, as a whole, “know” their sexual identity.
We suggest that sexual antagonism may appear in tissues with or without dsx or fru expression, but it will be resolved in those tissues that are already transcribing these genes (i.e., sexually differentiated) or that will end up recruiting these pathways. The overlap between tissues exhibiting sex-biased or sex-specific expression and dsx or fru expression makes perfect biological sense because the tissue first needs to trigger a sex-specific cascade to later transcribe a sex-specific gene. In other words, for a tissue to express a gene male (female)-specifically, it must first know it is a male (female) tissue. Therefore, we think that the sex determination pathways are important to understanding the evolution and resolution of the sexually antagonistic conflict.
The testis is a tissue where these sex determination pathways are triggered and is very different from other tissues. The testis must make sperm, and it is under strong selective pressures from sexual selection, parasite-related conflicts and segregation distortion to specialize and evolve quickly. Therefore, testis probably generates most of the antagonistic conflicts among tissues [59], and consequently, it is likely the most sexually antagonistic tissue, which explains the amount of sex-biased genes expressed there and the amount of duplicated genes. While control of parasite-related conflicts and segregation distortion will also occur in ovaries and this might exert strong selection in both testis and ovaries, sexual selection might be stronger in males [37, 62], and it is likely to lead to stronger selection and tissue antagonism in testes than in ovaries.
Conclusions and further analyses
In this review, we propose a model and introduce some available experimental data suggesting that gene duplication can act as a resolution mechanism for intralocus sexual conflict. Consistent with the possibility that these new duplicated genes resolve sexual conflicts, most of them exhibit sex-specific functions. Additionally, most of these genes not only have sex-specific, but also tissue-specific function. Interestingly, only a few tissues have the potential to establish a sexual cellular identity in Drosophila, and only those tissues will be able to evolve sex-biased or sex-specific expression. We suggest that these tissues, in particular the testis, are the source of sexually antagonistic conflict and also the place where the conflict will be resolved, leading to tissue-specific expression.
Several predictions emerge from the data, the model and our views presented in this article. We observe many housekeeping genes being duplicated and evolving testis-specific expression, and we predict that much of the sexually antagonistic variation will map to genes that are housekeeping genes involved in conflicts that do not have a sex-specific duplicate. However, this prediction remains to be tested. In addition, if sexually antagonistic conflicts are resolved through relocated sex-specific duplicates, then antagonistic variation will often map to a different location than sex-biased expression and the location of sex-biased expression will not be a good predictor of the location of sexually antagonistic genes (Box 3). This is in agreement with recent results [54] and in contrast with previous models and assumptions [10, 63, 64].
Theoretically, both the X chromosome and autosomes can support antagonistic variation (Box 2). However, the data seem to indicate that, in Drosophila, a higher proportion of antagonistic variation maps to the X chromosome [9, 54]. Under our model and hypothesis about what tissues contribute the most to the antagonism, the conflict will often be resolved through duplication of the antagonistic allele and evolution of testis-specific expression of the duplicate. Because the X chromosome holds most antagonistic alleles, most relocated genes will be generated from the X, contributing to explain the autosomal location of male-biased genes and the out-of-the-X pattern [27, 65].
According to our model, the initial sexual antagonism is erased after the duplication of the antagonistic allele, making this model difficult to prove; but some predictions can be tested as mentioned above (see also Box 2). However, what proportion of sexually antagonistic conflict is resolved through gene duplication remains to be determined and this will not be easy to estimate. Given the amount of male-specific duplicated genes ([24, 37-39, 41]; Box 2) and their features (i.e., a non-random set of genes are being duplicated sometimes recurrently and often from autosome to autosome, evolving testis-specific expression, often evolving under recurrent positive selection or becoming specialized and eventually being lost sometimes), it would seem that a significant part of gene duplications occur to resolve sexual conflicts. Importantly, a large part of the intralocus sexual antagonism is likely driven by recurrent selective pressures in male germline (sexual selection, parasite-related conflicts and segregation distortion), and hence the intralocus sexual conflict emerging from these conflicts might be resolved only transitorily (Box 3), explaining its persistence [52] and leading to the rapid turnover of male-specific duplicates [24] and male-biased expression [37, 41]. Therefore, observations made at a particular point in time likely represent an underestimation of how often intralocus sexual antagonism is resolved through gene duplication.
Acknowledgments
We would like to thank Jeff Demuth, Bill Rice, three anonymous reviewers for reading this work and providing feedback, and Jennifer Gage for proofreading the manuscript. This research was supported by UTA startup funds and grant GM071813 from the National Institutes of Health, USA to Esther Betrán.
Glossary
- Intralocus sexual antagonism
conflict that occurs when alleles of a gene (locus) encoding different isoforms are selected in opposite directions in males and in females
- Tissue antagonism
conflict that occurs when alleles of a gene (locus) encoding different isoforms have different effects in different tissues (i.e., the allele that functions better in a tissue is the allele that performs the worst in another tissue and vice versa). Tissue antagonism can lead to sexual antagonism, if the tissue specific advantage translates also in a sex specific advantage
- Gene duplication
process that leads to additional copies of genes in the genomes it can be DNA- or RNA-mediated if the mechanism involves only DNA or involves RNA respectively
- Paralogs
genes that originated through gene duplication
- Retroduplication
RNA-mediated duplication duplication mechanism that involves the retrotranscription of an mRNA and insertion in the genome
- Retrogene
gene that originated trough retroduplication
- Sperm individualization
process that leads to the formation of individual sperm cells from previously interconnected spermatides via cytoplasmic bridges
- Relocation
event of gene duplication to a different genomic location while the parental gene is preserved it can be DNA- or RNA-mediated
- Bidirectional regulatory regions
regulatory regions that can drive expression of flanking genes in both directions
- Meiotic sex chromosome inactivation (MSCI)
premature inactivation of the sex chromosome during meiosis in the germline of the heterogametic sex (XY males and ZW females)
- Subfunctionalization
neutral partition of the broad pattern of expression of a gene in two genes after gene duplication
- Dosage compensation (DC)
regulatory mechanism of the gene expression used in many XY organisms to achieve the same transcriptional level of the X-linked genes between sexes despite the presence of different number of sex chromosomes in males (XY) and females (XX)
- Hemizygosity
state of the X or Z chromosome in the heterogametic sex characterized by the presence of only one copy of the genes in that chromosome and by the observation of recessive effects of those genes
- Chromosome residency
time that a chromosome spends in males and females, which is the same in the case of autosomes. On the contrary, sex chromosomes (i.e., X and Z) spend twice as much time in the homogametic (i.e., XX and ZZ) than in the heterogametic sex (XY and ZW)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parisi M, et al. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5:R40. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Doorn GS. Intralocus sexual conflict. Ann N Y Acad Sci. 2009;1168:52–71. doi: 10.1111/j.1749-6632.2009.04573.x. [DOI] [PubMed] [Google Scholar]
- 3.Bonduriansky R, Chenoweth SF. Intralocus sexual conflict. Trends Ecol Evol. 2009;24:280–288. doi: 10.1016/j.tree.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Chippindale AK, et al. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc Natl Acad Sci U S A. 2001;98:1671–1675. doi: 10.1073/pnas.041378098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice WR. Sexually antagonistic genes: experimental evidence. Science. 1992;256:1436–1439. doi: 10.1126/science.1604317. [DOI] [PubMed] [Google Scholar]
- 6.Rice WR. The accumulation of sexually antagonistic genes as a selective agent promotig the evolution of reduced recombination between primitive sex chromosomes. Evolution. 1987;41:911–914. doi: 10.1111/j.1558-5646.1987.tb05864.x. [DOI] [PubMed] [Google Scholar]
- 7.Rice WR. Evolution of the Y sex chromosome in animals. BioScience. 1996;46:331–343. [Google Scholar]
- 8.van Doorn GS, Kirkpatrick M. Turnover of sex chromosomes induced by sexual conflict. Nature. 2007;449:909–912. doi: 10.1038/nature06178. [DOI] [PubMed] [Google Scholar]
- 9.Gibson JR, et al. The X chromosome is a hot spot for sexually antagonistic fitness variation. Proc R Soc Lond B Biol Sci. 2002;269:499–505. doi: 10.1098/rspb.2001.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 11.Gallach M, et al. Analyses of nuclearly-encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexual antagonistic conflict in Drosophila. Genome Biol Evol. 2010;2:835–850. doi: 10.1093/gbe/evq069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen JF. Separate sexes and the mitochondrial theory of ageing. J Theor Biol. 1996;180:135–140. doi: 10.1006/jtbi.1996.0089. [DOI] [PubMed] [Google Scholar]
- 13.Blumenstiel JP. Sperm competition can drive a male-biased mutation rate. J Theor Biol. 2007;249:624–632. doi: 10.1016/j.jtbi.2007.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pröschel M, et al. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics. 2006;174:893–900. doi: 10.1534/genetics.106.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proulx SR, Phillips PC. Allelic divergence precedes and promotes gene duplication. Evolution. 2006;60:881–892. [PubMed] [Google Scholar]
- 16.Spofford JB. Heterosis and evolution of duplications. Amer Natural. 1969;103:407–432. [Google Scholar]
- 17.Zhang F, et al. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danshina PV, et al. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol Reprod. 2010;82:136–145. doi: 10.1095/biolreprod.109.079699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krisfalusi M, et al. Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa. Biol Reprod. 2006;75:270–278. doi: 10.1095/biolreprod.105.049684. [DOI] [PubMed] [Google Scholar]
- 20.Vemuganti SA, et al. Three male germline-specific aldolase A isozymes are generated by alternative splicing and retrotransposition. Dev Biol. 2007;309:18–31. doi: 10.1016/j.ydbio.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Potrzebowski L, et al. The emergence of new genes on the young therian X. Trends Genet. 2010;26:1–4. doi: 10.1016/j.tig.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Belote JM, Zhong D. Duplicated proteasome subunit genes in Drosophila and their roles in spermatogenesis. Heredity. 2009;103:23–31. doi: 10.1038/hdy.2009.23. [DOI] [PubMed] [Google Scholar]
- 23.Bai Y, et al. Comparative Genomics Reveals a Constant Rate of Origination and Convergent Acquisition of Functional Retrogenes in Drosophila. Genome Biology. 2007;8:R11. doi: 10.1186/gb-2007-8-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tracy C, et al. Convergently recruited nuclear transport retrogenes are male biased in expression and evolving under positive selection in Drosophila. Genetics. 2010;184:1067–1076. doi: 10.1534/genetics.109.113522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Presgraves DC. Does genetic conflict drive rapid molecular evolution of nuclear transport genes in Drosophila? Bioessays. 2007;29:386–391. doi: 10.1002/bies.20555. [DOI] [PubMed] [Google Scholar]
- 26.Presgraves DC, Stephan W. Pervasive adaptive evolution among interactors of the Drosophila hybrid inviability gene, Nup96. Mol Biol Evol. 2007;24:306–314. doi: 10.1093/molbev/msl157. [DOI] [PubMed] [Google Scholar]
- 27.Betran E, et al. Retroposed New Genes Out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vibranovski MD, et al. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 2009;19:897–903. doi: 10.1101/gr.088609.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vinckenbosch N, et al. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai Y, et al. Evolutionary origin of regulatory regions of retrogenes in Drosophila. BMC Genomics. 2008;9:241. doi: 10.1186/1471-2164-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorus S, et al. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nature Genetics. 2006;38:1440–1445. doi: 10.1038/ng1915. [DOI] [PubMed] [Google Scholar]
- 32.Loppin B, et al. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr Biol. 2005;15:87–93. doi: 10.1016/j.cub.2004.12.071. [DOI] [PubMed] [Google Scholar]
- 33.Boutanaev AM, et al. Large clusters of co-expressed genes in the Drosophila genome. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- 34.Kalmykova AI, et al. Regulated chromatin domain comprising cluster of co-expressed genes in Drosophila melanogaster. Nucleic Acids Res. 2005;33:1435–1444. doi: 10.1093/nar/gki281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mank JE, et al. Pleiotropic constraint hampers the resolution of sexual antagonism in vertebrate gene expression. Am Nat. 2008;171:35–43. doi: 10.1086/523954. [DOI] [PubMed] [Google Scholar]
- 36.Casola C, et al. Nonallelic gene conversion in the genus Drosophila. Genetics. 2010;185:95–103. doi: 10.1534/genetics.110.115444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haerty W, et al. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics. 2007;177:1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hahn MW, et al. Gene Family Evolution across 12 Drosophila Genomes. PLoS Genetics. 2007;3:e197. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaessmann H. Origins, evolution and phenotypic impact of new genes. Genome Res. 2010;20:1313–1326. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li VC, et al. Molecular evolution of the testis TAFs of Drosophila. Mol Biol Evol. 2009;26:1103–1116. doi: 10.1093/molbev/msp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, et al. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450:233–337. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dubruille R, et al. Specialization of a Drosophila capping protein essential for the protection of sperm telomeres. Curr Biol. 2010;20:2090–2099. doi: 10.1016/j.cub.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 43.Arbeitman MN, et al. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- 44.Gnad F, Parsch J. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics. 2006;22:2577–2579. doi: 10.1093/bioinformatics/btl422. [DOI] [PubMed] [Google Scholar]
- 45.Mikhaylova LM, et al. Analysis of the Drosophila melanogaster testes transcriptome reveals coordinate regulation of paralogous genes. Genetics. 2008;179:305–315. doi: 10.1534/genetics.107.080267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- 47.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vicoso B, Charlesworth B. The deficit of male-biased genes on the D. melanogaster X chromosome is expression-dependent: a consequence of dosage compensation? J Mol Evol. 2009;68:576–583. doi: 10.1007/s00239-009-9235-4. [DOI] [PubMed] [Google Scholar]
- 49.Fry JD. The genomic location of sexually antagonistic variation: some cautionary comments. Evolution. 2010;64:1510–1516. doi: 10.1111/j.1558-5646.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellegren H, et al. Faced with inequality: chicken do not have a general dosage compensation of sex-linked genes. BMC Biol. 2007;5:40. doi: 10.1186/1741-7007-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schoenmakers S, et al. Female meiotic sex chromosome inactivation in chicken. PLOS Genet. 2009;5:e1000466. doi: 10.1371/journal.pgen.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pischedda A, Chippindale AK. Intralocus sexual conflict diminishes the benefits of sexual selection. PLoS Biol. 2006;4:e356. doi: 10.1371/journal.pbio.0040356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Connallon T, Knowles LL. Intergenomic conflict revealed by patterns of sex-biased gene expression. Trends Genet. 2005;21:495–499. doi: 10.1016/j.tig.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 54.Innocenti P, Morrow EH. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol. 2010;8:e1000335. doi: 10.1371/journal.pbio.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mank JE, et al. Ontogenetic complexity of sexual dimorphism and sex-specific selection. Mol Biol Evol. 2010;27:1570–1578. doi: 10.1093/molbev/msq042. [DOI] [PubMed] [Google Scholar]
- 56.Reinke V, et al. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 57.Sturgill D, et al. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rice WR, Chippindale AK. The evolution of hybrid infertility: perpetual coevolution between gender-specific and sexually antagonistic genes. Genetica. 2002;116:179–188. [PubMed] [Google Scholar]
- 59.Wu CI, Xu EY. Sexual antagonism and X inactivation - the SAXI hypothesis. TIGS. 2003;19:243–247. doi: 10.1016/s0168-9525(03)00058-1. [DOI] [PubMed] [Google Scholar]
- 60.Camara N, et al. The creation of sexual dimorphism in the Drosophila soma. Curr Top Dev Biol. 2008;83:65–107. doi: 10.1016/S0070-2153(08)00403-1. [DOI] [PubMed] [Google Scholar]
- 61.Robinett CC, et al. Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 2010;8:e1000365. doi: 10.1371/journal.pbio.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mallet MA, Chippindale AK. Inbreeding reveals stronger net selection on Drosophila melanogaster males: implications for mutation load and the fitness of sexual females. Heredity. 2011 doi: 10.1038/hdy.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mank JE, Ellegren H. Sex-linkage of sexually antagonistic genes is predicted by female, but not male, effects in birds. Evolution. 2009;63:1464–1472. doi: 10.1111/j.1558-5646.2009.00618.x. [DOI] [PubMed] [Google Scholar]
- 64.Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- 65.Emerson JJ, et al. Extensive Gene Traffic on the Mammalian X Chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]


