Abstract
Sphingolipids are structurally diverse and their metabolic pathways highly complex, which makes it difficult to follow all of the subspecies in a biological system, even using “lipidomic” approaches. This report describes a method to use transcriptomic data to visualize and predict potential differences in sphingolipid composition, and it illustrates its use with published data for cancer cell lines and tumors. In addition, several novel sphingolipids that were predicted to differ between MDA-MB-231 and MCF7 cells based on published microarray data for these breast cancer cell lines were confirmed by mass spectrometry. For the data that we were able to find for these comparisons, there was a significant match between the gene expression data and sphingolipid composition (P < 0.001 by Fisher's exact test). Upon considering the large number of gene expression datasets produced in recent years, this simple integration of two types of “omic” technologies (“transcriptomics” to direct “sphingolipidomics”) might facilitate the discovery of useful relationships between sphingolipid metabolism and disease, such as the identification of new biomarkers.
Keywords: lipidomics, pathway visualization, transcriptomics, cancer
The importance of sphingolipids for cell structure and regulation (1–3) has inspired the development of mass spectrometric methods to analyze key categories of these compounds (see Refs. 4–8 for examples). These cover hundreds of subspecies but still miss a large portion of the “sphingolipidome” due to its size and complexity (including the existence of many isomers and isobars) (9) and wide range in amounts. Therefore, it would be useful to have tools that draw on other sources of information, such as gene expression data, to direct attention toward particular subspecies that might otherwise be overlooked by mass spectrometry or other methods.
This article describes a pathway visualization tool to predict differences in sphingolipid composition using microarray data, then explores its utility using publicly available datasets for cancer cells in culture and tumors. Cancer was selected because sphingolipids are involved in many processes that are important in regulation of growth, survival, adhesion, and migration (10), and tumors often display alterations in sphingolipid composition (11, 12), some of which have been proposed as cancer markers (13–15). Furthermore, there have been a large number of microarray analyses of gene expression in cancer cell lines and tumors, and more are certain to be conducted; therefore, a tool that can utilize this data to direct attention to compounds that are potentially different in cancer might aid the understanding of the molecular changes associated with this disease and possibly uncover novel biomarkers.
There are ample precedents for this approach. For example, elevated expression of mRNA for GD3 synthase has been correlated with the amounts of GD3 in melanoma cell lines (16), and in the reverse direction, the finding of low amounts of C18-ceramide (Cer) in head and neck cancer led to the discovery of lower expression of CerS1 (17). Nonetheless, there is currently no accurate pathway map for the visualization of datasets for large categories of genes and metabolites involved in sphingolipid biosynthesis. Therefore, the goal of these studies was to develop tools for this purpose by using PathVisio v2 (18), an open access pathway browser, and updated and expanded maps to visualize data from analyses of gene expression and sphingolipid metabolites. A preliminary version of this tool was described earlier (19); this article provides more information and illustrates its application to cancer cell lines, tumors, and normal tissues, which found substantial agreement between gene expression and metabolite profiles.
MATERIALS AND METHODS
Materials
MCF7 cells were purchased from the ATCC (Manassas, VA), and MDA-MB-231 cells were provided by Dr. Nathan Bowen (School of Biology, Georgia Institute of Technology, Atlanta, GA). Cell culture medium was procured from Gibco (Carlsbad, CA), and FBS was obtained from Hyclone (Logan, UT). U-13C labeled palmitic acid was obtained from Cambridge Isotopes (Andover, MA). The mass spectrometry internal standards were procured from Avanti Polar Lipids (Alabaster, AL); other glycosphingolipid (GSL) standards were obtained from Sigma (St. Louis, MO) and Matreya (Pleasant Gap, PA). All other reagents and solvents were HPLC grade.
Modification of PathVisio v2 and construction of sphingolipid pathway maps
PathVisio v2 (18), open source Java software, was selected for the construction of pathway maps for different branches of sphingolipid biosynthesis and the visualization of differentially expressed genes. A tutorial on how to use Path Visio v2 and the tools described in this manuscript is available on the web site http://sphingolab.biology.gatech.edu/sphingoPathvisioTutorial1.html. To differentiate metabolites from genes, the former were displayed as circular objects, while the latter had a rectangular shape. New pathway maps using PathVisio v2 were constructed by altering Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway diagrams to include metabolite identifiers at the nodes, while the genes were updated to reflect new discoveries and changes in nomenclature. Some of the prominent changes in the sphingolipid pathway maps are as follows: the addition of ceramide synthase gene isoforms (CerS1-6) (20–24), dihydroceramide desaturase (DEGS1, DEGS2) (25), and α(1,2)fucosyltransferases (FUT1 and FUT2) (14). The layout of the backbone biosynthesis map was published previously (19) and is provided in supplementary Fig. I; it was modified to include branches for the synthesis of 3-ketodihydroceramides (26), sphingolipids with dihydroceramide backbones, and phyto-sphingolipids. These modified KEGG pathway maps, as well as ones for gangliosides and globo series GSL, can be obtained from www.sphingomaps.org or the Wikipathways database (27). A list of all the genes is provided in Table 1 and supplementary online data.
TABLE 1.
Genes involved in de novo sphingolipid biosynthesis in Fig. 1
| Gene ID | Gene Name | Ref. |
|---|---|---|
| SPTLC1, SPTLC2, SPTLC 3 | Serine palmitoyltransferase subunits (1, 2, and 3) | (52) |
| KDSR, 3KSR, FVT1 | 3-Ketosphinganine reductase | (53) |
| CerS1, CerS2, CerS3, CerS4CerS5, CerS6 | Ceramide synthase (1-6) | (20-24) |
| SPHK1, SPHK2 | Sphingosine kinase (1, 2) | (54) |
| SGPL1 | Sphingosine 1-phosohate lyase | (55) |
| DES1, DES2, DEGS1, 2 | Dihydroceramide desaturase (1 and 2) | (25) |
| ASAH1, ASAH2, ASAH3, ASAH3L, PCHA | Ceramidases (Acid, neutral, phytoceramidase) | (56-60) |
| SMS1,SMS2, SMSr, | Sphingomyelin synthase (1, 2, and atypical) | (40, 61) |
| UGCG, Gba | Glucosylceramide synthase | (62) |
| UGT8, CGT | Galactosylceramide synthase | (63) |
| CERK | Ceramide kinase | (64, 65) |
| CERT | Ceramide transporter | (42, 66) |
| SMPD1, SMPD2, SMPD3, SMPD4 | Sphingomyelinases (acidic, neutral) | (67, 68) |
| B4GALT6 | Lactosylceramide synthase | (69) |
| GAL3ST1 | galactose-3-O-sulfotransferase 1 | (70) |
| ELOVL 1-7 | Fatty acid CoA elongases (1-7) | (71) |
| SCD | Stearoyl CoA desaturase | (72) |
| ssSPTa, ssSPTb | Small subunits of SPT | (73) |
| ORMDL1, ORMDL 2, ORMDL 3 | Orosomucoid (1-3) | (74, 75) |
The table lists alternative gene abbreviations and complete names for genes encoding enzymes for different steps of de novo sphingolipid biosynthesis displayed in Fig. 1. Genes involved in the biosynthesis of glycosphingolipids are described in a table in the supplementary online data.
Selection of DNA microarray datasets and statistical analysis
Keyword searches were conducted on National Center for Biotechnology Information, Gene expression omnibus (NCBI GEO) (28) Array express (29) and Oncomine (30) for “cancer type” or “cancer cell line,” and the outcomes were examined to identify either expression datasets having multiple replicates of control and cancer tissues or all the cell lines to be compared. The Affymetrix expression files (.CEL file) for the selected datasets were downloaded and processed by Affymetrix expression console v1.1 (Affymetrix, Santa Clara, CA) using the MAS5 algorithm. For non-Affymetrix studies, the normalized and background-corrected expression datasets (GDS file, NCBI GEO format) were accessed using the GEOquery package (31) of Bioconductor v1.8 (32) and exported as tab-delineated text files. Sphingolipid-specific gene expression values were filtered using a Perl search algorithm and a list of preselected Affymetrix probe IDs or gene ID (see below). The gene expression fold change was determined using Excel (Microsoft, Seattle, WA), after which the dataset was formatted as a PathVisio input file (comma-separated values) with “gene ids,” “system code,” and “fold change.”
Selection of gene probes
To identify the best probes for sphingolipid genes, the sequences of all probes directed to individual transcripts were retrieved from the Affymetrix Netaffx tool (33) and compared with the mRNA using Blat search (34) in the University of California, Santa Cruz (UCSC) genome browser (35). The probe that recognized the catalytic transcript and hybridized nearest to its 3′ UTR was selected to represent the gene for pathway model visualization, and others were excluded. The procedure was repeated for all sphingolipid genes for Affymetrix chips HG-U95 A-E and HG-U133 plus 2 (supplementary online data).
Perl algorithm for extraction of expression values
A Perl script was written to search for and extract gene expression values for sphingolipid-related transcripts. The program transforms the list of probe IDs or gene IDs into an array, and then searches for similar objects in a normalized gene expression data file, where the rows represent probe IDs and columns individual experiments. The loop function efficiently extracts and writes the sphingolipid gene expression values into a text file, which is used to calculate gene fold changes. A copy of the Perl script and instructions for its use are provided in the supplementary online data.
Pathway visualization
In the PathVisio v2 browser, the appropriate gene and metabolite database was selected, and the expression dataset was created after importing the saved input file using the Import Expression Dataset tab. The color criteria for the expression dataset were specified using the Visualization Option function. Gene and metabolite expression changes were visualized on the modified sphingolipid pathway maps by opening the pathway diagram in PathVisio v2, and then selecting the saved expression dataset and visualization criteria. The pictures were prepared by exporting the images as PDF files; while changes to font colors were performed with CorelDraw X4 (Mountain View, CA) as summarized in the supplementary online data. A detailed tutorial for this tool (including how to download PathVisio software) is available (see the supplementary online data).
Estimation of the relative amounts of the sphingolipids for the samples used in the gene expression comparisons
The MCF7 and MDA-MB-231 cells were grown in 100 mm tissue culture dishes using RPMI 1640 medium containing 5% FBS under conditions described previously (36). When the cells reached 80-90% confluency, they were washed once with PBS solution, before being scraped into screw capped glass test tubes for sphingolipid extraction as previously described (37). An aliquot of the cells was retained from each dish to measure the protein concentration by the bicinchoninic acid (BCA) assay (Pierce Biotechnology, Rockford, IL). During stable isotope-labeling studies, the media was supplemented with 0.1 mM [U-13C]palmitic acid complexed with equimolar fatty acid free BSA (Calbiochem, Gibbstown, NJ). After the times described for each experiment, the sphingolipids were analyzed by LC ESI-MS/MS using ABI 3000 and ABI 4000 LC-tandem mass spectrometers (Applied Biosystems, Foster City, CA), as previously described (37).
When available, published quantitative data for metabolites were used to compute fold changes. In some instances, only photographs of TLC plates were provided, and in those cases, crude estimates were obtained from the relative densities obtained using Fluorchem 5500 (Alpha Innotech, San Leandro, CA). The sources of these data are provided in the text.
RESULTS AND DISCUSSION
Summary of the sphingolipid biosynthetic pathway as the basis for construction of maps to visualize gene expression and metabolite data in a pathway context
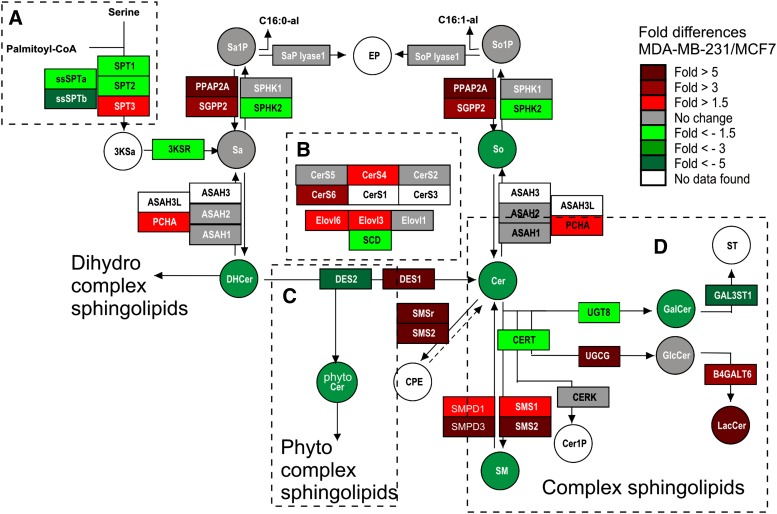
Fig. 1 displays the pathway map for the initial steps of sphingolipid biosynthesis that we constructed for use with PathVisio v2 (18). Metabolites are in circles, and the genes associated with each step of the pathway are in boxes. The heatmap coloring of Fig. 1 is a comparison of MDA-MB-231 and MCF7 cells, which will be discussed later in this article (Fig. 2–5). The gene annotations for the individual steps of the pathway are listed in Table 1, and additional details are given in supplementary Fig. I and recent reviews (9, 19). An expanded form of the pathway has been provided to www.wikipathways.com (27) and can be downloaded from that site.
Fig. 1.
Visualization of transcription and metabolite differences in early steps of sphingolipids biosynthesis between MDA-MB-231 and MCF7 cells using PathVisio v2 with an expanded pathway map. The figure depicts sphingolipid genes (rectangles) and metabolites (circles) that participate in early steps of sphingolipid biosynthesis prepared using PathVisio v2 (for more detail, see supplementary Fig. I) (18). The updated and expanded pathway map was overlaid by the gene expression fold change difference between MDA-MB-231 and MCF7 cells obtained from a previous study (36). Variations in sphingolipid composition (shaded circles) between the two cell lines were determined by LC-ESI-MS/MS as previously described (4) and compared with gene expression as highlighted by circumscribed dash boxes (A-D) and discussed in Fig. 3–6. The box and circle shades represent the degree of upregulation and downregulation indicated by the color chart.
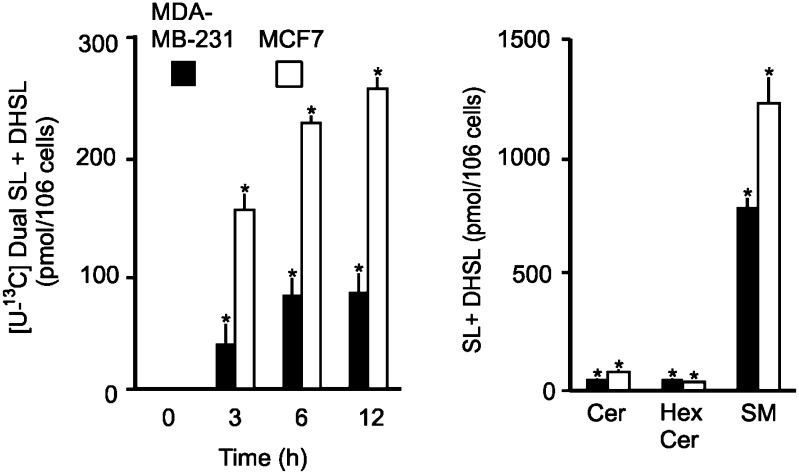
Fig. 2.
Comparison of de novo sphingolipid biosynthesis between MDA-MB-231 and MCF7 cells with gene expression of serine palmitoyltransferase. The two cell lines were grown as previously described (36) and incubated with and without 0.1 mM [13C]palmitic acid as the BSA complex for 0, 3, and 6 h. A: The sphingolipids labeled in sphingoid base and fatty acid (Dual) were measured by LC-ESI-MS/MS (4). B: In parallel experiments, unlabeled sphingolipids were also measured for both cell types. A: Sum of Dual [13C]palmitate-labeled complex sphingolipids with SL (Cer) and DHSL (DHCer) backbone. B: Sum of unlabeled complex SL and DHSL in ceramide (Cer), monohexosylceramide (HexCer), and sphingomyelin (SM). The significance, indicated by an asterisk, was determined by a t-test at P < 0.05 for n = 3 (labeling study) and n = 6 (for unlabeled cells).
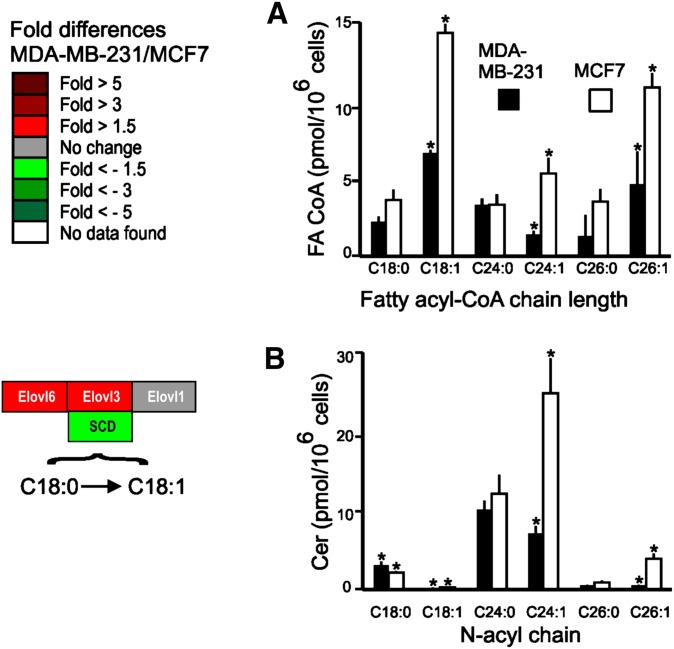
Fig. 3.
Comparison of the composition of fatty acyl CoA and ceramide chain lengths for MDA-MB-231 and MCF7 cells. MDA-MB-231 and MCF7 cells were grown as previously described (36). The fatty acyl-CoA (FA-CoA) and ceramides (Cer) were measured by LC-ESI-MS/MS (37, 84). A: Composition of saturated and monounsaturated FA-CoA chain lengths. B: N-acyl chain distribution of Cer corresponding to the FA-CoA measured in A. The significance, indicated by an asterisk, was determined by a t-test at P < 0.05 for n = 6.
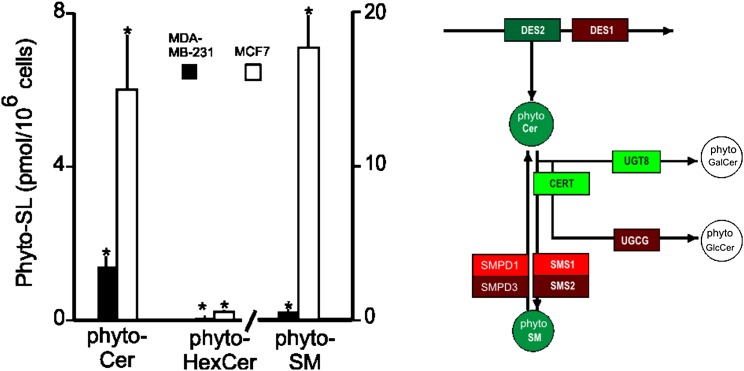
Fig. 4.
Comparison of the phyto-sphingolipids of MDA-MB-231 and MCF7 cells. LC-ESI-MS/MS comparison of phyto-sphingolipids (phyto-SL; i.e., with a 4-hydroxysphinganine backbone), phyto-ceramide (phyto-Cer), phyto-monohexosylceramide (phyto-HexCer), and phyto-sphingomyelin (phyto-SM) (note different scale) for MDA-MB-231 and MCF7 cells. The significance, indicated by an asterisk, was determined by a t-test at P < 0.05 for n = 6.
Fig. 5.
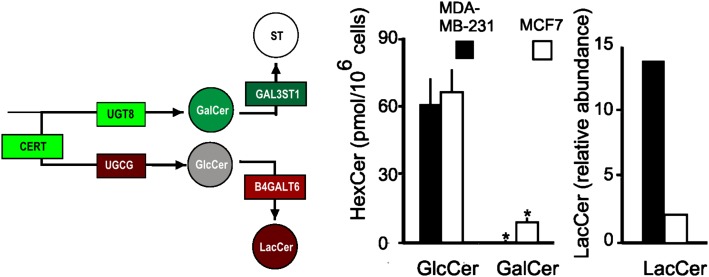
Comparison of monohexosylceramides and lactosylceramide of MDA-MB-231 and MCF7 cells. Results from LC-ESI-MS/MS analysis of GluCer and GalCer of MDA-MB-231 and MCF7 cells as previously described (4). The significance of differences, indicated by an asterisk, was determined by a t-test at P < 0.05 for n = 6. LacCer was estimated by densitometric analysis of LacCer data from a previous study (48).
As displayed, de novo sphingolipid biosynthesis is initiated in the upper-left corner (Fig. 1, dashed box A) by the condensation of serine and palmitoyl-CoA by serine palmitoyltransferase (SPT) to produce 3-ketosphinganine (3KSa). The five genes for SPT represent the three genes known to code for functional proteins (SPT1-3 from SPTLC 1, 2, and 3) and two shorter transcripts (ssSPTa and ssSPTb) that have been suggested to regulate SPT (38). The 3KSa is reduced to sphinganine (Sa), which is at a branch point where it can be acylated by ceramide synthases (CerS, in dashed box B2) or phosphorylated by sphingosine kinase (SPHK1 or 2) to Sa 1-phosphate (Sa1P), which can be terminally degraded to hexadenanal (C16:0-al) and ethanolamine phosphate (EP). The acylation products, dihydroCer (DHCer), are at another branch point (dashed box C) and are desaturated to Cer by DES1 (and possibly DES2), hydroxylated to 4-hydroxyDHCer (phyto-Cer) by DES2 (25), or converted to a more complex dihydrosphingolipids, as will be discussed later. DHCer can also be hydrolyzed by ceramidases (ASAH 1-3, 3L, and PCHA). For most cells, the desaturation branch is presumed to predominate because Cer is the major lipid backbone of most complex sphingolipids (9, 39).
Cer is also at a branch point (illustrated in dashed box D for Cer) for the biosynthesis of sphingomyelin (SM) (40) (and a recently reported Cer-phosphoethanolamine) (41), glucosylCer (GlcCer), galactosylCer (GalCer), or Cer 1-phosphate (Cer1P). Shown in this scheme is CERT, an additional protein that transports Cer to the trans-Golgi for biosynthesis of SM, CerP, and possibly some GlcCer (42). These reactions sometimes take place with DHCer and phyto-Cer, but for simplicity, these additional steps have not been included in Fig. 1. Pathway maps can be customized for PathVisio v2 at the discretion of the user, which allows inclusion of additional steps, such as genes coding for the enzymes of sphingolipid turnover (see Fig. 1 for the sphingomyelinases but not the glycosidases for GlcCer and GalCer), and for updating as new genes and metabolites are discovered.
This particular scheme displays the next metabolic step for GlcCer, glycosylation of GlcCer to lactosylCer (LacCer), after which the pathway branches into five so-called root structure series (globo-series, isoglobo-series, ganglio-series, lacto-series and neo-lacto-series) (9), and for GalCer, which is primarily converted to sulfatides (ST). Separate pathway maps are shown for two subcategories of more complex glycosphingolipids, gangliosides (Fig. 6), and globo-series neutral glycosphingolipids (Fig. 7) (these have also been made available through Wikipathways) (27). Background references for the genes for these steps are given in the supplementary online data. As noted before, users of PathVisio v2 can update and expand these pathways as new information is obtained.
Fig. 6.
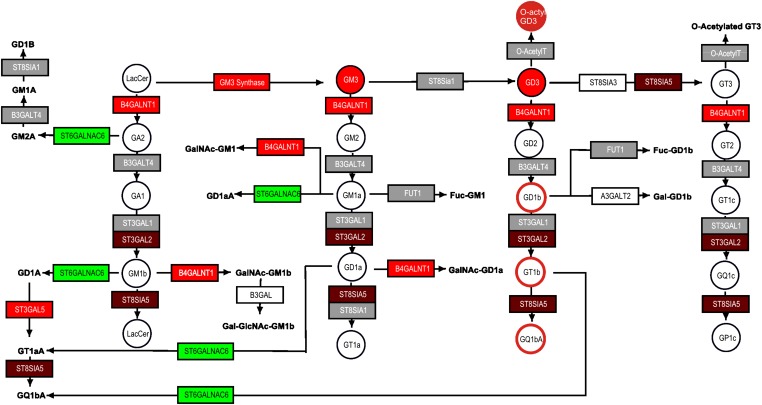
Comparison of microarray data and metabolites for ganglioside biosynthesis for invasive ductal breast carcinoma and normal ductal tissue. Gene expression differences between the two breast cancer stages were visualized using data from a previous study (43) and updated ganglioside biosynthetic maps in PathVisio v2 (18). Ganglioside measurements are from reports using similar tumors (circles with a solid shade) (49) and patient serum (circles with colored edges) (15). The color scale for the difference between MDA-MB-231/MCF7 cells is the same as in Fig. 1.
Fig. 7.
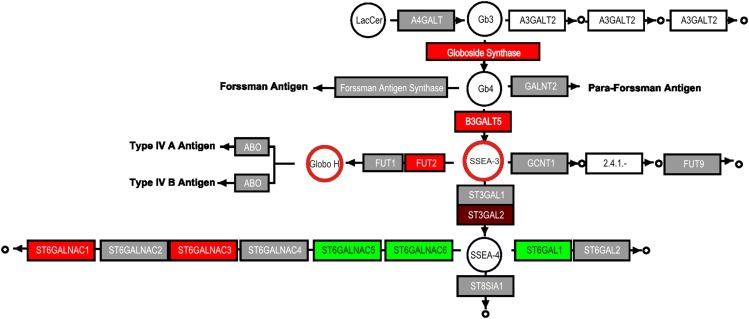
Comparison of microarray data and corresponding metabolites for globo-series glycosphingolipid biosynthesis for invasive ductal breast carcinoma and normal ductal tissue. Comparison of gene expression differences between normal and invasive ductal carcinoma (IDC) from a previous study (43) using updated KEGG pathway maps for globo-series biosynthesis to metabolite changes previously reported using flow cytometry (14). The color scale for the difference between MDA-MB-231/MCF7 cells is the same as in Fig. 1.
Preparation of data for visualization in sphingolipid gene expression pathway diagrams
Before discussing the heatmap differences in these figures, some comments will be made about the datasets and the formatting of the data for visualization using PathVisio v2. Cancer was selected as the source of the datasets because published gene expression data are available for cancer cells, normal cells, and tumor cells (36, 43, 44) from public microarray repositories (28, 29). Platform-specific tools were used to normalize raw expression values for samples within studies as described under “Methods,” and a Perl script was written (see supplementary online data) to sort out expression values for transcripts involved in sphingolipid biosynthesis. Fold differences were computed either by taking the ratio between two conditions (such as two cell lines or cancer stages) or by normalizing the individual gene expression by the average among all samples (in the case of multiple cell lines), and then visualized using the sphingolipid biosynthesis pathway maps and PathVisio v2.
The presence of multiple probes for individual transcripts and a large number of transcripts posed a challenge during the analysis. To address the former, probes specific to the 3′ UTR of the mRNA were chosen by a Blat search (34) using the UCSC genome browser (35). A list of the optimum probes for sphingolipid biosynthesis in human HG-U133 Plus2 and HG-U95 Affymetrix gene chips is provided in the supplementary online data. In case of the latter, a search algorithm was written in Perl (supplementary online data) to sort out the expression values for the optimum probes using the list of preselected probes.
Rules for correlating mRNA and metabolite abundance
To study the correlation between transcript and metabolite amounts, certain rules were followed based on biochemical understanding: a) Only the de novo biosynthesis steps beginning with the condensation of serine and palmitoyl-CoA were evaluated; the reverse steps were usually not included; b) For isoforms that are known to display distinct specificities, such as SPTLC3, the CerS isoforms (CerS1-6), ELOVL 1-6, SCD, and DES2 (versus DES1), the changes in individual transcripts were compared with their corresponding sphingolipid subspecies; c) Enzymatic steps represented by multiple gene isoforms without known differences in the specificities of their product enzyme were compared with the total metabolite amount (in addition, the step was considered “upregulated” if at least one gene isoform was elevated, the step was considered “downregulated” if all isoforms were downregulated, or the step was considered “unchanged” if all isoforms were unchanged or there was a mix of unchanged and reduced); and d) correlations were only counted when there was data for both the relative gene expression and accompanying metabolites, which is obviously a small subset of all of the potential molecules.
To test the utility of this technique, gene expression datasets from breast cancer cell lines were compared first, then normal breast cells and tumor cells were examined as described in the following sections.
Comparison of gene expression and sphingolipid differences between MCF7 and MDA-MB-231 breast cancer cells
Examination of the apparent3 gene expression differences between the highly metastatic ER-negative MDA-MB-231 and ER-positive MCF7 cell lines (Fig. 1) leads to a number of relatively self-evident predictions, including that MDA-MB-231 cells might have: a) lower de novo sphingolipid biosynthesis due to lower mRNA abundance of SPTLC1 and SPTLC2 (Fig. 1, dashed box A), whereas higher expression of SPTLC3 might indicate an elevation of sphingolipids with a 16-carbon sphingoid base backbone; b) lower proportions of Cer with unsaturated fatty acyl chains (e.g., C24:1 and C26:1) due to lower SCD, the enzyme that converts C18:0-CoA to C18:1-CoA, which is subsequently elongated to C24:1- and C26:1-CoA (45); c) lower phyto-sphingolipids in MDA-MB-231 cells due to lower DES2 (Fig. 1, dashed box C); and d) lower GalCer production but higher GlcCer and/or LacCer due to lower UGT8 and higher UDP-glucose ceramide glucosyltransferase (UGCG) and B4GALT6 (Fig. 1, dashed box D). In the interest of space, the complete figures comparisons of mRNA abundance between the two cells for other branches of de novo sphingolipid and glycosphingolipid biosynthesis are displayed in supplementary Fig. II.
To test the prediction that MDA-MB-231 cells have lower de novo sphingolipid biosynthesis, cells were incubated with [U-13C]palmitic acid, and the backbone and fatty acid-labeled sphingolipids (i.e., unambiguously biosynthesized de novo) were quantified by LC ESI-MS/MS. At each time point, there was less incorporation of 13C-palmitate into the sphingolipids of MDA-MB-231 cells than in those of MCF7 cells. Differences in the composition of these cell lines was also seen in LC ESI-MS/MS analysis of the unlabeled cells (shown in Fig. 2B), with MDA-MB-231 cells having 48.8 ± 4.1 Cer/106 cells and 783 ± 40 pmol SM/106 cells (Fig. 2B, black bars), which was significantly lower than in MCF7 cells, which had 79.8 ± 11.1 pmol Cer/106 cells (P < 0.001) and 1,220 ± 110 pmol SM/106 cells (P < 0.001) (Fig. 2B, white bars). In contrast, the amounts of unlabeled HexCer were higher in MDA-MB-231 cells, 50.3 + 3.4 pmol HexCer/106 cells, than in MCF7 cells, 36.9 ± 5.9 pmol HexCer/106 cells (P < 0.001). Overall, however, both the 13C-labeling and the SL amounts in MDA-MB-231 cells were lower than those in MCF7 cells, which were consistent with the apparent differences in SPT1 and SPT2 (Fig. 1).
Another apparent difference between MDA-MB-231 and MCF7 cells is a higher expression of SPTLC3, which has been proposed to metabolize a shorter chain (d16:1) (46, 47). Although standards are not yet available for a rigorous quantitation of d16:1-containing sphingolipids, preliminary analysis of the Cer subspecies of these cells by LC ESI-MS/MS (4) indicated that MDA-MB-231 cells had a higher proportion of d16:1-Cer (1.9 ± 0.4 pmol/106 cells) than did MCF7 cells (1.1 ± 0.2 pmol/106 cells) (P < 0.05, n = 3), which is agreement with the apparently higher SPTLC3 expression in MDA-MB-231 cells (Fig. 1).
To test the prediction from the microarray data that MDA-MB-231 cells would have lower amounts of SL with unsaturated fatty acids (due to production of less mono-unsaturated fatty acyl-CoAs because SCD expression is lower) (Fig. 1, dashed box B), the FA-CoAs and the chain-length distribution of the Cer were determined for the two cell lines. As shown in Fig. 3, MDA-MB-231 cells had significantly lower amounts of all of the major mono-unsaturated fatty acyl-CoAs (i.e., per 106 MDA-MB-231 cells: 6.3 ± 0.22 pmol of C18:1, 1.4 ± 0.2 pmol of C24:1, and 4.6 ± 2.1 pmol C26:1; compared with MCF7 cells: 14.3 ± 0.6 pmol C18:1/106 cells, P < 0.001; 5.3 ± 1 pmol C24:1/106, P < 0.001; and 10.8 ± 0.9 pmol C26:1/106 cells, P < 0.001). In some cases, the corresponding saturated FA-CoA was also somewhat lower (Fig. 3A). MDA-MB-231 cells also have significantly lower amounts of Cer with mono-unsaturated fatty acids (Fig. 3B) (the quantities for MDA-MB-231 versus MCF7 cells were: 24.8 ± 4.5 pmol versus 7.1 ± 1.0 pmol C24:1-Cer/106 cells, P < 0.001; and 3.9 ± 0.6 pmol versus 0.5 ± 0.1 pmol C26:1-Cer/106 cells, P < 0.001). In contrast, there were fewer differences in the Cer with saturated fatty acids (Fig. 3B). All together, these findings are consistent with the apparently lower expression of SCD in the MDA-MB-231 cells.
Likewise, the lower DES2 expression (Fig. 1) is reflected in lower amounts of phyto-sphingolipids in MDA-MB-231 cells (Fig. 4). Per 106 cells, the amounts in MDA-MB-231 cells (1.4 ± 0.3 pmol phyto-Cer, 0.06 ± 0.02 pmol phyto-HexCer, and 0.6 ± 0.1 pmol phyto-SM) are all lower than the amounts in MCF7 cells (6 ± 1.4 pmol phyto-Cer, P < 0.001; 0.21 ± 0.04 pmol phyto-HexCer, P < 0.001; and 17.6 ± 1.9 pmol phyto-SM, P < 0.001).
Shown in Fig. 5 are the data relevant to the branch point for biosynthesis of GalCer, which would be predicted to be lower for MDA-MB-231 cells due to lower expression of UGT8, versus GlcCer, which might be higher due to the higher UGCG in MDA-MB-231 cells. Consistent with these predictions, MDA-MB-231 cells had barely detectable GalCer (0.6 ± 0.6 pmol/106 cells), whereas MCF7 cells had 9 ± 2 pmol GalCer/106 cells (P < 0.001). In contrast, the two cell lines contain essentially the same amounts of GlcCer (61 ± 11 pmol GlcCer/106 MDA-MB-231 cells and 66 ± 10 pmol GlcCer /106 MCF7 cells, P = 0.41), despite the apparent differences in UGCG expression. This might be due to consumption of GlcCer by the next step of the pathway as B4GALT6 expression also appears to be higher for MBA-MB-231 cells, and LacCer has been reported to be higher for MDA-MB-231 compared with MCF7 cells (Fig. 5, right panel) (48).
Other predictions might be made from the microarray data (Fig. 1), but they would be more tenuous. For example, there are differences in some of the CerS that imply that the chain length subspecies of MDA-MB-231 and MCF7 cells might differ (beyond what has already been addressed above); however, two of the members (CerS1 and CerS3) were not represented in this data set, which adds unknown variables. As another example, mRNAs for both SM synthases (SMS1 and SMS2) appear to be higher for MDA-MB-231 cells, but sphingomyelinases (SMPD1 and SMPD3) are also elevated, and a transporter that is thought to be involved in Cer trafficking to Golgi for SM synthesis (CERT) is lower. On the basis of the lower SM amounts in MDA-MB-231 cells (Fig. 2), it appears that one or both of the latter factors predominate(s).
Correlation of gene predictions and metabolite analysis in breast tumor tissues
Visualization of downstream steps of sphingolipid metabolism is also possible, as shown in Fig. 6 for ganglioside biosynthesis using published data from invasive ductal carcinoma (IDC) breast tumors and normal ductal tissue (43). This dataset predicts that IDC have higher expression of GM3 synthase (which makes GM3 from LacCer), B4GALNT1 (which adds GalNAc to LacCer to produce GA2, and to GM3 to produce GM2, etc., as shown), ST3gal2 (which catalyzes the transfer of sialic acid from CMP-sialic acid to GA1 to produce GM1b, to GM1a for GD1a, etc.) and ST8SIA5 (which is responsible for biosynthesis of GT3, GT1a, GQ1b, etc.). We have not found data to evaluate all of these predictions, but published results for the sphingolipid composition of IDC tissue versus normal tissue (49) noted higher quantities of GM3 (49) as well as higher GD3, and N- and O-acetyl GD3 and GT3 (49), which might be due to elevation of GM3 because the mRNAs for the proximal enzymes (e.g., ST8Sia1 and O-AcetylT in Fig. 6) did not appear to be higher than for normal tissue. It is perhaps relevant that sera from patients with IDC have been found to have significant elevations in GD1b, GT1b, and GQ1b (highlighted in Fig. 6 as circles with colored edges) compared with healthy volunteers (15), which is also consistent with the gene expression data. It would be interesting to know whether this gene expression profile might direct investigators to additional differences in glycosphingolipid biosynthesis that might have functional relevance in this cancer or perhaps serve as biomarkers.
Fig. 7 shows a comparison of the available gene expression data for steps in the biosynthesis of globo-series glycosphingolipids for IDC versus normal tissue. From the pathway depiction, higher globoside synthase, B3GalT5, and Fut2 would be predicted to elevate Gb4, SSEA-3, and Globo-H antigen, and the latter two (highlighted in Fig. 7 as circles with colored edges) have been reported to be elevated in 77% and 61%, respectively, of breast cancer tissues (14, 50).
Comparison of the frequency of gene and sphingolipid alterations in cancers
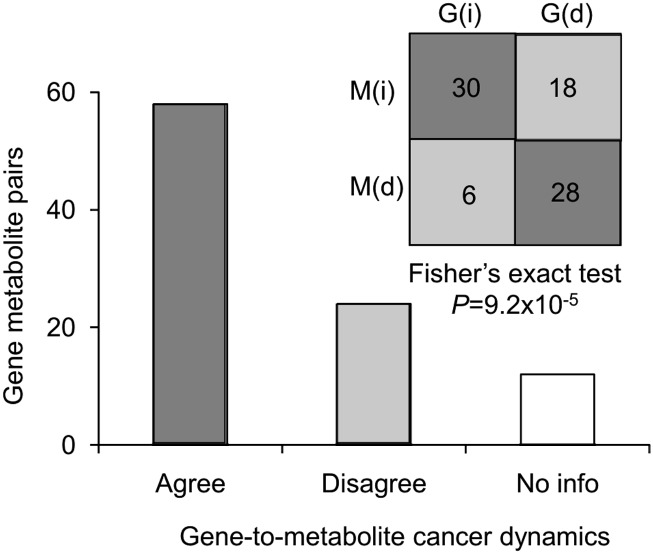
There is no comprehensive dataset that allows rigorous comparison of the correlation between gene expression data and sphingolipid composition; however, we have analyzed the frequency for the data at hand (Table 2): nine gene expression datasets for which corresponding metabolite data are available, for a total of 82 observations, including the present study. Evaluation of positive and negative correlations between gene expression differences and metabolite amounts revealed 30 instances in which both increased, 28 in which both decreased, for a total of 58 positive correlations (Fig. 8 and Table 2). In 24 cases, genes and metabolite changes showed opposing outcomes (a negative correlation). In 12 instances, the metabolites were different, but the respective mRNA changes were less than ±1.5-fold. Thus, although we do not have data for all of the genes and metabolites of this pathway, for the ones we were able to locate there was a 70.7% correlation between gene expression and sphingolipid differences, which was statistically significant at P < 0.001 by Fisher's exact test (Fig. 8).
TABLE 2.
Comparison of gene expression and glycosphingolipid differences for nine cancer datasets
| Cancer Type | Gene Expression Ref. | Metabolite Ref. | G(i)M(i) | G(d)M(d) | G(i)M(d) | G(d)M(i) |
|---|---|---|---|---|---|---|
| MDA-MB231 / MCF7 | (36) | Current analysis and (48) | 4 | 8 | 4 | 2 |
| IDC / normal ductal | (43) | (14, 15, 49, 50) | 5 | 1 | ||
| Gliosarcoma | (76) | (77) | 2 | 1 | 1 | |
| GBM / white matter | (76) | (78) | 1 | 3 | 2 | 2 |
| Serious ovarian | (79) | (13, 80) | 2 | 1 | ||
| Mucinous ovarian | (79) | (13, 80) | 4 | 1 | ||
| SSEA3 / globo-H | (36) | (81) | 3 | 7 | 5 | |
| Melanoma cells | (16) | (16) | 6 | 9 | 5 | |
| Melanoma | (82) | (83) | 3 | |||
| Total | 30 | 28 | 18 | 6 |
The table compares the agreement G(i)M(i) and G(d)M(d), and disagreement G(i)M(d) and G(i)M(d), between gene expression and sphingolipid differences for nine cancer datasets. G, gene expression difference; M, metabolite difference; (i), higher ratio; and (d), lower ratio. The total for each category were used to determine the statistical significance as indicated in Fig. 8.
Fig. 8.
Statistical comparison of gene expression differences with metabolite amounts. Results from a survey comparing nine different gene expression datasets with corresponding sphingolipids. G(i) and G(d) indicate an increase and decrease in gene levels, while M(i) and M(d) indicate increase and decrease in metabolites. The significance of agreement and disagreement between genes and metabolites was measured by Fisher's exact test (P = 0.01).
Although many factors other than mRNA amount are well known to regulate metabolic pathways (e.g., rates of translation, post translational modification, protein turnover, etc.), this degree of correlation suggests that it would still be worthwhile to use a pathway depiction of gene expression data to direct attention to particular sphingolipids for subsequent analysis by a method such as mass spectrometry. To illustrate the wide range of similarities and differences among different cancer cell lines, pathway maps based on published gene expression data (36) for the 59 cancer cells lines in the NCI 60-cell line drug screen (NCI-60, http://discover.nci.nih.gov/cellminer/) have been provided in the supplementary online data. There is clearly a rich amount of diversity that warrants exploration.
SUMMARY
These studies illustrate how use of pathway maps to visualize gene expression data can facilitate the prediction of metabolic changes and direct attention to particular sphingolipids for analysis. Two examples of this in our studies were the observation of elevated SPTLC3 in MDA-MB-231 cells versus MCF7 cells, which directed our attention toward the possibility that shorter chain (d16:1) sphingoid bases are present and that lower DES2 implied that MDA-MB-231 cells have less phyto-sphingolipids. In a separate study, we recently used this pathway map to note that epithelial ovarian cancer cells (collected by laser capture microdissection) displayed elevated expression of mRNAs for the steps in sulfatide biosynthesis; the presence of sulfatides was confirmed by LC ESI-MS/MS and tissue-imaging mass spectrometry (51). Considering the already large, and continually growing, body of data on differences in gene expression in cancer and other diseases, these pathway maps should be useful in accelerating the discovery of interesting new ways that sphingolipids are involved, and possibly in identifying new disease biomarkers. As lipidomic data sets become more plentiful, there might also be occasions where the display of the metabolite data in PathVisio could assist investigators in the discovery of interesting aspects of regulation of sphingolipid metabolism at transcription or other levels.
Supplementary Material
Acknowledgments
The authors thank Sarah Singh for working with the online visualization tool and helping explore and validate its utility.
Footnotes
Abbreviations:
- 3KSa
- 3-ketosphinganine
- BCA
- bicinchoninic acid
- Cer
- ceramide
- CerP
- ceramide 1-phosphate
- CerS
- ceramide synthase (isoforms 1-6)
- CERT
- Cer transporter
- DEGS
- dihydroceramide desaturase
- DHCer
- dihydroceramide
- DH
- dihydro- backbone
- ELOVL
- fatty acyl-CoA elongase
- EP
- ethanolamine phosphate
- Fut
- fucosyltransferase
- Fut1/Fut2
- alpha(1,2)fucosyltransferase
- GalCer
- galactosylCer
- Gb3 or CTH
- ceramide trihexoside
- Gb4
- ceramide tetrahexoside (globoside)
- GlcCer
- glucosylCer
- GSL
- glycosphingolipid
- HexCer
- monohexosylCer
- HNSCC
- head and neck small cell carcinoma
- IDC
- invasive ductal carcinoma
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- LacCer
- lactosylceramide
- LCB
- long chain base
- phyto-Cer
- phytoceramide
- phyto-HexCer
- monohexosyl phytoceramide
- phyto-SM
- phytosphingomyelin
- Sa
- sphinganine
- Sa1P
- sphinganine 1-phosphate
- SCD
- stearyol-CoA desaturase
- SL
- sphingolipid
- SM
- sphingomyelin
- SPTLC1
- serine palmitoyltransferase 1
- SPTLC2
- serine palmitoyltransferase 2
- SPTLC3
- serine palmitoyltransferase 3
- SPHK
- sphingosine kinase (isoforms 1 and 2)
- So
- sphingosine
- So1P
- sphingosine 1-phosphate
- SSEA-3
- stage-specific embryonic antigen 3
- SSEA-4
- stage-specific embryonic antigen 4
- ST
- sulfatide
- UGCG
- UDP-glucose ceramide glucosyltransferase
- UGT1
- glucosylceramide synthase
- UGT8
- galactosylceramide synthase
This work by was supported by the National Institutes of Health Grant U54 GM-069338 (Lipid MAPS Consortium “Glue” Grant). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of i) one table with accompanying text, ii) online tutorial screens, iii) two figures, and iv) 59 heat maps.
The fatty acyl-CoA elongases and desaturases that biosynthesize the cosubstrates for CerS are also shown in this diagram and in supplementary Fig. I.
These differences are referred to as “apparent” because microarray data are not quantitative and should be validated by another method before drawing mechanistic conclusions. However, the purpose of this study was not to establish the validity of the gene expression data per se, but rather to explore whether publicly available microarray data can aid sphingolipid analysis by directing attention to possible differences.
REFERENCES
- 1.Hannun Y. A., Obeid L. M. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9: 139–150. [DOI] [PubMed] [Google Scholar]
- 2.Maceyka M., Milstien S., Spiegel S. 2009. Sphingosine-1-phosphate: the Swiss army knife of sphingolipid signaling. J. Lipid Res. 50(Suppl): S272–S276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez P. H., Schnaar R. L. 2009. Gangliosides in cell recognition and membrane protein regulation. Curr. Opin. Struct. Biol. 19: 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaner R. L., Allegood J. C., Park H., Wang E., Kelly S., Haynes C. A., Sullards M. C., Merrill A. H., Jr 2009. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 50: 1692–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haynes C. A., Allegood J. C., Park H., Sullards M. C. 2009. Sphingolipidomics: methods for the comprehensive analysis of sphingolipids. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2696–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han X., Jiang X. 2009. A review of lipidomic technologies applicable to sphingolipidomics and their relevant applications. Eur. J. Lipid Sci. Technol. 111: 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirsch S., Zarei M., Cindric M., Muthing J., Bindila L., Peter-Katalinic J. 2008. On-line nano-HPLC/ESI QTOF MS and tandem MS for separation, detection, and structural elucidation of human erythrocytes neutral glycosphingolipid mixture. Anal. Chem. 80: 4711–4722. [DOI] [PubMed] [Google Scholar]
- 8.Serb A., Schiopu C., Flangea C., Sisu E., Zamfir A. D. 2009. Top-down glycolipidomics: fragmentation analysis of ganglioside oligosaccharide core and ceramide moiety by chip-nanoelectrospray collision-induced dissociation MS2–MS6. J. Mass Spectrom. 44: 1434–1442. [DOI] [PubMed] [Google Scholar]
- 9.Merrill A. H., Jr, Wang M. D., Park M., Sullards M. C. 2007. (Glyco)sphingolipidology: an amazing challenge and opportunity for systems biology. Trends Biochem. Sci. 32: 457–468. [DOI] [PubMed] [Google Scholar]
- 10.Hakomori S. I. 2008. Structure and function of glycosphingolipids and sphingolipids: recollections and future trends. Biochim. Biophys. Acta. 1780: 325–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakomori S. 2002. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc. Natl. Acad. Sci. USA. 99: 10231–10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hakomori S. 2001. Tumor-associated carbohydrate antigens defining tumor malignancy: basis for development of anti-cancer vaccines. Adv. Exp. Med. Biol. 491: 369–402. [DOI] [PubMed] [Google Scholar]
- 13.Makhlouf A. M., Fathalla M. M., Zakhary M. A., Makarem M. H. 2004. Sulfatides in ovarian tumors: clinicopathological correlates. Int. J. Gynecol. Cancer. 14: 89–93. [DOI] [PubMed] [Google Scholar]
- 14.Chang W. W., Lee C. H., Lee P., Lin J., Hsu C. W., Hung J. T., Lin J. J., Yu J. C., Shao L. E., Yu J., et al. 2008. Expression of Globo H and SSEA3 in breast cancer stem cells and the involvement of fucosyl transferases 1 and 2 in Globo H synthesis. Proc. Natl. Acad. Sci. USA. 105: 11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiesner D. A., Sweeley C. C. 1995. Circulating gangliosides of breast-cancer patients. Int. J. Cancer. 60: 294–299. [DOI] [PubMed] [Google Scholar]
- 16.Yamashiro S., Okada M., Haraguchi M., Furukawa K., Lloyd K. O., Shiku H. 1995. Expression of alpha 2,8-sialyltransferase (GD3 synthase) gene in human cancer cell lines: high level expression in melanomas and up-regulation in activated T lymphocytes. Glycoconj. J. 12: 894–900. [DOI] [PubMed] [Google Scholar]
- 17.Senkal C. E., Ponnusamy S., Rossi M. J., Bialewski J., Sinha D., Jiang J. C., Jazwinski S. M., Hannun Y. A., Ogretmen B. 2007. Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol. Cancer Ther. 6: 712–722. [DOI] [PubMed] [Google Scholar]
- 18.van Iersel M. P., Kelder T., Pico A. R., Hanspers K., Coort S., Conklin B. R., Evelo C. 2008. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 9: 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merrill A. H., Jr, Stokes T. H., Momin A., Park H., Portz B. J., Kelly S., Wang E., Sullards M. C., Wang M. D. 2009. Sphingolipidomics: a valuable tool for understanding the roles of sphingolipids in biology and disease. J. Lipid Res. 50(Suppl): S97–S102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laviad E. L., Albee L., Pankova-Kholmyansky I., Epstein S., Park H., Merrill A. H., Jr, Futerman A. H. 2008. Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 283: 5677–5684. [DOI] [PubMed] [Google Scholar]
- 21.Mizutani Y., Kihara A., Igarashi Y. 2005. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 390: 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizutani Y., Kihara A., Igarashi Y. 2006. LASS3 (longevity assurance homologue 3) is a mainly testis-specific (dihydro)ceramide synthase with relatively broad substrate specificity. Biochem. J. 398: 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riebeling C., Allegood J. C., Wang E., Merrill A. H., Jr, Futerman A. H. 2003. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J. Biol. Chem. 278: 43452–43459. [DOI] [PubMed] [Google Scholar]
- 24.Venkataraman K., Riebeling C., Bodennec J., Riezman H., Allegood J. C., Sullards M. C., Merrill A. H., Jr, Futerman A. H. 2002. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J. Biol. Chem. 277: 35642–35649. [DOI] [PubMed] [Google Scholar]
- 25.Omae F., Miyazaki M., Enomoto A., Suzuki M., Suzuki Y., Suzuki A. 2004. DES2 protein is responsible for phytoceramide biosynthesis in the mouse small intestine. Biochem. J. 379: 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng W., Kollmeyer J., Symolon H., Momin A., Munter E., Wang E., Kelly S., Allegood J. C., Liu Y., Peng Q., et al. 2006. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim. Biophys. Acta. 1758: 1864–1884. [DOI] [PubMed] [Google Scholar]
- 27.Pico A. R., Kelder T., van Iersel M. P., Hanspers K., Conklin B. R., Evelo C. 2008. WikiPathways: pathway editing for the people. PLoS Biol. 6: e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett T., Troup D. B., Wilhite S. E., Ledoux P., Rudnev D., Evangelista C., Kim I. F., Soboleva A., Tomashevsky M., Edgar R. 2007. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic Acids Res. 35: D760–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkinson H., Sarkans U., Shojatalab M., Abeygunawardena N., Contrino S., Coulson R., Farne A., Lara G. G., Holloway E., Kapushesky M., et al. 2005. ArrayExpress--a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 33: D553–D555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhodes D. R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A. M. 2004. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 6: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sean D., Meltzer P. S. 2007. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 23: 1846–1847. [DOI] [PubMed] [Google Scholar]
- 32.Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu G., Loraine A. E., Shigeta R., Cline M., Cheng J., Valmeekam V., Sun S., Kulp D., Siani-Rose M. A. 2003. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 31: 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kent W. J. 2002. BLAT--the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shankavaram U. T., Reinhold W. C., Nishizuka S., Major S., Morita D., Chary K. K., Reimers M. A., Scherf U., Kahn A., Dolginow D., et al. 2007. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol. Cancer Ther. 6: 820–832. [DOI] [PubMed] [Google Scholar]
- 37.Sullards M. C., Allegood J. C., Kelly S., Wang E., Haynes C. A., Park H., Chen Y., Merrill A. H., Jr 2007. Structure-specific, quantitative methods for analysis of sphingolipids by liquid chromatography-tandem mass spectrometry: “inside-out” sphingolipidomics. Methods Enzymol. 432: 83–115. [DOI] [PubMed] [Google Scholar]
- 38.Han G., Gupta S. D., Gable K., Niranjanakumari S., Moitra P., Eichler F., Brown R. H., Jr, Harmon J. M., Dunn T. M. 2009. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl. Acad. Sci. USA. 106: 8186–8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruett S. T., Bushnev A., Hagedorn K., Adiga M., Haynes C. A., Sullards M. C., Liotta D. C., Merrill A. H., Jr 2008. Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols. J. Lipid Res. 49: 1621–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaoka S., Miyaji M., Kitano T., Umehara H., Okazaki T. 2004. Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J. Biol. Chem. 279: 18688–18693. [DOI] [PubMed] [Google Scholar]
- 41.Ternes P., Brouwers J. F., van den Dikkenberg J., Holthuis J. C. 2009. Sphingomyelin synthase SMS2 displays dual activity as ceramide phosphoethanolamine synthase. J. Lipid Res. 50: 2270–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanada K., Kumagai K., Tomishige N., Yamaji T. 2009. CERT-mediated trafficking of ceramide. Biochim. Biophys. Acta. 1791: 684–691. [DOI] [PubMed] [Google Scholar]
- 43.Turashvili G., Bouchal J., Baumforth K., Wei W., Dziechciarkova M., Ehrmann J., Klein J., Fridman E., Skarda J., Srovnal J., et al. 2007. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 7: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoeflich K. P., O'Brien C., Boyd Z., Cavet G., Guerrero S., Jung K., Januario T., Savage H., Punnoose E., Truong T., et al. 2009. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin. Cancer Res. 15: 4649–4664. [DOI] [PubMed] [Google Scholar]
- 45.Kishimoto Y., Radin N. S. 1963. Biosynthesis of nervonic acid and its homologues from carboxyl-labeled oleic acid. J. Lipid Res. 4: 444–447. [PubMed] [Google Scholar]
- 46.Hornemann T., Penno A., Rutti M. F., Ernst D., Kivrak-Pfiffner F., Rohrer L., von Eckardstein A. 2009. The SPTLC3 subunit of serine palmitoyltransferase generates short chain sphingoid bases. J. Biol. Chem. 284: 26322–26330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han G., Gupta S. D., Gable K., Niranjanakumari S., Moitra P., Eichler F., Brown R. H., Harmon J. M., Dunn T. M. 2009. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl. Acad. Sci. USA. 106: 8186–8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nohara K., Wang F., Spiegel S. 1998. Glycosphingolipid composition of MDA-MB-231 and MCF-7 human breast cancer cell lines. Breast Cancer Res. Treat. 48: 149–157. [DOI] [PubMed] [Google Scholar]
- 49.Marquina G., Waki H., Fernandez L. E., Kon K., Carr A., Valiente O., Perez R., Ando S. 1996. Gangliosides expressed in human breast cancer. Cancer Res. 56: 5165–5171. [PubMed] [Google Scholar]
- 50.Menard S., Tagliabue E., Canevari S., Fossati G., Colnaghi M. I. 1983. Generation of monoclonal antibodies reacting with normal and cancer cells of human breast. Cancer Res. 43: 1295–1300. [PubMed] [Google Scholar]
- 51.Liu Y., Chen Y., Momin A., Shaner R., Wang E., Bowen N. J., Matyunina L. V., Walker L. D., McDonald J. F., Sullards M. C., et al. 2010. Elevation of sulfatides in ovarian cancer: an integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol. Cancer. 9: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hornemann T., Richard S., Rutti M. F., Wei Y., von Eckardstein A. 2006. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J. Biol. Chem. 281: 37275–37281. [DOI] [PubMed] [Google Scholar]
- 53.Kihara A., Igarashi Y. 2004. FVT-1 is a mammalian 3-ketodihydrosphingosine reductase with an active site that faces the cytosolic side of the endoplasmic reticulum membrane. J. Biol. Chem. 279: 49243–49250. [DOI] [PubMed] [Google Scholar]
- 54.Liu H., Sugiura M., Nava V. E., Edsall L. C., Kono K., Poulton S., Milstien S., Kohama T., Spiegel S. 2000. Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. J. Biol. Chem. 275: 19513–19520. [DOI] [PubMed] [Google Scholar]
- 55.Van Veldhoven P. P., Gijsbers S., Mannaerts G. P., Vermeesch J. R., Brys V. 2000. Human sphingosine-1-phosphate lyase: cDNA cloning, functional expression studies and mapping to chromosome 10q22(1). Biochim. Biophys. Acta. 1487: 128–134. [DOI] [PubMed] [Google Scholar]
- 56.Mao C., Xu R., Szulc Z. M., Bielawski J., Becker K. P., Bielawska A., Galadari S. H., Hu W., Obeid L. M. 2003. Cloning and characterization of a mouse endoplasmic reticulum alkaline ceramidase: an enzyme that preferentially regulates metabolism of very long chain ceramides. J. Biol. Chem. 278: 31184–31191. [DOI] [PubMed] [Google Scholar]
- 57.Koch J., Gartner S., Li C. M., Quintern L. E., Bernardo K., Levran O., Schnabel D., Desnick R. J., Schuchman E. H., Sandhoff K. 1996. Molecular cloning and characterization of a full-length complementary DNA encoding human acid ceramidase. Identification of the first molecular lesion causing Farber disease. J. Biol. Chem. 271: 33110–33115. [DOI] [PubMed] [Google Scholar]
- 58.El Bawab S., Roddy P., Qian T., Bielawska A., Lemasters J. J., Hannun Y. A. 2000. Molecular cloning and characterization of a human mitochondrial ceramidase. J. Biol. Chem. 275: 21508–21513. [DOI] [PubMed] [Google Scholar]
- 59.Xu R., Jin J., Hu W., Sun W., Bielawski J., Szulc Z., Taha T., Obeid L. M., Mao C. 2006. Golgi alkaline ceramidase regulates cell proliferation and survival by controlling levels of sphingosine and S1P. FASEB J. 20: 1813–1825. [DOI] [PubMed] [Google Scholar]
- 60.Mao C., Xu R., Szulc Z. M., Bielawska A., Galadari S. H., Obeid L. M. 2001. Cloning and characterization of a novel human alkaline ceramidase. A mammalian enzyme that hydrolyzes phytoceramide. J. Biol. Chem. 276: 26577–26588. [DOI] [PubMed] [Google Scholar]
- 61.Huitema K., van den Dikkenberg J., Brouwers J. F., Holthuis J. C. 2004. Identification of a family of animal sphingomyelin synthases. EMBO J. 23: 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ichikawa S., Sakiyama H., Suzuki G., Hidari K. I., Hirabayashi Y. 1996. Expression cloning of a cDNA for human ceramide glucosyltransferase that catalyzes the first glycosylation step of glycosphingolipid synthesis. Proc. Natl. Acad. Sci. USA. 93: 4638–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bosio A., Binczek E., Le Beau M. M., Fernald A. A., Stoffel W. 1996. The human gene CGT encoding the UDP-galactose ceramide galactosyl transferase (cerebroside synthase): cloning, characterization, and assignment to human chromosome 4, band q26. Genomics. 34: 69–75. [DOI] [PubMed] [Google Scholar]
- 64.Lamour N. F., Stahelin R. V., Wijesinghe D. S., Maceyka M., Wang E., Allegood J. C., Merrill A. H., Jr, Cho W., Chalfant C. E. 2007. Ceramide kinase uses ceramide provided by ceramide transport protein: localization to organelles of eicosanoid synthesis. J. Lipid Res. 48: 1293–1304. [DOI] [PubMed] [Google Scholar]
- 65.Sugiura M., Kono K., Liu H., Shimizugawa T., Minekura H., Spiegel S., Kohama T. 2002. Ceramide kinase, a novel lipid kinase. Molecular cloning and functional characterization. J. Biol. Chem. 277: 23294–23300. [DOI] [PubMed] [Google Scholar]
- 66.Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. 2003. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 426: 803–809. [DOI] [PubMed] [Google Scholar]
- 67.Schuchman E. H., Suchi M., Takahashi T., Sandhoff K., Desnick R. J. 1991. Human acid sphingomyelinase. Isolation, nucleotide sequence and expression of the full-length and alternatively spliced cDNAs. J. Biol. Chem. 266: 8531–8539. [PubMed] [Google Scholar]
- 68.Hofmann K., Tomiuk S., Wolff G., Stoffel W. 2000. Cloning and characterization of the mammalian brain-specific, Mg2+-dependent neutral sphingomyelinase. Proc. Natl. Acad. Sci. USA. 97: 5895–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takizawa M., Nomura T., Wakisaka E., Yoshizuka N., Aoki J., Arai H., Inoue K., Hattori M., Matsuo N. 1999. cDNA cloning and expression of human lactosylceramide synthase. Biochim. Biophys. Acta. 1438: 301–304. [DOI] [PubMed] [Google Scholar]
- 70.Ogawa D., Shikata K., Honke K., Sato S., Matsuda M., Nagase R., Tone A., Okada S., Usui H., Wada J., et al. 2004. Cerebroside sulfotransferase deficiency ameliorates L-selectin-dependent monocyte infiltration in the kidney after ureteral obstruction. J. Biol. Chem. 279: 2085–2090. [DOI] [PubMed] [Google Scholar]
- 71.Jakobsson A., Westerberg R., Jacobsson A. 2006. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog. Lipid Res. 45: 237–249. [DOI] [PubMed] [Google Scholar]
- 72.Miyazaki M., Ntambi J. M. 2003. Role of stearoyl-coenzyme A desaturase in lipid metabolism. Prostaglandins Leukot. Essent. Fatty Acids. 68: 113–121. [DOI] [PubMed] [Google Scholar]
- 73.Han G., Gupta S. D., Gable K., Niranjanakumari S., Moitra P., Eichler F., Brown R. H., Jr, Harmon J. M., Dunn T. M. 2009. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl. Acad. Sci. USA. 106: 8186–8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Han S., Lone M. A., Schneiter R., Chang A. 2010. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. USA. 107: 5851–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., Weissman J. S. 2010. Orm family proteins mediate sphingolipid homeostasis. Nature. 463: 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bredel M., Bredel C., Juric D., Duran G. E., Yu R. X., Harsh G. R., Vogel H., Recht L. D., Scheck A. C., Sikic B. I. 2006. Tumor necrosis factor-alpha-induced protein 3 as a putative regulator of nuclear factor-kappaB-mediated resistance to O6-alkylating agents in human glioblastomas. J. Clin. Oncol. 24: 274–287. [DOI] [PubMed] [Google Scholar]
- 77.Vukelic Z., Kalanj-Bognar S., Froesch M., Bindila L., Radic B., Allen M., Peter-Katalinic J., Zamfir A. D. 2007. Human gliosarcoma-associated ganglioside composition is complex and distinctive as evidenced by high-performance mass spectrometric determination and structural characterization. Glycobiology. 17: 504–515. [DOI] [PubMed] [Google Scholar]
- 78.Sung C. C., Pearl D. K., Coons S. W., Scheithauer B. W., Johnson P. C., Yates A. J. 1994. Gangliosides as diagnostic markers of human astrocytomas and primitive neuroectodermal tumors. Cancer. 74: 3010–3022. [DOI] [PubMed] [Google Scholar]
- 79.Hendrix N. D., Wu R., Kuick R., Schwartz D. R., Fearon E. R., Cho K. R. 2006. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 66: 1354–1362. [DOI] [PubMed] [Google Scholar]
- 80.Kiguchi K., Takamatsu K., Tanaka J., Nozawa S., Iwamori M., Nagai Y. 1992. Glycosphingolipids of various human ovarian tumors: a significantly high expression of I3SO3GalCer and Lewis antigen in mucinous cystadenocarcinoma. Cancer Res. 52: 416–421. [PubMed] [Google Scholar]
- 81.Schrump D. S., Furukawa K., Yamaguchi H., Lloyd K. O., Old L. J. 1988. Recognition of galactosylgloboside by monoclonal antibodies derived from patients with primary lung cancer. Proc. Natl. Acad. Sci. USA. 85: 4441–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Riker A. I., Enkemann S. A., Fodstad O., Liu S., Ren S., Morris C., Xi Y., Howell P., Metge B., Samant R. S., et al. 2008. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med. Genomics. 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ravindranath M. H., Muthugounder S., Presser N. 2008. Ganglioside signatures of primary and nodal metastatic melanoma cell lines from the same patient. Melanoma Res. 18: 47–55. [DOI] [PubMed] [Google Scholar]
- 84.Haynes C. A., Allegood J. C., Sims K., Wang E. W., Sullards M. C., Merrill A. H., Jr 2008. Quantitation of fatty acyl-coenzyme As in mammalian cells by liquid chromatography-electrospray ionization tandem mass spectrometry. J. Lipid Res. 49: 1113–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.