Abstract
The concept of a CpG island methylator phenotype, or CIMP, quickly became the focus of several colorectal cancer studies describing its clinical and pathological features after its introduction in 1999 by Toyota and colleagues. Further characterization of CIMP in tumors lead to widespread acceptance of the concept, as expressed by Shen and Issa in their 2005 editorial, “CIMP, at last.” Since that time, extensive research efforts have brought great insights into the epidemiology and prognosis of CIMP+ tumors and other epigenetic mechanisms underlying tumorigenesis. With the advances in technology and subsequent cataloging of the human methylome in cancer and normal tissue, new directions in research to understand CIMP and its role in complex biological systems yield hope for future epigenetically based diagnostics and treatments.
1. Introduction
In the October 29, 2010 issue, Science turned the spotlight on epigenetics—a term that encompasses histone modification, nucleosome location, noncoding RNA, and DNA methylation. Epigenetic processes do not involve changes to DNA sequence but rather are self-propagating molecular signatures that are potentially reversible, unlike changes in genetic information [1]. DNA methylation is the most widely studied epigenetic marker [2]. The discovery of global DNA hypomethylation in human tumors was followed by the identification of hypermethylated tumor-suppressor genes and recently, inactivation of microRNA (miRNA) by DNA methylation also has been described [3–5]. Growing interest in epigenetic systems stems from an inability to determine causative genetic variants in many disorders. It is hoped that a better understanding of these systems may provide insight into our understanding of complex diseases such as cancer.
Approximately half of all protein-encoding genes in the human genome contain CG-rich regions in their promoters or CpG islands. Aberrant DNA methylation, in the form of hypermethylation of CpG islands, results in repression of transcription in tumor suppressor genes. For example, inactivation of the mismatch repair gene MLH1 by promoter methylation is the molecular basis for microsatellite instability in sporadic microsatellite unstable colon cancers [6]. This phenomenon of tumor alteration via epigenetic silencing associated with dense hypermethylation of CpG islands, and their complex interplay with modifications in histone structure, provides an alternate mechanism to genetic inactivation of tumor suppressor genes via loss or mutation [2]. The presence of widespread CpG island methylation in a tumor is termed the CpG island methylator phenotype, or CIMP, and this paper is focused on this specific aspect of epigenetics.
2. CIMP in Colorectal Cancer
The role of CIMP in colorectal carcinogenesis was originally postulated in 1999 by Toyota et al. [7]. Their pioneering study distinguished between age-related and cancer-related methylation and defined CIMP in terms of the latter. Recognition of CIMP as a phenomenon in colorectal cancer is relatively recent; more than a decade earlier, in 1988, Vogelstein and colleagues developed a model that hypothesized that colorectal neoplasia occurs from a series of genetic alterations that includes activation of oncogenes and inactivation of tumor suppressor genes [8]. The concept of an epigenetic etiology in cancer introduced in the late 1990s was met with some controversy and resistance in the field of carcinogenesis [6, 9, 10]. However, the existence of CIMP has since gained wide acceptance, as the epidemiology characterizing this epigenetic alteration and its utility in understanding carcinogenic pathways support its significance in colorectal cancer biology [11–14].
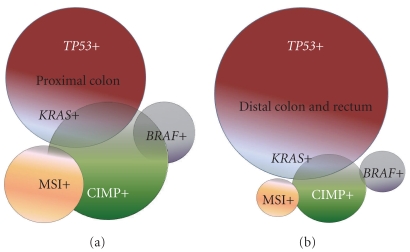
Most sporadic microsatellite unstable colon tumors are CIMP positive, whereas CIMP is uncommon in Lynch syndrome-associated cancers which exhibit microsatellite instability (MSI), indicative of distinct underlying molecular processes [13, 15]. Based on a number of relatively large case-control and prospective cohort studies, ~30–40% of sporadic proximal-site colon cancers are CIMP positive, compared to 3–12% of distal colon and rectal cancers [16–21] as illustrated in Figure 1. Thus CIMP is significantly more frequent in tumors of the proximal colon, and this is independent of MSI status. CIMP is also associated with BRAF mutations in both microsatellite stable and unstable colon cancers [11, 18, 20]. CIMP is observed in proximal hyperplastic (serrated) polyps, suggesting this lesion may be a precursor to unstable tumors (and perhaps stable tumors) in the CIMP high pathway [22]. Once thought to lack potential for malignant progression, hyperplastic polyps are now considered to represent a heterogeneous group, most of which harbor BRAF mutations and some of which exhibit epigenetic alterations (both uncommon in colorectal adenomas). A subset of hyperplastic polyps has been defined by architectural features and renamed sessile serrated polyps (or sessile serrated sessile adenomas). Most of these polyps are right-sided and many show CIMP, supporting the notion that they may be a precursor lesion for CIMP high tumors [23, 24].
Figure 1.
In colorectal cancer, CIMP+ occurs more frequently in tumors of the proximal colon (Figure 1(a)) and less frequently in tumors of the distal colon and rectum (Figure 1(b)). An approximate distribution of genetic and epigenetic tumor alterations is shown.
Some case-control and cohort studies have reported a poor prognosis associated with CIMP in combination with microsatellite stable tumors [19, 25–27], although this may reflect the co-occurrence of BRAF V600E mutations, which have been associated with significantly poorer survival in colon cancer [28, 29]. Relatively minor effects of CIMP on prognosis suggest that the effect of mutations in BRAF on survival in stable tumors is not dependent on CIMP [16, 28]. Indeed, Ogino et al. reported that CIMP-high appears to be an independent predictor of a low colon cancer-specific mortality [30]. These results suggest the need for a large sample size to determine the relative contributions of BRAF and CIMP on prognosis.
3. Characterization of CIMP
In contrast to the relatively straightforward determination of MSI tumor status, a consensus as to the optimal panel of CpG sites for CIMP determination is only starting to take shape (Table 1). Different panels may yield slightly different results, although a strong relationship to the presence of a BRAF V600E mutation is consistently observed with all panels. The so-called “classic” panel of Park et al. utilized to assess CIMP status consists of CpG sites in MLH1, CDKN2A (p16), and methylated in tumors (MINTS) 1, 2, and 31 [31]. It has been suggested that there are two general types of CIMP in sporadic tumors: CIMP high, related to BRAF mutations and MLH1 methylation; and CIMP low, related to KRAS mutations [12, 32, 33]. Tumors characterized as CIMP positive (CIMP+) based on the classic panel include both CIMP high and CIMP low categories; therefore, a subset of CIMP+ associates with BRAF and another with KRAS mutations, somewhat surprising given mutations in these genes are typically mutually exclusive since both are members of the ras signal transduction pathway [6, 11].
Table 1.
A history of CIMP panels used to assess CpG island methylation in colorectal cancer.
| Study | CIMP panel markers | Notes |
|---|---|---|
| Toyota et al. [7] | CDKN2A (p16), MINT1, MINT2, MINT12, MINT17, MINT25, MINT27, MINT31, MLH1, THBS1 | Pioneering work to identify markers that distinguish CIMP from age-related methylation |
| Park et al. [31] | CDKN2A, MINT1, MINT2, MINT31, MLH1 | So-called “classic” or traditional panel |
| Weisenberger et al. [13] | CACNA1G, IGF2, NEUROG1, RUNX3, SOCS1 | “New” panel based on stepwise screen of 195 markers |
| Ogino et al. [34] | CACNA1G, CDKN2A, CRABP1, MLH1, NEUROG1 | Selected markers to distinguish high-level from low-level methylation |
| Shen et al. [33] | CIMP1: MINT1, MLH1, RIZ1, TIMP3, BRAF mutation; CIMP2: MINT2, MINT27, MINT31, Megalin, KRAS mutation | Examined 27 CpG sites, proposed optimal epigenetic and genetic markers to identify CIMP1, CIMP2, or CIMP- |
| Tanaka et al. [35] | CACNA1G, CDKN2A, CHFR, CRABP1, HIC1, IGF2, IGFBP3, MGMT, MINT1, MINT31, NEUROG1, p14, RUNX3, SOCS1, WRN | Correlation structures of markers and CIMP differ by KRAS and BRAF status |
| Ang et al. [36] | Total of 202 CpG sites differentially methylated between tumor and normal | Comprehensive DNA methylation profiling in 807 cancer genes |
| Kaneda and Yagi [37] | Group 1: IGF2, LOX, MINT1, MINT2, MINT31, MLH1, RUNX3, SOCS1; Group 2: ADAMTS1, DUSP26, EDIL3, ELMO1, FBN2, HAND1, IGFBP3, NEUROG1, RASSF2, STOX2, THBD, UCHL1 | Comprehensive DNA epigenotyping of genomewide regions indentified two groups (high and intermediate to low methylation) |
Based on a systematic screen of 195 CpG sites and an unsupervised two-dimensional cluster analysis, Weisenberger et al. proposed a robust alternative to the classic panel to classify CIMP+ tumors consisting of CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1 [13]; CIMP as defined by this panel did not show a relationship to KRAS. Using quantitative real-time PCR, Ogino et al. [34] selected a panel of markers to distinguish high from low levels of methylation including MLH1 and CDKN2A, and three markers that differ from the classic panel: CACNA1G, CRABP1, and NEUROG1. Shen et al. examined genetic markers (BRAF, KRAS) and epigenetic markers at 27 promoter-associated CpG sites using clustering analysis to identify two distinct CIMP+ groups: CIMP1, characterized by MSI+ and BRAF mutations, and MINT1, MLH1, RIZ1, TIMP3 methylation; and CIMP2, characterized by KRAS mutations and methylation of several MINT markers [33].
Using structural equation modeling to construct causal models of CIMP and locus-specific CpG island methylation and a large database of colorectal cancers, Tanaka et al. showed the correlation structures of 16 methylation markers and CIMP status differed between BRAF mutated, KRAS mutated, and wildtype BRAF/KRAS tumors [35]. They suggested a possible role of these mutations differentially modifying the propensity for locus-specific methylation at the cellular level. To examine the question of whether or not BRAF V600E plays a causal role in the development of CIMP, Hinoue et al. determined 100 CIMP-associated CpG sites and examined changes in DNA methylation in eight stably transfected clones over multiple passages [38]. They observed that BRAF was not sufficient to induce CIMP in their system.
In contrast to evaluation of relatively small sets of CIMP markers, comprehensive DNA methylation profiling and unsupervised hierarchical clustering were recently used to identify several CpG sites that were differentially methylated between tumor and normal tissue [36]. Using a similar approach, the use of two methylation panels as classifiers of colorectal cancer has been proposed: the first to identify highly methylated tumors (strongly correlated with BRAF) and a second to distinguish between intermediate (associated with KRAS) and low methylation groups [37]. Since epigenetic therapy is in clinical use or trials for several cancers, efficient methods for epigenetic profiling are needed; Kondo and Issa provide a summary of available profiling techniques and their features [39]. As approaches to CIMP characterization in colorectal cancer continue to evolve, it is clear that BRAF and KRAS oncogene mutations will continue to refine any definition of CIMP. Although characterization of CIMP status depends on methylation markers and criteria used, classification of tumors by both CIMP and MSI status recently proposed by Jass [40] and further refined by Ogino and Goel [14] has become an increasingly common strategy to define the pathological and clinical features of colorectal cancer.
4. Epidemiology of CIMP
4.1. Characteristics
Relationships between CIMP and clinicopathologic features of colorectal tumors that have been widely reported include proximal location, older age at diagnosis, female gender, poor tumor differentiation, MSI (CIMP high cancer), BRAF mutations, KRAS mutations (microsatellite stable cancer), and wildtype TP53 [11, 13, 16, 17, 34, 41]. Based on a large Australian cohort, English et al. reported that individuals of southern European ethnicity had lower risk of CIMP and BRAF mutation than those with origins in northern Europe [21].
Using actual data and a classification tree method to visualize carcinogenic pathways, Slattery et al. suggested that unique mutational pathways to colon and rectal cancer likely exist [18]. This method describes the probability of developing various alterations in proximal colon, distal colon, or rectal tumors given previously acquired mutations. Using bootstrap resampling, the probabilities of developing specific mutations differed across tumor sites. For example, the estimated proportion of tumors that will develop methylation at CpG sites decreased as one goes from proximal colon to rectal cancers. Regardless of site, a methylation pathway in which BRAF is subsequently acquired independent of MSI or MLH1 methylation was predicted. This work supports previous observations that link CIMP and BRAF mutation, independent of MSI status [11, 12].
4.2. Smoking
The presence of methylation in human malignancies bears a relationship to a history of cigarette smoking. Cigarette smoking has been associated with CpG island methylation within the bronchial epithelium of smokers and in lung cancer, and activation of the aromatic hydrocarbon receptor by cigarette smoke has been associated with CpG methylation [42–44]. A significant relationship has been reported between cigarette smoking and CIMP (and closely related mutations in BRAF) in colon and rectal carcinomas in both prospective cohort and case-control studies [45–48]. Interestingly, the relationship between smoking and CIMP provides an explanation for the previously observed association between cigarette smoking and MSI, as most of these tumors also exhibit CIMP [49]. Evidence also suggests that cigarette smoking (related to CIMP and BRAF) may be associated with hyperplastic polyps rather than adenomatous polyps; as mentioned above, a subset of hyperplastic polyps has been hypothesized to be the precursor to CIMP high colorectal carcinomas [50].
4.3. Other Risk Factors
S Adenosylmethionine (SAM), the universal donor of methyl groups in humans, and S Adenosylhomocysteine (SAH), the product of and an inhibitor of DNA methyltransferase (DNMT) enzymes, provide connections between folate metabolism and DNA methylation [51]. It has been hypothesized that diets low in folate and high in alcohol intake may disturb DNA methylation, which may result in global DNA hypomethylation concurrently with a greater risk of cancers with CpG island methylation [52, 53]. In contrast, other studies have shown that global DNA hypomethylation is inversely correlated with CIMP and may represent different pathways to colorectal cancer [54, 55]. Studies generally do not support a unique role for alcohol and folate in CIMP+ tumors [56, 57], although genetic polymorphisms in MTHFR 1298A > C (rs1801131), interacting with diet, and TCN2 776G > C (rs1801198) may be involved in the development of highly CpG-methylated colon and rectal cancers [58–60]. Conversely, MTHFR 1298A > C was not associated with CIMP+ tumors in the Netherlands cohort study [61]. Polymorphisms in DNA repair genes have been implicated in CIMP-positive colon cancer [62, 63]. A promoter polymorphism in MLH1 (−93G > A) was associated with CIMP, MLH1 methylation, and BRAF mutations in unstable sporadic colon tumors and not in stable tumors, suggesting the genetic variant may be acting at a relatively late stage of carcinogenesis to drive CIMP-positive tumors down the microsatellite instability pathway [63].
Overexpression of DNMT3B has been shown to be a risk factor for the development of CIMP in colorectal cancer [64, 65]. DNMT3B is important in establishing and maintaining genomic methylation patterns, and overexpression in mice can induce tumors with methylation in specific CpG islands. Recent findings indicate that DNA methylation changes occur sequentially during tumor progression, and DNMT3B expression levels increase during this progression [66].
The future of CIMP in colorectal cancer research may well take place in the evolving trans- and interdisciplinary field of “molecular pathological epidemiology” outlined by Ogino et al, which is designed to elucidate how genetic factors and lifestyle exposures interact with specific molecular subtypes of cancer [67]. Hughes et al. reported that severe caloric restriction was associated with decreased risk of developing a tumor characterized by CIMP. This study provides a potential link between early life conditions and epigenetic changes that later influence colorectal cancer development [68]. The work of Slattery et al. regarding differences in the etiologies of rectal-site and colon-site tumors, and the influence of genetic factors in the inflammatory pathway in the etiology of CIMP in both, is an example of this approach [69, 70].
5. Emerging Trends in CIMP Research
Although aberrant DNA methylation of promoter CpG islands in cancer genes as well as repressive chromatin are frequently involved in gene inactivation during tumorigenesis, the mechanisms underlying CIMP are poorly understood. Patterns of hypermethylation are specific to tumor type, and it is unclear why certain regions become hypermethylated; however, mapping of the human methylome as a result of technological advances has expanded our understanding of epigenetic mechanisms [71]. Inactivation of particular genes may confer a growth advantage, resulting in clonal selection [72]. Another possibility is that long-range epigenetic silencing by DNA methylation can span chromosome regions of 1 Mb in colorectal cancer, resembling the loss of heterozygosity often observed in tumors [73]. In a large cohort of sporadic colorectal cancers, Wong et al. reported a strong relationship between long-range silencing of chromosome region 3p22 and CIMP+ tumors [74].
Recent findings suggest that most DNA methylation alterations in colon cancer occur in CpG island shores, sequences up to 2 kb distant from CpG islands [75]. Hypermethylated CpG shores appear closer to their associated CpG islands, while hypomethylated shores occur further away from their associated islands and resemble noncolon normal tissues. These findings are consistent with an epigenetic progenitor model of cancer, in which epigenetic alterations affecting tissue-specific differentiation are the predominant mechanism by which epigenetic changes cause cancer. Alternative transcription may be a function of differential DNA methylation during differentiation, and one role of altered methylation in cancer may be to disrupt regulatory control of specific promoter usage [75].
Previous studies suggest a general model in which genes reposition away from the heterochromatin when activated and gravitate to heterochromatin when silenced [76]. However, Easwaran et al. demonstrated that aberrant silencing of cancer-related genes occurred without requirement for their being positioned at heterochromatic domains using immunostaining for active/repressive chromatin marks and fluorescence in-situ hybridization in CRC cell lines. Furthermore, CpG hypermethylation, even associated with long-range silencing of nearby genes, occurred independently of their heterochromatic or euchromatic location [77]. These findings have important implications for understanding relationships between gene expression patterns and nuclear organization in cancer.
Another area under investigation is the understanding of mechanisms underlying which tumor suppressor genes are targeted for inactivation in cancer. Studies suggest a stem cell origin linked to epigenetic control of gene expression patterns in precursor cells regulated by constituent proteins in PcG repressive complexes including Polycomb Repressive Complex PRC1 [78, 79]. It was subsequently shown that sustained expression of the PRC1 protein, CBX7 along with other proteins, targets gene promoters in a progenitor-like embryonic tumor cell resulting in a cell population that models epigenetic characteristics of adult cancer (including aberrant CpG-island methylation) via inhibited response to differentiation cues [80].
DNA methylation markers have potential clinical use as diagnostic and prognostic tools. Hypermethylation of CpG islands can serve as a biomarker of cancer cells in tumor biopsies and other specimens. For example, quantitative assessment of methylation in CIMP-specific promoters of MLH1, WRN, and other DNA-repair genes in colon tumors, in comparison to paired normal tissues, may predict response to treatment [2]. Profiles of miRNA expression also differ between tumor and normal tissues; silencing of miR-124a in colon cancer cells activates expression of the oncogene CDK6 [5]. Continued research involving detailed DNA methylomes in healthy and diseased tissues will help distinguish causal epigenetic alterations from “bystander changes” which are a consequence of cellular processes [71].
Unlike mutations in DNA sequence, epigenetic alterations such as CpG Island hypermethylation are potentially reversible by “reawakening” silenced tumor suppressor genes. Two nucleoside DNA methylation inhibitors, azacitidine and decitabine, are used clinically in low doses to treat myelodysplastic syndrome, providing proof of principal for epigenetic therapy [81]. Structurally, these agents mimic cytosine; during cell replication, fake cytosines replace real cytosines in growing DNA strands and then trap DNA methyltransferases to interfere with the ability of these enzymes to reproduce existing methylation in new cells. While inhibiting DNA methylation is a targeted molecular approach to therapy, downstream effects on neoplastic behavior are nonspecific and may result in cytotoxic cell death, making predictions of clinical outcomes difficult [81]. Clinical trials are being extended to test DNMT inhibitors in solid tumors of the breast, lung, and colon, in combination with histone deacetylase (HDAC) inhibitors which provide synergistic benefits in cell studies [82]. Other treatment avenues on the horizon include induced cellular programming to guide development of epigenetic-modifying drugs [83].
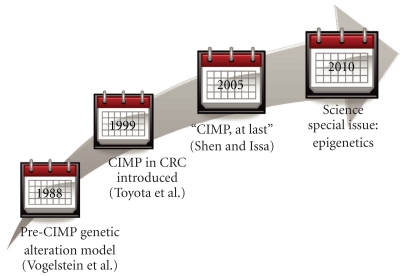
It has been a little over a decade since the concept of a CpG island methylator phenotype in colorectal cancer was introduced, and subsequent focus of several studies on describing the clinical and pathological features of CIMP as well as its characterization in tumors has supported widespread acceptance of the role of DNA methylation in cancer (Figure 2). The past few years have brought substantial insights as to the mechanisms underlying the CIMP pathway in cancer, and the future development of diagnostics and treatments based on our understanding of this epigenetic alteration are an exciting development in epigenetic research.
Figure 2.
A decade of epigenetic research in colorectal cancer (CRC) has led to widespread recognition and acceptance of the CpG Island Methylator Phenotype.
References
- 1.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330(6004):612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteller M. Molecular origins of cancer: epigenetics in cancer. New England Journal of Medicine. 2008;358(11):1148–1096. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 4.Greger V, Passarge E, Hopping W, Messmer E, Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Human Genetics. 1989;83(2):155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 5.Lujambio A, Ropero S, Ballestar E, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Research. 2007;67(4):1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 6.Samowitz WS. The CpG island methylator phenotype in colorectal cancer. Journal of Molecular Diagnostics. 2007;9(3):281–283. doi: 10.2353/jmoldx.2007.070031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JPJ. CpG island methylator phenotype in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. New England Journal of Medicine. 1988;319(9):525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 9.Issa JPJ, Shen L, Toyota M. CIMP, at last. Gastroenterology. 2005;129(3):1121–1124. doi: 10.1053/j.gastro.2005.07.040. [DOI] [PubMed] [Google Scholar]
- 10.Laird PW. Cancer epigenetics. Human Molecular Genetics. 2005;14(1):R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 11.Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129(3):837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Jass JR. Colorectal cancer: a multipathway disease. Critical Reviews in Oncogenesis. 2006;12(3-4):273–287. doi: 10.1615/critrevoncog.v12.i3-4.50. [DOI] [PubMed] [Google Scholar]
- 13.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nature Genetics. 2006;38(7):787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 14.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. Journal of Molecular Diagnostics. 2008;10(1):13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGivern A, Wynter CVA, Whitehall VLJ, et al. Promoter hypermethylation frequency and BRAF mutations distinguish hereditary non-polyposis colon cancer from sporadic MSI-H colon cancer. Familial Cancer. 2004;3(2):101–107. doi: 10.1023/B:FAME.0000039861.30651.c8. [DOI] [PubMed] [Google Scholar]
- 16.Hawkins N, Norrie M, Cheong K, et al. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122(5):1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- 17.Van Rijnsoever M, Grieu F, Elsaleh H, Joseph D, Iacopetta B. Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut. 2002;51(6):797–802. doi: 10.1136/gut.51.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slattery ML, Curtin K, Wolff RK, et al. A comparison of colon and rectal somatic DNA alterations. Diseases of the Colon and Rectum. 2009;52(7):1304–1311. doi: 10.1007/DCR.0b013e3181a0e5df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barault L, Charon-Barra C, Jooste V, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Research. 2008;68(20):8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 20.Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS ONE. 2008;3(11, article e3698) doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.English DR, Young JP, Simpson JA, et al. Ethnicity and risk for colorectal cancers showing somatic BRAF V600E mutation or CpG island methylator phenotype. Cancer Epidemiology Biomarkers and Prevention. 2008;17(7):1774–1780. doi: 10.1158/1055-9965.EPI-08-0091. [DOI] [PubMed] [Google Scholar]
- 22.Vaughn CP, Wilson AR, Samowitz WS. Quantitative evaluation of CpG island methylation in hyperplastic polyps. Modern Pathology. 2010;23(1):151–156. doi: 10.1038/modpathol.2009.150. [DOI] [PubMed] [Google Scholar]
- 23.Baretton GB, Autschbach F, Baldus S. Histopathological diagnosis and differential diagnosis of colorectal serrated polys: findings of a consensus conference of the working group “Gastroenterological pathology of the German Society of Pathology”. Pathologe. 2011;32(1):76–82. doi: 10.1007/s00292-010-1365-3. [DOI] [PubMed] [Google Scholar]
- 24.Torlakovic EE, Gomez JD, Driman DK, et al. Sessile serrated adenoma (SSA) vs. traditional serrated adenoma (TSA) American Journal of Surgical Pathology. 2008;32(1):21–29. doi: 10.1097/PAS.0b013e318157f002. [DOI] [PubMed] [Google Scholar]
- 25.Shen L, Catalano PJ, Benson ALB, O’Dwyer P, Hamilton SR, Issa JPJ. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil-based chemotherapy. Clinical Cancer Research. 2007;13(20):6093–6098. doi: 10.1158/1078-0432.CCR-07-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino S, Meyerhardt JA, Kawasaki T, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Archiv. 2007;450(5):529–537. doi: 10.1007/s00428-007-0398-3. [DOI] [PubMed] [Google Scholar]
- 27.Dahlin AM, Palmqvist R, Henriksson ML, et al. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clinical Cancer Research. 2010;16(6):1845–1855. doi: 10.1158/1078-0432.CCR-09-2594. [DOI] [PubMed] [Google Scholar]
- 28.Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Research. 2005;65(14):6063–6070. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 29.Kim JH, Shin SOH, Kwon HJ, Cho NY, Kang GH. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Archiv. 2009;455(6):485–494. doi: 10.1007/s00428-009-0857-0. [DOI] [PubMed] [Google Scholar]
- 30.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58(1):90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SJ, Rashid A, Lee JH, Kim SG, Hamilton SR, Wu TT. Frequent CpG island methylation in serrated adenomas of the colorectum. American Journal of Pathology. 2003;162(3):815–822. doi: 10.1016/S0002-9440(10)63878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. Journal of Molecular Diagnostics. 2006;8(5):582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(47):18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55(7):1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka N, Huttenhower C, Nosho K, et al. Novel application of structural equation modeling to correlation structure analysis of CpG island methylation in colorectal cancer. American Journal of Pathology. 2010;177(6):2731–2740. doi: 10.2353/ajpath.2010.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ang PW, Loh M, Liem N, et al. Comprehensive profiling of DNA methylation in colorectal cancer reveals subgroups with distinct clinicopathological and molecular features. BMC Cancer. 2010;10, article 227 doi: 10.1186/1471-2407-10-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaneda A, Yagi K. Two groups of DNA methylation markers to classify colorectal cancer into three epigenotypes. Cancer Science. 2011;102(1):18–24. doi: 10.1111/j.1349-7006.2010.01712.x. [DOI] [PubMed] [Google Scholar]
- 38.Hinoue T, Weisenberger DJ, Pan F, et al. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PloS one. 2009;4(12):p. e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondo T, Issa JP. DNA methylation profiling in cancer. Expert Review of Proteomics. 2010;12, article e23 doi: 10.1017/S1462399410001559. [DOI] [PubMed] [Google Scholar]
- 40.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50(1):113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 41.Toyota M, Ohe-Toyota M, Ahuja N, Issa JPJ. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(2):710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dertinger SD, Silverstone AE, Gasiewicz TA. Influence of aromatic hydrocarbon receptor-mediated events on the genotoxicity of cigarette smoke condensate. Carcinogenesis. 1998;19(11):2037–2042. doi: 10.1093/carcin/19.11.2037. [DOI] [PubMed] [Google Scholar]
- 43.Belinsky SA, Palmisano WA, Gilliland FD, et al. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Research. 2002;62(8):2370–2377. [PubMed] [Google Scholar]
- 44.Ray SS, Swanson HI. Dioxin-induced immortalization of normal human keratinocytes and silencing of p53 and p16. Journal of Biological Chemistry. 2004;279(26):27187–27193. doi: 10.1074/jbc.M402771200. [DOI] [PubMed] [Google Scholar]
- 45.Samowitz WS, Albertsen H, Sweeney C, et al. Association of smoking, CpG island methylator phenotype, and V600E BRAF mutations in colon cancer. Journal of the National Cancer Institute. 2006;98(23):1731–1738. doi: 10.1093/jnci/djj468. [DOI] [PubMed] [Google Scholar]
- 46.Curtin K, Samowitz WS, Wolff RK, Herrick J, Caan BJ, Slattery ML. Somatic alterations, metabolizing genes and smoking in rectal cancer. International Journal of Cancer. 2009;125(1):158–164. doi: 10.1002/ijc.24338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Limsui D, Vierkant RA, Tillmans LS, et al. Cigarette smoking and colorectal cancer risk by molecularly defined subtypes. Journal of the National Cancer Institute. 2010;102(14):1012–1022. doi: 10.1093/jnci/djq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozek LS, Herron CM, Greenson JK, et al. Smoking, gender, and ethnicity predict somatic BRAF mutations in colorectal cancer. Cancer Epidemiology Biomarkers and Prevention. 2010;19(3):838–843. doi: 10.1158/1055-9965.EPI-09-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slattery ML, Curtin K, Anderson K, et al. Associations between cigarette smoking, lifestyle factors, and microsatellite instability in colon tumors. Journal of the National Cancer Institute. 2000;92(22):1831–1836. doi: 10.1093/jnci/92.22.1831. [DOI] [PubMed] [Google Scholar]
- 50.Morimoto LM, Newcomb PA, Ulrich CM, Bostick RM, Lais CJ, Potter JD. Risk factors for hyperplastic and adenomatous polyps: evidence for malignant potential? Cancer Epidemiology Biomarkers and Prevention. 2002;11(10 I):1012–1018. [PubMed] [Google Scholar]
- 51.Ulrich CM. Nutrigenetics in cancer research—folate metabolism and colorectal cancer. Journal of Nutrition. 2005;135(11):2698–2702. doi: 10.1093/jn/135.11.2698. [DOI] [PubMed] [Google Scholar]
- 52.James SJ, Melnyk S, Pogribna M, Pogribny IP, Caudill MA. Elevation in S-Adenosylhomocysteine and DNA hypomethylation: potential epigenetic mechanism for homocysteine-related pathology. Journal of Nutrition. 2002;132(8):2361S–2366S. doi: 10.1093/jn/132.8.2361S. [DOI] [PubMed] [Google Scholar]
- 53.Kawakami K, Ruszkiewicz A, Bennett G, Moore J, Watanabe GO, Iacopetta B. The folate pool in colorectal cancers is associated with DNA hypermethylation and with a polymorphism in methylenetetrahydrofolate reductase. Clinical Cancer Research. 2003;9(16 I):5860–5865. [PubMed] [Google Scholar]
- 54.Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. International Journal of Cancer. 2008;122(12):2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Estécio MRH, Gharibyan V, Shen L, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS ONE. 2007;2(5, article e399) doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slattery ML, Curtin K, Sweeney C, et al. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. International Journal of Cancer. 2007;120(3):656–663. doi: 10.1002/ijc.22342. [DOI] [PubMed] [Google Scholar]
- 57.Van Engeland M, Weijenberg MP, Roemen GMJM, et al. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer Research. 2003;63(12):3133–3137. [PubMed] [Google Scholar]
- 58.Curtin K, Slattery ML, Ulrich CM, et al. Genetic polymorphisms in one-carbon metabolismml: associations with CpG island methylator phenotype (CIMP) in colon cancer and the modifying effects of diet. Carcinogenesis. 2007;28(8):1672–1679. doi: 10.1093/carcin/bgm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Curtin K, Samowitz W, Ulrich C, et al. Nutrients in folate-mediated, one-carbon metabolism and risk of rectal tumors in men and women. Nutrition and Cancer. 2011;31(1) doi: 10.1080/01635581.2011.535965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hazra A, Fuchs CS, Kawasaki T, Kirkner GJ, Hunter DJ, Ogino S. Germline polymorphisms in the one-carbon metabolism pathway and DNA methylation in colorectal cancer. Cancer Causes and Control. 2010;21(3):331–345. doi: 10.1007/s10552-009-9464-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Vogel S, Wouters KAD, Gottschalk RWH, et al. Genetic variants of methyl metabolizing enzymes and epigenetic regulators: associations with promoter CpG island hypermethylation in colorectal cancer. Cancer Epidemiology Biomarkers and Prevention. 2009;18(11):3086–3096. doi: 10.1158/1055-9965.EPI-09-0289. [DOI] [PubMed] [Google Scholar]
- 62.Curtin K, Samowitz WS, Wolff RK, et al. MSH6 G39E polymorphism and CpG island methylator phenotype in colon cancer. Molecular Carcinogenesis. 2009;48(11):989–994. doi: 10.1002/mc.20566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Samowitz WS, Curtin K, Wolff RK, et al. The MLH1 −93G > A promoter polymorphism and genetic and epigenetic alterations in colon cancer. Genes Chromosomes and Cancer. 2008;47(10):835–844. doi: 10.1002/gcc.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Linhart HG, Lin H, Yamada Y, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes and Development. 2007;21(23):3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nosho K, Shima K, Irahara N, et al. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clinical Cancer Research. 2009;15(11):3663–3671. doi: 10.1158/1078-0432.CCR-08-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ibrahim AE, Arends MJ, Silva AL, et al. Sequential DNA methylation changes are associated with DNMT3B overexpression in colorectal neoplastic progression. Gut. 2011;60(4):444–508. doi: 10.1136/gut.2010.223602. [DOI] [PubMed] [Google Scholar]
- 67.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60(3):397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hughes LAE, van den Brandt PA, de Bruïne AP, et al. Early life exposure to famine and colorectal cancer risk: a role for epigenetic mechanisms. PLoS ONE. 2009;4(11, article e7951) doi: 10.1371/journal.pone.0007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Slattery ML, Lundgreen A, Herrick JS, Wolff RK. Genetic variation in RPS6KA1, RPS6KA2, RPS6KB1, RPS6KB2, and PDK1 and risk of colon or rectal cancer. Mutation Research. 2011;706(1-2):13–20. doi: 10.1016/j.mrfmmm.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slattery ML, Curtin K, Poole EM, et al. Genetic variation in C-reactive protein (CRP) in relation to colon and rectal cancer risk and survival. International Journal of Cancer. 2011;128(11):2726–2734. doi: 10.1002/ijc.25721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Portela A, Esteller M. Epigenetic modifications and human disease. Nature Biotechnology. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 72.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Human Molecular Genetics. 2007;16(1):R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 73.Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nature Genetics. 2006;38(5):540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 74.Wong JJ, Hawkins NJ, Ward RL, Hitchins MP. Methylation of the 3p22 region encompassing MLH1 is representative of the CpG island methylator phenotype in colorectal cancer. Modern Pathology. 2011;24:396–411. doi: 10.1038/modpathol.2010.212. [DOI] [PubMed] [Google Scholar]
- 75.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature Genetics. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Malik P, Zuleger N, Schirmer EC. Nuclear envelope influences on genome organization. Biochemical Society Transactions. 2010;38(1):268–272. doi: 10.1042/BST0380268. [DOI] [PubMed] [Google Scholar]
- 77.Easwaran HP, Van Neste L, Cope L, et al. Aberrant silencing of cancer-related genes by CpG hypermethylation occurs independently of their spatial organization in the nucleus. Cancer Research. 2010;70(20):8015–8024. doi: 10.1158/0008-5472.CAN-10-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nature Genetics. 2007;39(2):237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nature Genetics. 2007;39(2):232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 80.Mohammad HP, Cai Y, McGarvey KM, et al. Polycomb CBX7 promotes initiation of heritable repression of genes frequently silenced with cancer-specific DNA hypermethylation. Cancer Research. 2009;69(15):6322–6330. doi: 10.1158/0008-5472.CAN-09-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Issa JPJ, Kantarjian HM. Targeting DNA methylation. Clinical Cancer Research. 2009;15(12):3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaiser J. Epigenetic drugs take on cancer. Science. 2010;330(6004):576–578. doi: 10.1126/science.330.6004.576. [DOI] [PubMed] [Google Scholar]
- 83.Ohm JE, Mali P, Van Neste L, et al. Cancer-related epigenome changes associated with reprogramming to induced pluripotent stem cells. Cancer Research. 2010;70(19):7662–7673. doi: 10.1158/0008-5472.CAN-10-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]