Abstract
Increased serum apolipoprotein (apo)B and associated LDL levels are well-correlated with an increased risk of coronary disease. ApoE–/– and low density lipoprotein receptor (LDLr)–/– mice have been extensively used for studies of coronary atherosclerosis. These animals show atherosclerotic lesions similar to those in humans, but their serum lipids are low in apoB-containing LDL particles. We describe the development of a new mouse model with a human-like lipid profile. Ldlr CETP+/– hemizygous mice carry a single copy of the human CETP transgene and a single copy of a LDL receptor mutation. To evaluate the apoB pathways in this mouse model, we used novel short-interfering RNAs (siRNA) formulated in lipid nanoparticles (LNP). ApoB siRNAs induced up to 95% reduction of liver ApoB mRNA and serum apoB protein, and a significant lowering of serum LDL in Ldlr CETP+/– mice. ApoB targeting is specific and dose-dependent, and it shows lipid-lowering effects for over three weeks. Although specific triglycerides (TG) were affected by ApoB mRNA knockdown (KD) and the total plasma lipid levels were decreased by 70%, the overall lipid distribution did not change. Results presented here demonstrate a new mouse model for investigating additional targets within the ApoB pathways using the siRNA modality.
Keywords: low density lipoprotein cholesterol, low density lipoprotein receptor, cholesteryl ester transfer protein, short-interfering RNA
Coronary atherosclerosis is the most prevalent disease in industrialized societies. Although numerous advances have been made in understanding the underlying causes of atherosclerosis and treatment thereof, this condition still remains the leading cause of death in the Western world. The most important risk factor for atherosclerosis is hyperlipidemia (1). Development of atherosclerosis correlates with high levels of low density lipoprotein cholesterol (LDL). As a result, several therapies have been developed for management of LDL levels. Among these, statins are most widely used (2). However, there is a range of statin response in humans, and a subset of familial hyperlipidemia patients is unresponsive to statins, prompting the development of additional therapies.
Mouse models have been widely used for studies of atherosclerosis and hyperlipidemia. Since wild-type mice have very high anti-atherogenic high density lipoprotein (HDL) and low pro-atherogenic LDL lipoprotein levels, genetic manipulations have been necessary for development of mouse models of coronary atherosclerosis. Development of these animal models started with the discovery of human mutations in individuals with lipoprotein disorders. The best-known examples include mutations in the LDL receptor (LDLr) in association with familial hypercholesterolemia (FH) and mutations in ApoE (including ApoE Leiden) in cases of Type III hyperlipoproteinemia (HLP) (3–5). As a result, LDLr–/–, Apo E–/–, and ApoE3-Leiden transgenic mice have been widely used as mouse models of atherosclerosis (6–9). These animals show a range of changes in serum lipids in response to different diets and exhibit atherosclerotic lesions very similar to the lesions found in humans. However, these animals have serum lipid profiles that are very different from those of normal healthy humans that make them less than optimal for use in studies of lipid homeostasis.
Here we describe the development of a mouse model with a human-like lipid profile for use in preclinical studies of coronary atherosclerosis. These mice were genetically engineered to contain one copy of human cholesteryl ester transfer protein (CETP+/–) and are hemizygous for a LDL receptor mutation (Ldlr+/–). Ldlr CETP+/– hemizygous mice have serum lipid levels very similar to those of healthy humans, which make them suitable for investigation of lipid changes in response to different treatment regimens, and are very similar in lipid composition to ApoE3-Leiden/CETP transgenic mice (10, 11). We have used Ldlr CETP+/– mice to explore the effect of targeting ApoB mRNA in the liver using chemically modified siRNAs. ApoB is the main lipoprotein required for synthesis and secretion of VLDL particles from the liver (12). Levels of ApoB protein, LDL, and total cholesterol (TC) are highly correlated with increased risk for atherosclerosis. Patients with FH, who show reduced uptake of apoB-bound LDL from the circulation, are at high risk for development of coronary heart disease and atherosclerosis (13, 14). Contrarily, humans with very low levels of plasma apoB reported in particular cases of familial hypobetalipoproteinemia (FHBL) are at a reduced risk for coronary atherosclerosis (15). Because targeting of ApoB has proven difficult with conventional small molecule approaches, it presents an attractive target for development of a putative RNAi-based therapeutic.
RNA interference (RNAi) is a regulatory sequence-dependent RNA silencing mechanism that uses small double-stranded RNA (dsRNA) molecules to direct gene silencing in a homology-based manner (16). These molecules, also known as short-interfering RNAs (siRNA), recruit a RNA-induced silencing complex (RISC) to the target mRNA and eventually lead to site-specific cleavage of the target mRNA and its subsequent degradation (17). RNAi-mediated gene silencing has been extensively used for target validation as it enables fast and relatively inexpensive screens without the need to generate knockout (KO) animals. We used chemically modified siRNAs in a mouse model with a human-like lipid profile to interrogate ApoB pathways. We demonstrated that LNP-formulated siRNAs can be successfully used in Ldlr CETP+/– hemizygous mice to achieve hepatic ApoB mRNA knockdown and that this reduction in ApoB mRNA levels results in significant reductions in serum ApoB protein, changes in genes in the lipid and fatty acid pathways, significant and prolonged reductions in serum total cholesterol, triglycerides, and LDL levels, as well as correlative hepatic steatosis.
MATERIALS AND METHODS
siRNA synthesis and characterization
Chemically modified siRNAs used in these studies were synthesized and characterized as previously described (18–20). ApoB lead siRNAs used in these experiments are listed in Table 1 (all in the 5′-3′ direction). siRNA sequences contained the following chemical modifications added to the 2′ position of the ribose sugar when indicated: deoxy (d), 2′ fluoro (flu), or 2′ O-methyl (ome). Modification abbreviations are given immediately preceding the base to which they were applied. Passenger strands are blocked with an inverted abasic nucleotide on the 5′ and 3′ ends (iB). Nontargeting control siRNAs (nt controls) used in the experiments are listed in Table 2 (all in the 5′-3′ direction). Nontargeting siRNA sequences contained the same chemical modifications as ApoB siRNAs described above.
TABLE 1.
List of lead ApoB siRNAs used for in vitro and in vivo screens
| siRNA | Guide Strand Sequence | Passenger Strand Sequence |
|---|---|---|
| ApoB:(19) | rU;rU;rU;fluC;omeA;omeA;fluU;fluU;omeG;fluU;omeA;fluU;omeG;fluU;omeG;omeA;omeG;omeA;omeG;omeU;omeU | iB;fluC;fluU;fluC;fluU;fluC;dA;fluC;dA;fluU;dA;fluC;dA;dA;fluU;fluU;dG;dA;dA;dA;dT;dT;iB |
| ApoB:(1446) | rU;rU;rU;fluU;omeG;fluC;fluU;fluU;fluC;omeA;fluU;fluU;omeA;fluU;omeA;omeG;omeG;omeA;omeG;omeU;omeU | iB;fluC;fluU;fluC;fluC;fluU;dA;fluU;dA;dA;fluU;dG;dA;dA;dG;fluC;dA;dA;dA;dA;dT;dT;iB |
| ApoB:(9216) | rA;rA;rU;fluU;fluU;fluU;fluU;fluC;omeA;omeA;omeA;omeG;fluU;fluU;fluC;fluC;omeA;omeA;fluU;omeU;omeU | iB;dA;fluU;fluU;dG;dG;dA;dA;fluC;fluU;fluU;fluU;dG;dA;dA;dA;dA;dA;fluU;fluU;dT;dT;iB |
| ApoB:(9331) | rU;rA;rG;fluU;fluU;omeA;fluU;fluU;fluC;omeA;omeG;omeG;omeA;omeA;omeG;fluU;fluC;fluU;omeA;omeU;omeU | iB;fluU;dA;dG;dA;fluC;fluU;fluU;fluC;fluC;fluU;dG;dA;dA;fluU;dA;dA;fluC;fluU;dA;dT;dT;iB |
| ApoB:(10168) | rU;rU;rG;omeG;fluU;omeA;fluU;fluU;fluC;omeA;omeG;fluU;omeG;fluU;omeG;omeA;fluU;omeG;omeA;omeU;omeU | iB;fluU;fluC;dA;fluU;fluC;dA;fluC;dA;fluC;fluU;dG;dA;dA;fluU;dA;fluC;fluC;dA;dA;dT;dT;iB |
| ApoB:(9514) | rA;rU;rU;fluU;fluC;omeA;omeG;omeG;omeA;omeA;fluU;fluU;omeG;fluU;fluU;omeA;omeA;omeA;omeG;omeU;omeU | iB;fluC;fluU;fluU;fluU;dA;dA;fluC;dA;dA;fluU;fluU;fluC;fluC;fluU;dG;dA;dA;dA;fluU;dT;dT;iB |
TABLE 2.
List of nt control siRNAs used for in vitro and in vivo screens
| siRNA | Guide Strand Sequence | Passenger Strand Sequence |
|---|---|---|
| nt control 1 | rU;rU;rA;fluC;omeA;fluU;fluU;omeA;omeA;omeA;omeG;fluU;fluC;fluU;omeG;fluU;fluU;omeG;fluU;omeU;omeU | iB;dA;fluC;dA;dA;fluC;dA;dG;dA;fluC;fluU;fluU;fluU;dA;dA;fluU;dG;fluU;dA;dA;dT;dT;iB |
| nt control 2 | fluC;fluC;fluU;omeG;omeA;omeA;omeG;omeA;omeG;omeA;omeG;fluU;fluU;omeA;omeA;omeA;rA;rG;rA;omeU;omeU | iB;fluU;fluC;fluU;fluU;fluU;fluU;dA;dA;fluC;fluU;fluC;fluU;fluC;fluU;fluU;fluC;dA;dG;dG;dT;dT;iB |
Encapsulation of siRNA
siRNAs were encapsulated into two different types of liposomes. Liposome 201 (LNP201) with a lipid composition of CLinDMA (2-{4-[ (3b)-cholest-5-en-3-yloxy]butoxy}-N,N-dimethyl-3-[(9Z,12Z)-octadeca-9,12-dien-1-yloxy]propan-1-amine), cholesterol, and PEG-DMG (monomethoxy(polyethyleneglycol)-1,2-dimyristoylglycerol) in 50.3:44.3:5.4 molar ratio, respectively, was prepared as previously described (21, 22). Liposome OCD was made using the cationic lipid CLinDMA (2-{4-[(3b)-cholest-5-en-3-yloxy]octyl}-N,N-dimethyl-3-[(9Z,12Z)-octadeca-9,12-dien-1-yloxy]propan-1-amine), cholesterol, and PEG-DMG (monomethoxy(polyethyleneglycol)-1,2-dimyristoylglycerol) in 60:38:2 molar ratio, respectively. siRNAs were incorporated into the LNPs with high encapsulation efficiency by mixing siRNA in citrate buffer with an ethanolic solution of the lipid mixture, followed by a stepwise diafiltration process. Cholesterol was purchased from Northern Lipids (Northern Lipids, Inc., Canada), PEG-DMG was purchased from NOF Corporation (NOF Co., Japan), and CLinDMA was synthesized by Merck and Co, Inc. The encapsulation efficiency of the particles was determined using a SYBR Gold fluorescence assay in the absence and presence of triton, and the particle size measurements were performed using a Wyatt DynaPro plate reader (Wyatt Technology Co.). The siRNA and lipid concentrations in the LNP were quantified by a HPLC method developed inhouse, using a PDA and ELSD detector, respectively.
Cell culture studies
Mouse hepatoma Hepa1-6 cells were grown in Dulbecco's modified Eagle medium (Mediatech, Cat # 10-013-CV) with 4 mM L-glutamine adjusted to contain 1.5 g/l sodium bicarbonate and 4.5 g/l glucose and supplemented with 10% of fetal bovine serum, 100 µg/ml of streptomycin, and 100 U/ml penicillin. Cells were plated (5 × 103 cells/well) in a 96-well microtiter plates and incubated overnight at 37°C/5%CO2. A total of 49 siRNA sequences were screened in the initial screening. Transfections were performed using Lipofectamine RNAiMAX reagent as per manufacturer's instructions (Invitrogen, Cat # 13778-150). Final siRNA concentration was 10 nM per well. For dose response curves, cells were plated, cultured, and transfected as described for the initial screening. The final concentrations of each siRNA dose curve were 6-fold dilutions starting at 30 nM for a 12-point curve. Cell lysates were prepared using Biomek FX liquid handler and Cells-to-Ct Bulk Lysis Reagents (ABI, Cat # 4391851C) according to the manufacturer's instructions. cDNA was generated using Ambion Cells-to-Ct Kit (ABI, Cat # 4368813). Taqman qPCR analysis was done on ABI 7900 Real-Time PCR System. Reactions were set up using TaqMan Gene Expression Master Mix (ABI, Cat # 4370074) and commercially available Taqman probes and primers (Applied Biosystems mouse GAPDH, Cat # 4352339E; mouse ApoB, Assay ID Mm01545154_g1). Data analysis was performed as described below.
Generation of Ldlr CETP+/+ and Ldlr CETP+/− mice
Mice homozygous for the Ldlr mutation (Ldlr–/–, stock number 002077) were purchased from the Jackson Laboratory (The Jackson Laboratory). Mice homozygous for the human CETP transgene (CETP+/+, model number 1003), driven by the human ApoA1 promoter, were purchased from Taconic Farms (Taconic Farms, Inc.) through a renewable crossbreeding agreement. Ldlr–/– mice were crossed with CETP+/+ mice, and the resulting hemizygous intercross generation were bred together to identify and produce a homozygous Ldlr–/– CETP+/+ breeding colony. Genotyping for the Ldlr–/– mutation was done according to the Jackson Laboratory protocol, and genotyping for CETP+/+ gene expression was accomplished with a quantitative TaqMan assay. For the production of Ldlr CETP+/– hemizygous mice, female C57BL/6NTac (B6) mice from Taconic Farms were bred to male homozygous Ldlr–/– CETP+/+ mice. All animal studies were conducted at Sirna Therapeutics or Merck Research Laboratories with the approval by Sirna's and Merck's Institutional Animal Care and Use Committees (IACUC). Both facilities are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Analysis of in vivo ApoB mRNA knockdown
In vivo efficacy studies were conducted in Balb/C mice obtained from Harlan Laboratories (Harlan Laboratories, Inc.). Animals weighed approximately 20-25 g at the time of the study. Mice were dosed intravenously via tail vein injections with a single 3 mg/kg dose of LNP-encapsulated siRNAs and euthanized at different times following dosing. Cohorts for controls and each siRNA tested consisted of five animals. Blood and liver samples were collected immediately following euthanization. Total RNA was isolated from liver tissue using the RNeasy 96 Tissue Kit (Qiagen, Cat # 74881) according to the manufacturer's instructions. All RNA samples were treated with DNase I (Qiagen, Cat # 79254) on a column for 15 min at room temperature. Final RNA eluted was quantified and normalized to a concentration of 50 ng/μl. First strand cDNA was generated from 0.1 µg of total RNA using the MultiScribe™ Reverse Transcriptase and Random Primers contained in a High-Capacity cDNA Reverse Transcription Kit (ABI, Cat # 4368813). Taqman qPCR analysis was done on an ABI 7900 Real-Time PCR System as described for cell culture studies.
Taqman data analysis
Taqman data analysis was done using standard methods on an ABI 7900 Real-Time PCR System. Within each experiment, the baseline was set in the exponential phase of the amplification curve. The instrument assigned a cycle threshold (Ct) value based on the intersection point of the baseline with the amplification curve. The expression level of the gene of interest and percent knockdown (KD) were calculated using the comparative Ct method:
ΔCt = Ct Target − Ct GAPDH
ΔΔCt = ΔCt (Target siRNA) − ΔCt (Nontargeting Control)
Relative expression level = 2−ΔΔCt
% knockdown = 100 × (1 − 2−ΔΔCt)
The mRNA knockdown (KD) was calculated relative to a nontargeting control (nt control) siRNA in each experiment.
Serum lipid analysis
Ldlr CETP+/– hemizygous mice were fed Purina 5021 or an equivalent 9% fat diet (Research Diets) for at least two weeks prior to the onset of the study. For lipid analysis, serum samples were obtained via retro-orbital sinus both prior to dosing and just prior to euthanization. Following euthanization, liver samples and additional serum samples were collected as well. Cohorts for controls and each siRNA tested consisted of six to eight animals. To prevent lipid degradation in the serum, 2 µl of lipase inhibitor, (Sigma, Cat # D9286-1G) diluted to a working concentration of 200 mM in 100% ethanol, was added to every 100 µl of serum. LDL, HDL, and total cholesterol serum levels were determined using standard biochemical methods.
Serum HDL levels were determined using enzymatic colorimetric assay (HDL cholesterol E kit, WAKO Diagnostics, Cat # 431-52501). In this assay, 20 µl of precipitating reagent was added to 20 µl of serum, vortexed, and incubated at room temperature for 5 min. The serum was then spun at 13K rpm for 5 min to remove any precipitated nonHDL lipid particles. Then 10 µl of the serum supernatant was transferred to a flat-bottom, 96-well plate (Costar, Cat # 3596) containing 90 µl of dPBS. A standard curve was generated by diluting Cholesterol-E standard solution (WAKO Diagnostics, Cat # 43917501) per manufacturer's instructions. Then 100 µl of WAKO reagent solution was added to the serum and incubated at 37°C for 35 min. The OD600 was then determined on Spectramax plate reader (Molecular Devices, Inc.), and the amount of HDL particles was calculated using a standard curve. Serum TC levels were determined using the Total Cholesterol E Kit by WAKO Diagnostics (Cat # 43917501). An amount of 5 µl of serum was added to a flat-bottom, 96-well plate, followed by 95 µl of dPBS. The standard curve was generated as outlined in the HDL assay. Then 100 µl of WAKO reagent was added to each well of the flat-bottom plate and incubated at 37°C for 35 min. The optical density (O.D.) readings were determined at 600 nm using a Spectramax plate reader.
The LDL levels were calculated either indirectly as nonHDL fraction by subtracting HDL from TC (nonHDL includes LDL, VLDL, and chylomicron fractions) or directly using the Equal LDL Direct Select Cholesterol Reagent (Genzyme Diagnostics, Cat # 7122) according to the manufacturer's instructions. In brief, an eight-point standard curve (10, 8, 6, 4, 2, 1, 0.5, 0 µg) was generated using the Equal LDL Liquid Select Cholesterol Calibrator Serum (Genzyme Diagnostics, Cat # 7272) and added to a Costar 3957 plate in 70 µl aliquots. Lipase inhibitor-treated serum samples were also added to the Costar 3957 plate in duplicates of 10 µl each. Then 60 µl of McGal 0.9% NaCl Saline (B. Braun Medical, Cat # R5201-01) was added to each well containing serum sample. Next 200 µl of Reagent 1 found in Equal LDL Direct Select Cholesterol Reagent (Genzyme Diagnostics, Cat # 7122) was added to each well, gently mixed, and spun at 4,680 rpm for 30 min. A 265 µl of sample was used for O.D. readings at 600 nm using a Spectramax plate reader to determine background levels. Then 100 µl of Reagent 2 (Equal LDL Direct Select Cholesterol Reagent; Genzyme Diagnostics, Cat # 7122) was added to each well of the Costar 3596 plate and incubated at room temperature for 15 min. The O.D. readings were determined at 600 nm using Spectramax plate reader. Background levels were subtracted from sample readings before calculating LDL levels.
To observe lipoprotein profiles, EDTA plasma filtered and treated with lipase and protease inhibitors were analyzed by fast-performance liquid chromatography (FPLC) gel filtration using a Superose-6 size exclusion column (24 ml, GE LifeSciences, Inc.) attached to a Dionex HPLC system (Dionex Co.). A 25 µl aliquot of pooled plasma was injected onto the column and eluted with PBS(–/–) buffer containing 1 mM EDTA at a flow rate of 0.4 ml/min. The column effluent was mixed with a commercially available enzymatic colorimetric cholesterol detection reagent (Total Cholesterol E, Wako Diagnostics) delivered at a rate of 0.2 ml/min. After passing through a 1,500 µl knitted reaction coil (∼15 min) that was maintained in a 37°C heated chamber, the reaction mixture was read at 600 nm absorbance using a photodiode array detector. The first peak of cholesterol eluted from the column was attributed to chylomicron/VLDL, the second peak to LDL, and the third to HDL; the area under each peak was calculated using Chromeleon software™ provided with the HPLC (Dionex Co.). To calculate the cholesterol concentration for each lipoprotein fraction, the ratio of the corresponding peak area to total peak area was multiplied by the total cholesterol concentration measured in the sample as determined using the Total Cholesterol E Kit by Wako.
Liver triglyceride analysis
Total triglycerides were determined from snap-frozen mouse livers after extraction using the Bligh and Dyer method (23). Approximately 80-100 mg of liver were homogenized in PBS (pH 7.4) using a Qiagen MixerMill homogenizer, twice for 3 min each with a frequency setting of 30 in Eppendorf tubes containing a Qiagen steel bead (Qiagen, Cat # 69989). The resulting homogenate (0.5 ml) was then diluted with 2.3 ml saline. Total protein was precipitated by adding 7 ml of methanol and 3.5 ml of chloroform, mixing the solution by inversion, and storing on ice for 10 min. After centrifuging the samples for 20 min at 2,000 rpm at 4°C, the supernatants were transferred to a clean tube with Teflon-lined screw caps (Kimble, Cat # 45066A-20125), then two phases were generated by adding 3.5 ml of saline and 3.5 ml of chloroform. The samples were then centrifuged as above, and 2 ml of the lower (organic) phase were transferred to screwcap tubes (Kimble, Cat # 45066A-13100) containing 1% triton X-100 in chloroform, mixed and dried under argon with low heat. The samples were then resolubilized in 0.25 ml distilled water, and aliquots were assayed for triglyceride levels using a GPO-PAP reagent (Roche Applied Science, Cat # 1488872). For a 96-well format, 10-40 μl of standards, samples, and controls were incubated with 200 μl of reagent for 10 min at room temperature, and then read at 505 nm. Control Serum Level 1 (Wako Diagnostics, Cat # 410-00101) aliquots were included to assure assay accuracy.
For Fig. 4, ∼250 mg of liver was isolated using the Folch method, and high-performance liquid chromatography was performed as described by Burrier et al. (24, 25). Briefly, an isocratic mobile phase (98% hexane: 1.2% isopropanol) was run through a Zorbax Sil silica column (Agilent Technologies) at a flow rate of 2 ml/min. Lipids were detected by absorbance (206 nm) from a 5 µl injection and quantified by calculating the area under the curve (AUC) using a Beckman System Gold Detector (Beckman Coulter, Inc.). Triglyceride concentrations were determined in reference to a standard curve generated using Nonpolar Lipid Mix-B (Matreya, Cat # 1130).
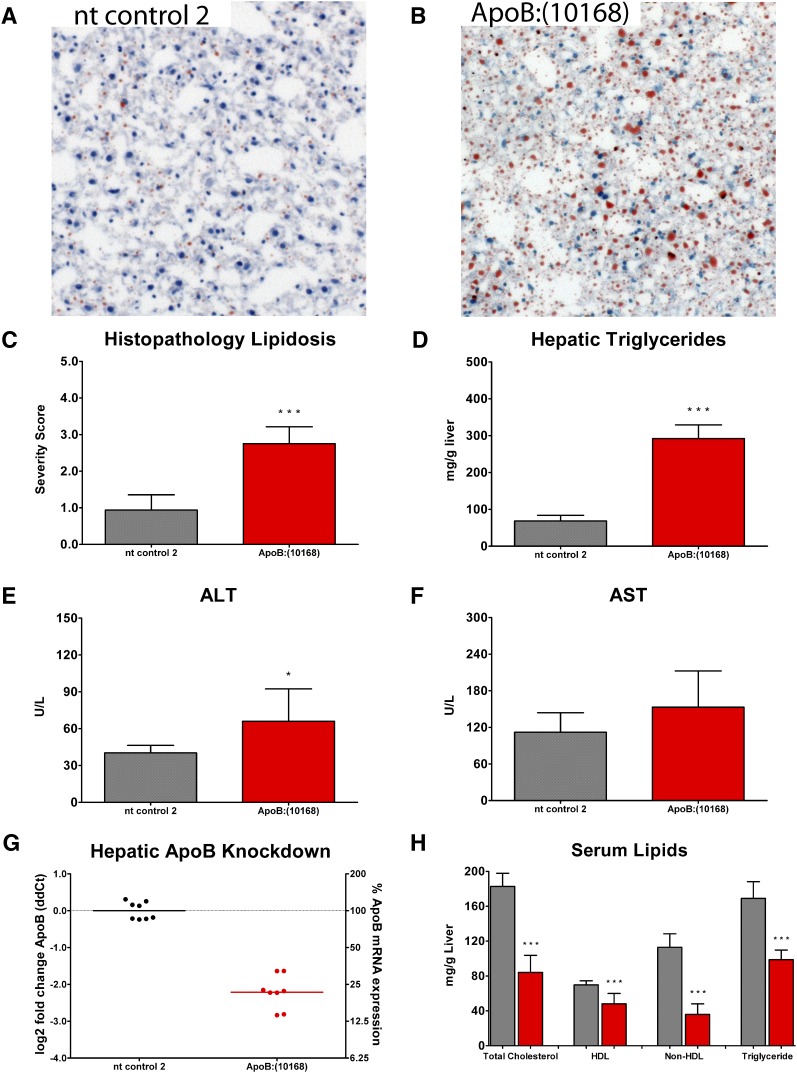
Fig. 4.
A, B: Representative images for Oil Red O-stained liver sections 15 days following a singe 3 mg/kg dose of OCD-ApoB:(10168) or nontargeting siRNA control 2 (nt control 2) reveal an increase in lipid accumulation (red) following OCD-ApoB siRNA treatment relative to the nt control siRNA group. C: Lipid accumulation was scored using a subjective scoring system (0-4), with 0 representing normal, 1 a minimal change, 2 a mild change, 3 a moderate change, and 4 representing a marked increase in lipid accumulation. D: An increase in hepatic lipids was associated with an elevation in hepatic triglyceride levels for the OCD-ApoB group. E, F: Elevated hepatic triglycerides are associated with a slight increase in serum ALT (P < 0.05) and AST (not significant) levels. G: The relative expression of ApoB was reduced in the liver. H: Serum lipids (TC, HDL, nonHDL, and TGs) decreased. In C, D, E, F, and H, bars represent the mean ± SD for N = 8. In G, individual animals (circles) with the group mean (bars) are shown.
Serum measurements for ApoA1, Pcsk9, #8232and ApoB proteins
Serum ApoA1 and proprotein convertase subtilisin-kexin type 9 (PCSK9) levels were measured using separate murine-specific ELISAs. For ApoA1 ELISA, black plates (ThermoLabsystems, Cat # 3925) were coated with 50 μl per well of 1 μg/ml Goat polyclonal Anti-mouse ApoA1 (Rockland, Cat # 600-101-196) diluted in PBS containing 0.6 mM EDTA (Sigma, Cat # 03690-100ml). For PCSK9 ELISA, 4HBX plates (ThermoLabsystems, Cat # 3855) were coated with 50 µl per well of 5 µg/ml H23 Antibody (Merck provided stock) diluted in DPBS (Cellgro, Cat # 21-031-0CV). Plates were incubated overnight at 4°C with shaking. The coating solution was then removed, and 200 µl per well of blocking buffer (1× TBST with 1% BSA) was added to the plates and incubated at room temperature for 1 h. The plates were then washed three times, with 1× TBST (pH 8.0, 0.05% Tween-20). An ApoA1 standard curve was generated using purified mouse HDL (obtained from pooled plasma from LDLR knockout mice) diluted in ApoA1 assay buffer (DPBS with 1% BSA and 1% Tween-20) starting from 7.5 µg/ml and diluted 2-fold for a 12-point curve. A PCSK9 standard curve was generated using purified mouse PCSK9 protein diluted in PCSK9 assay buffer (DPBS with 1% BSA) starting at 1 µg/ml and diluted 2-fold for a 12-point curve.
Test serum samples were either diluted 1:2000 in ApoA1 assay buffer or 1:8 in PCSK9 assay buffer. Then 50 µl of the standards and diluted samples were added to the plates and incubated at room temperature for 1 h with shaking. The plates were washed three times with 1× TBST. For the ApoA1 ELISA, 50 µl of 1 µg/ml biotinylated 2° antibody (Rockland, Cat # 600-101-196) diluted in blocking buffer was added to the plate. For the PCSK9 ELISA, 50 μl of a 1 μg/ml biotinylated B20 2° antibody (Merck provided stock) was diluted in Perkin Elmer assay buffer (Perkin Elmer, Cat # 1244-111) and added to the plate. The plates were then incubated at room temperature with shaking for 1 h. Plates were washed three times with 1× TBST. Then 75 µl of Streptavidin/Europium solution (Perkin Elmer, Cat # 1244-360) diluted 1:1000 in DELFIA assay buffer (Perkin Elmer, Cat # 1244-111) was added to the plates and incubated at room temperature for 20 min with shaking. Plates were washed three times with 1× TBST, and 100 μl of DELFIA Enhance (Perkin Elmer Cat # 1244-105) was added to the plates. The plates were incubated at room temperature for 30 min, and then read on a Perkin Elmer EnVision 2103 multilabel reader using the Europium settings.
Serum ApoB protein levels were measured by quantifying the GFEPTLEALFGK peptide of ApoB using ultra-performance liquid chromatography (UPLC)-MS/MS according to Lassman et al. (personal communication). Briefly, 4 µl of serum was diluted with 138 µl of 50 mM ammonium bicarbonate (pH 8.0), 50 µl of 80 nM internal standard ApoB peptides, and 10 µl of 10% sodium deoxycholate. Samples were reduced with dithiothreitol for 30 min at 60°C, alkylated with iodoacetamide for 60 min at 25°C in the dark, and digested overnight with 3 µg trypsin (1:50 serum proteins). To stop digestion, 10 µl of 20% formic acid was added to precipitate the sodium deoxycholate. Samples were then centrifuged for 15 min at 15,800 rcf, and 120 µl of the supernatant was removed for LC/MS analysis. Serum ApoB levels in the samples were then analyzed on a Waters Acquity UPLC and Xevo triple quadrupole mass spectrometer (Waters). The gradient was 95%A (0.1% formic acid in water)/5%B (0.1% formic acid in acetonitrile), ramped to 80%A at 1 min, 65%A at 4 min, and 5%A at 5 min. A Phenomenex Kinetex C18 50 × 2.1 mm 1.7 μm column (Phenomenex, Inc.) maintained at 50°C was used at a flow rate of 0.7 ml/min. ApoB peptide concentration was calculated by dividing the area under the curve for the analyte by the area of its internal standard and multiplying by the internal standard concentration. The concentration of ApoB was then converted to and reported as µg/dl.
Histology and hematology
Flash-frozen mouse liver samples were embedded in O.C.T. compound (Tissue Tek, Cat # 4583). Four micron sections were cut and stained with either hematoxylin and eosin (H and E) (Anatech, Cat # 812 and Cat # 832) or Oil Red O (Sigma, Cat # O-0625). A board-certified veterinary pathologist reviewed the slides for both inflammation and lipidosis, scoring the sections using a subjective scoring system (0-4), where 0 represented normal (or no observable effect), and 4 represented the most severe effect. Serum liver enzyme (ALT, alanine aminotransferase, AST, aspartate aminotransferase, and LDH, lactate dehydrogenase) levels were measured using 80 µl of serum and an ACE Alera Clinical Chemistry System (Alfa Wassermann, Inc.) according to the manufacturer's protocol.
LC/MS detection of total lipids in plasma
Plasma samples were extracted using the Bligh and Dyer (23) method for lipid profiling. Nonnaturally occurring internal standard solution containing cholesteryl ester (CE) 17:0, phosphatidylcholine (PC) 34:0, triglyceride (TG) 51:0, and free fatty acid (FFA) 17:0 (Sigma Aldrich, Co.) were added to each sample prior to extraction at a final concentration of 1 µg/ml. The inlet system consisted of an Acquity UPLC (Waters). Lipid extracts were injected onto a 1.8 µm particle 100 × 2.1 mm id Waters Acquity HSS T3 column (Waters); the column was maintained at 55°C for the lipid analysis. The flow rate used for these experiments was 0.4 ml/min. A binary gradient system was used. Acetonitrile (Burdick and Jackson) and water with 10 mM ammonium acetate (Sigma Aldrich, Co.) (40:60, v/v) was used as eluent A. Eluent B consisted of acetonitrile and isopropanol (Burdick and Jackson), both containing 10 mM ammonium acetate (10:90, v/v). A linear gradient was performed over a 15 min total run time. During the initial portion of the gradient, it was held at 60% A and 40% B. For the next 10 min, the gradient was ramped in a linear fashion to 100% B and held at this composition for 2 min. Thereafter the system was switched back to 60% B and 40% A and equilibrated for an additional 3 min.
The liquid chromatography system described was directly coupled to a hybrid quadrupole orthogonal time of flight mass spectrometer (SYNAPT G2 HDMS; Waters, MS Technologies, UK). Electrospray positive and negative ion ionization (ESI) modes were used. In both ESI modes, a capillary voltage and cone voltage of ±2 kV and ±30 V, respectively, were used. The desolvation source conditions were as follows: for the desolvation gas, 700 l/h was used, and the desolvation temperature was kept at 450°C.
The LC/MS data acquired was processed semiquantitatively and qualitatively using the manufacturer's software (MarkerLynx; Waters). The data was automatically deconvoluted into a table containing exact mass (m/z values) and retention time pairs. The entire data set was normalized to the total peak areas and scaled using the Paretto algorithm. Multivariate analysis (MVA) was used to differentiate lipids that were statistically significantly different in the nt control siRNA and ApoB siRNA groups. Principal component analysis (PCA) was used to visualize group differences. A more stringent MVA was employed to eliminate background peaks and select statistically relevant lipids, which were downregulated in the siRNA ApoB KD treatment group by the use of orthogonal partial least squares-discrimination analysis (OPLS-DA) and the S-plot statistical tool (26). S-plot allows for covariance measurement and correlation between lipids that are either upregulated or downregulated within and/or between the groups. Once the differences between the two sample groups were identified (nt control versus ApoB KD), the normalized lipid data, including the differences between the two groups, was further submitted to statistical analysis. All data is presented ± SEM. Differences between the groups were computed by Student's t-test (GraphPad Prism; GraphPad Software, Inc.). Posttest analysis for quantifiable variables was conducted using the Mann-Whitney algorithm. P < 0.05 was considered statistically significant for all the data derived from the experiments.
Lipid nomenclature used throughput this article follows the standard LIPID MAPS classification system (27, 28).
Molecular profiling of liver pathways
Liver tissue was collected and homogenized in 2 ml of Trizol (Invitrogen, Cat # 15596-026). After extraction with 0.4 ml of chloroform, RNA was extracted with SV Total RNA extraction kit (Promega, Cat # Z3100) followed by DNase I treatment (Qiagen, Cat # 79254) and purification using the RNeasy Kit (Qiagen, Cat # 74106). RNA was assayed for quality (Agilent Bioanalyzer; Agilent Technologies, Inc.) and yield (Ribogreen). RNA was amplified and labeled using a custom automated version of the RT/IVT protocol and reagents provided by Affymetrix (Affymetrix, Inc.). Hybridization, labeling, and scanning were according to Affymetrix. All samples were processed independently. Merck/Affymetrix mouse 1.0 custom arrays monitoring 38,384 individual transcripts (25,846 Entrez genes) were used. A custom definition file is available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/GPL9734). NCBI build 37 mapping definitions matched probesets in the array with transcripts (29). Raw intensity was normalized using the robust multiarray average algorithm (30). Prefiltering removed transcripts not detected (marked as “absent” by using the MAS5 algorithm with P > 0.05) in 50% or more of replicates in all treatment groups, which were not considered further (31). ANOVA (P < 0.05) between ApoB siRNA and PBS identified differentially expressed genes (the “gene signature”). A 1.2-fold change threshold (ApoB siRNA versus PBS) of the in silico combined biological replicates selected robust changes in gene expression. Raw data has been stored in the GEO database with reference number GSE23088.
Enrichment for biological processes was performed by comparing each gene signature against the public gene collections Gene Ontology, KEGG, Swissprot, and Panther families. Enrichment P values (hypergeometric distribution) were corrected for multiple testing by using Bonferroni correction. Pathway analysis was performed using Ingenuity Pathway Analysis.
Custom graphical representations of lipid metabolic pathways were generated using Ingenuity Path Designer (Ingenuity® Systems). Genes and metabolites are represented as nodes, using various shapes that represent the functional class of the gene product. Arrows between metabolite nodes represent the flow of a metabolic process (solid line) or a translocation across a membrane (dotted line). The gene nodes next to the arrows represent the gene product responsible for the metabolic process. All arrows and relationships are supported by data in the literature or textbook; some metabolic processes are simplified for illustrative purpose. The difference in gene expression calculated as the log10 to the ratio of the PBS gene expression relative to OCD-ApoB:(10168) gene expression was overlaid onto the graphical representation in Ingenuity Path Designer. The intensity of the node color indicates the degree of upregulation (red) or downregulation (green).
RESULTS
Ldlr CETP+/− hemizygous mice have a human-like lipid profile
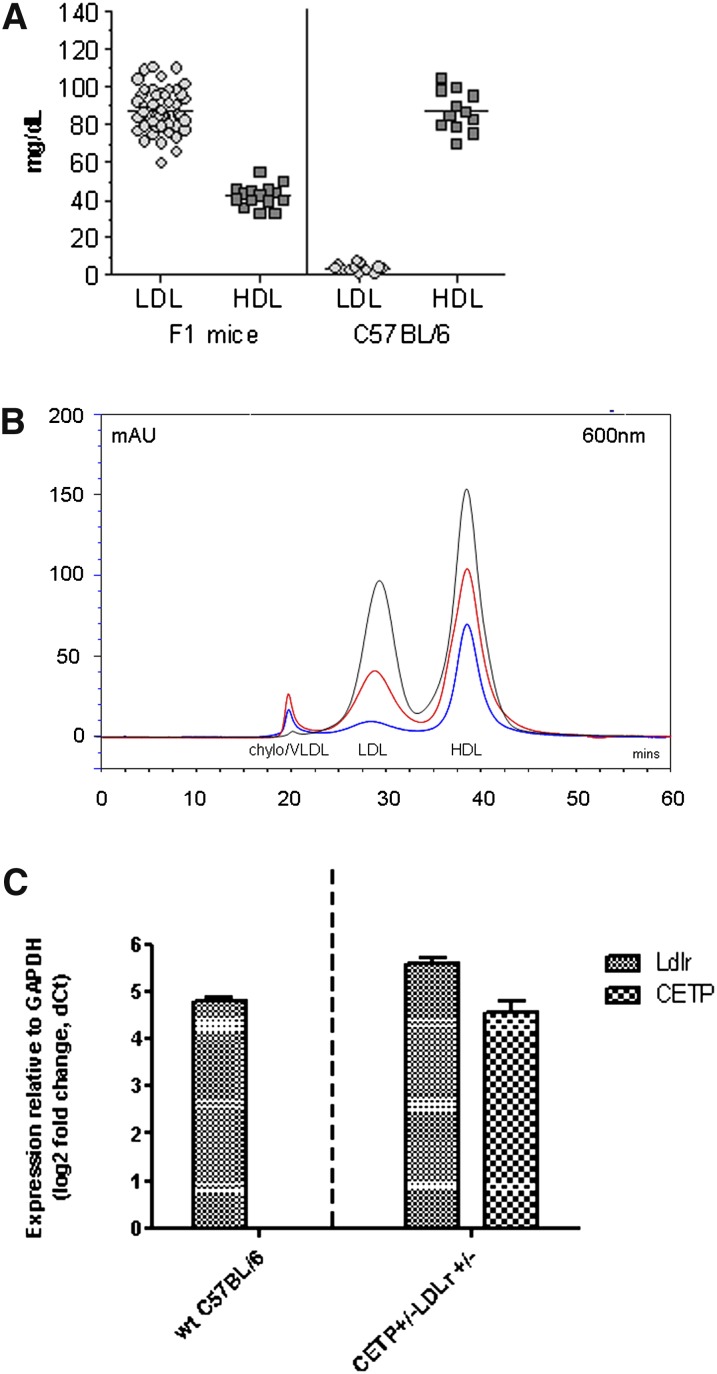
We developed a mouse model with a human-like lipid profile for use in preclinical studies of coronary atherosclerosis. These mice were genetically engineered to contain one copy of human cholesteryl ester transfer protein (CETP+/–), driven by the human ApoA1 promoter, and they are hemizygous for the LDL receptor mutation (Ldlr+/–) as described in Materials and Methods. Unlike other mouse models, these mice contain a significant level of plasma LDL cholesterol, even on a low-fat diet (Fig. 1A). FPLC analysis (Fig. 1B) shows that Ldlr CETP+/– hemizygous mice have a serum lipid profile very similar to that of healthy humans, which makes these mice suitable for investigation of lipid changes in response to different treatment regimens. Taqman analysis revealed that CETP mRNA levels in the liver homogenates of Ldlr CETP+/– hemizygous mice are approximately 5 dCts (dCt = Ct Target – Ct GAPDH), and expression level of Ldlr in the Ldlr CETP+/– hemizygous animals is roughly 50% that of wild-type C57Bl/6 mice (Fig. 1C). A similar mouse model has also been generated (32) on the Ldlr+/– background, where the human CETP transgene is driven by its natural flanking region. In this model, the plasma total cholesterol and CETP activities increased as the hepatic cholesterol production rate was impaired.
Fig. 1.
A: A new mouse model with human-like lipoprotein profiles obtained by genetically induced LDL phenotype that was enhanced by mild increases in dietary fat in Ldlr CETP+/– hemizygous mice. Contrary to wild-type mice whose serum lipids are predominantly HDL particles, serum lipids in Ldlr CETP+/– hemizygous mice have a significant contribution of LDL cholesterol (LDL) and reduced levels of HDL cholesterol (HDL). B: FPLC lipoprotein profile of C57Bl/6 (blue) and Ldlr CETP+/– hemizygous (red) mice. Plasma (25 µl) was injected and separated using a Sepharose 6 size-exclusion column, and total cholesterol in each lipoprotein particle was measured using an in-line detection system (see Materials and Methods). Larger particles, Chylo/VLDL (chylomicrons and very low-density lipoprotein) are detected from the column first, then remnant and LDL (low-density lipoprotein) particles, followed by smaller-sized HDL (high-density lipoprotein) particles. As shown, lipid profile of Ldlr CETP+/– hemizygous mice (red) more closely resembles that of humans (black) than does the lipid profile of C57Bl/6 mice (blue). C: Relative expression levels of mouse Ldlr and human CETP transcripts in liver homogenates from wild-type (wt) C57Bl/6 mice and Ldlr CETP+/– hemizygous mice. Expression was determined relative to GAPDH levels. As expected, liver Ldlr levels in the Ldlr CETP+/– hemizygous mice were roughly half that of the wt C57Bl/6 mice. No expression of human CETP transgene was detected in wt animals. (Ct = cycle threshold; dCt = Ct Target – Ct GAPDH). Bars represent the mean ± SD for N = 3.
Identification of efficacious and potent ApoB siRNAs
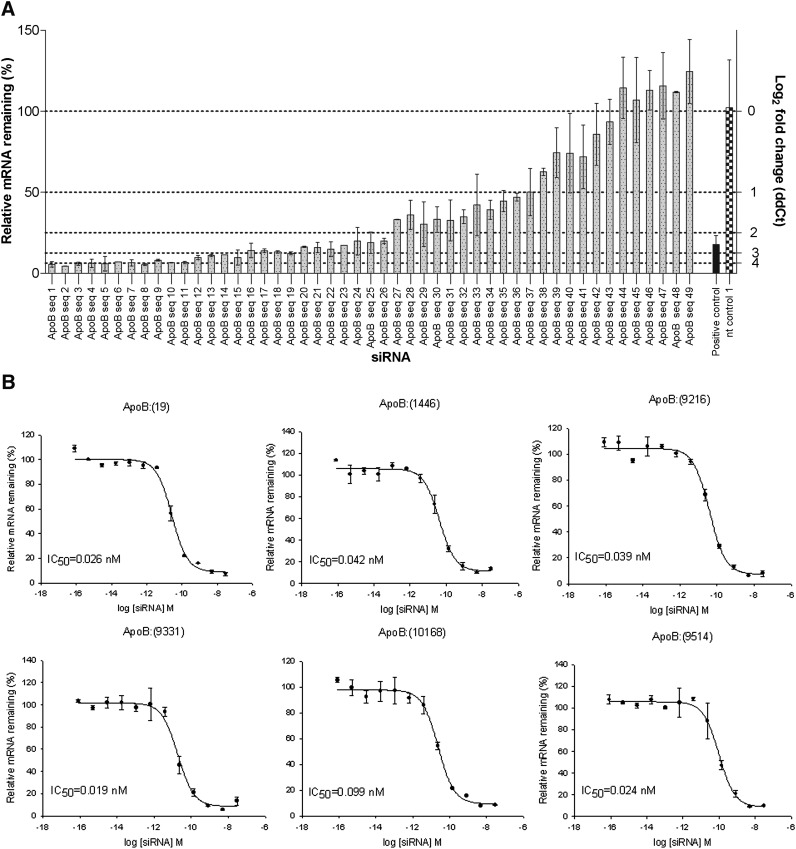
A total of 49 siRNAs were designed to target the mouse ApoB transcript. In vitro screening of siRNAs was done in mouse Hepa1-6 cells, and 28 siRNAs (57%) showed greater than 80% ApoB knockdown (Fig. 2A). The top six sequences were tested in dose-response experiments to verify the initial observed mRNA reduction and to determine IC50 values for each siRNA. Calculated IC50 values for all sequences were in subnanomolar range (Fig. 2B).
Fig. 2.
A: ApoB siRNA screen in mouse Hepa 1-6 cells. An amount of 49 siRNAs designed against mouse ApoB were tested for ApoB mRNA knockdown at 24 h post transfection. A total of 28 siRNAs (57%) showed greater than 80% target knockdown. nt control 1, nontargeting siRNA control 1. Bars represent a mean of two biological replicas ± SD. B: Dose response curves of ApoB siRNAs in mouse Hepa 1-6 cells. The dose response for the top six ApoB siRNAs was evaluated to determine their potency. Calculated IC50 values for all siRNAs were found to be in the subnanomolar range. Average of two biological replicas ± SD is shown.
In vivo ApoB KD results in potent lowering of serum LDL and increase in hepatic TG content
Initial in vivo efficacy studies were performed in BALB/c mice. The six lead siRNA sequences [ApoB:(19), ApoB:(1446), ApoB:(9216), ApoB:(9331), ApoB:(10168), and ApoB:(9514)] were packaged into lipid nanoparticle (LNP) consisting of the cationic lipid CLinDMA (LNP201). All six sequences showed significant reduction in ApoB mRNA levels, over 95% at 24 hr and over 90% at 72 h (supplemental Fig. I).
Changes in serum lipid profile following ApoB mRNA knockdown were investigated in Ldlr CETP+/– hemizygous mice. The steady-state cholesterol levels in these mice resemble that of normal healthy humans as they have a higher serum LDL component than wild-type BALB/c or C57Bl/6 animals, providing a more suitable model for serum lipid phenotyping (Fig. 1). Three ApoB sequences [ApoB:(9331), ApoB:(10168), and ApoB:(9514)] were selected from the initial six for testing in the transgenic model based on their in vitro efficacy, IC50 values, and in vivo ApoB mRNA KD in BALB/c mice. Preliminary efficacy studies with administration of a single 3 mg/kg dose of three independent LNP201-siRNAs targeting ApoB resulted in 58-80% reduction in liver ApoB mRNA levels and 47-56% reduction in serum LDL levels at 72 h (data not shown). Reduction in serum LDL levels was very uniform among LNP201-ApoB siRNA-treated Ldlr CETP+/– hemizygous mice and showed a dose-dependency (supplemental Fig. II). Moreover, trends in serum LDL reductions were very similar at days 1 and 7 post siRNA dosing, in agreement with sustained ApoB liver mRNA KD.
Further characterization of ApoB:(10168) was done using LNP OCD, a second-generation cationic lipid developed by Merck and Co. LNP OCD shows greater in vivo efficacy than LNP201 (greater target gene KD; compare 0.3 mg/kg dose in Fig. 5 and in supplemental Table I). As expected, hepatic ApoB mRNA targeting with OCD-siRNA resulted in significant ApoB mRNA KD, accompanied by lowering of serum total cholesterol (TC) and nonHDL levels that was sustained over a period of three weeks (Fig. 3). Lowering of nonHDL cholesterol was expected due to the fact that ApoB is critical for chylomicron, VLDL, and LDL formation. Serum HDL levels were lowered as well, but to a lesser extent than those of TC or nonHDL.
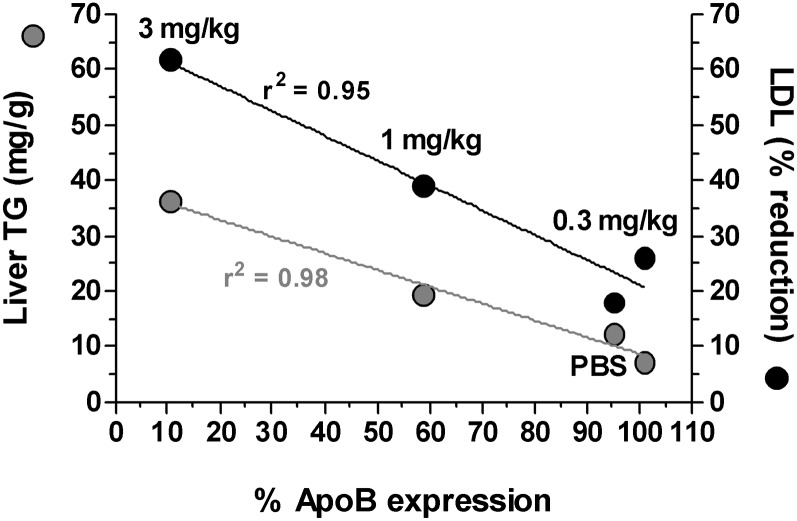
Fig. 5.
The extent of ApoB knockdown correlates with serum LDL reductions and liver triglyceride increases. Ldlr CETP+/– mice were dosed with a single 3 mg/kg dose of LNP201-ApoB:(10168). Liver ApoB mRNA knockdown, TG content, and serum LDL changes were determined 1 week post siRNA dosing. Gray circles represent the liver triglyceride content (as mg TG/g liver tissue), and the black circles represent the percentage (%) LDL lowering. The mean of 6 animals per group is shown.
Fig. 3.
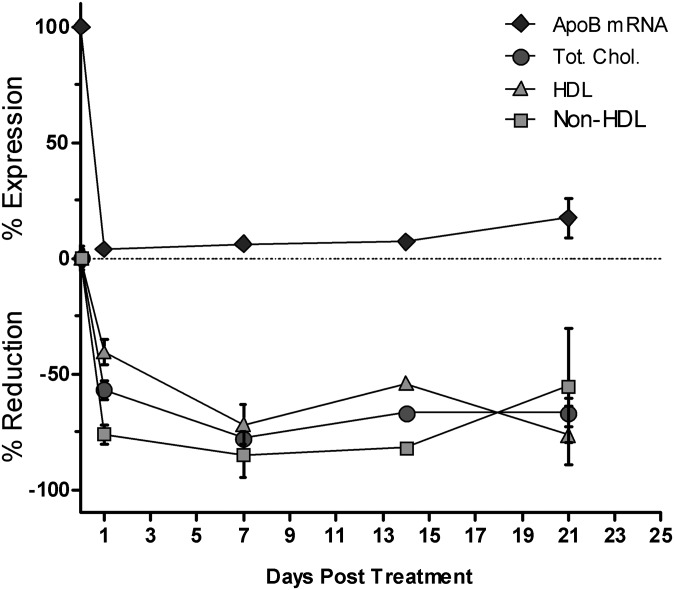
Three-week single dose time-course study in Ldlr CETP+/– mice. Animals were dosed intravenously with a single 3 mg/kg dose of OCD-ApoB:(10168). ApoB liver mRNA knockdown, and serum total cholesterol, HDL, and nonHDL were determined at days 1, 7, 14, and 21 post siRNA dosing. Both ApoB mRNA knockdown and serum lipid lowering were maintained over the course of three weeks. Mean for N = 8 ± SD is shown.
A reduction in serum cholesterol was associated with increased lipid accumulation within the liver 15 days post a single 3 mg/kg dose of OCD-ApoB siRNA (Fig. 4). An increase in the lipidosis severity score for Oil Red O-stained liver sections corresponded to an increase in liver TG levels (Fig. 4A–D). This was associated with a slight elevation in both serum ALT (P < 0.05) and AST (not significant) levels (Fig. 4E, F). LDH levels were not affected (data not shown). Dose-response experiments revealed that the increase in liver fat content is closely correlated with ApoB mRNA KD and serum LDL lowering. The greater the reduction of liver ApoB mRNA level, the larger the serum LDL lowering and the larger the increase of liver TG content (Fig. 5). These observations suggest that in this mouse model an increase in liver TG, and thus development of fatty liver, is tightly linked to ApoB liver mRNA levels. This fatty liver phenotype is reversible. We examined liver triglyceride content following a single 3 mg/kg dose of LNP-ApoB siRNA at weekly intervals over a five-week period and found that fatty liver phenotype resolves as the ApoB mRNA expression returns to normal (data not shown). The induction of liver steatosis by ApoB siRNA is consistent with others using this model (Ason et al., unpublished observations).
ApoB KD results in reductions of plasma ApoA1, ApoB, and Pcsk9
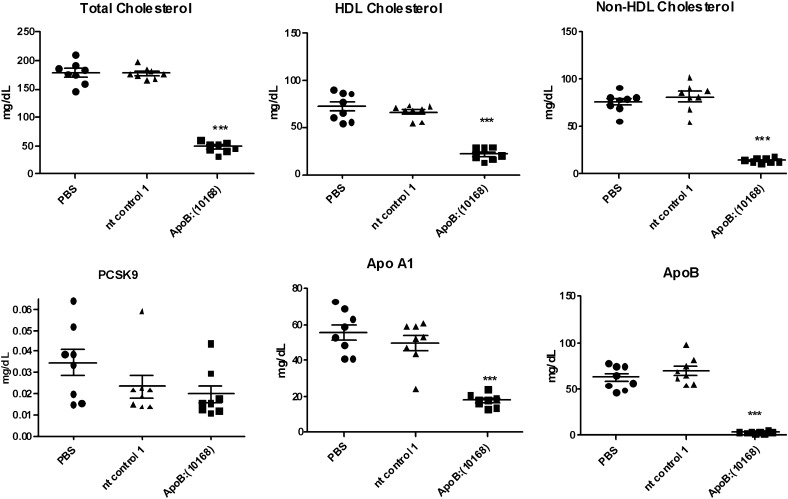
Consistent with the lowering of serum lipids, ApoB KD resulted in significant reductions in serum ApoA1, ApoB, and Pcsk9 protein levels (Fig. 6). The effects on ApoA1 and Pcsk9 are most likely physiological.
Fig. 6.
Changes in serum lipids and ApoA1, ApoB, and Pcsk9 serum protein levels following ApoB siRNA treatment in Ldlr CETP+/– mice. Animals were dosed with a single 3 mg/kg dose intravenously of either OCD-ApoB:(10168) or nontargeting control (nt control) siRNA-LNP. Serum lipids and serum protein levels were determined at day 5 following siRNA dosing (***P < 0.0001).
To expand upon the observation of serum HDL and ApoA1 lowering, we performed a siRNA dose titration experiment, which showed that the serum HDL reduction correlates with liver ApoB mRNA knockdown (supplemental Table I). Maximum serum HDL lowering of 56% was observed with liver ApoB mRNA KD of 94%. Similar trends in serum HDL lowering correlating with ApoB mRNA reduction were confirmed in C57Bl/6 mice (data not shown) and reported by others (Ref. 33 and personal communication). We also determined that the same HDL-lowering phenotype is observed with a second independent ApoB siRNA ApoB:(9514) in both C57Bl/6 and Ldlr CETP+/– hemizygous mice, suggesting that this observation is not due to off-targeting effects (data not shown).
ApoB KD reduces expression of genes involved in hepatic lipid synthetic pathways
To study the effects of ApoB knockdown in vivo, we examined global gene expression from livers of Ldlr CETP+/– hemizygous mice treated with ApoB siRNAs [ApoB:(10168) and ApoB:(9514)] compared with PBS-treated mice. Microrray analysis was performed with an Affymetrix Merck custom array containing 38,384 probe sets (25,846 Entrez genes). A total of 1,365 differentially expressed sequences were identified between ApoB siRNA and PBS control with a fold-change higher than 1.2. Our previous analysis showed that PBS and nt control siRNA yielded similar results. The molecular profile for ApoB:(10168) had a slightly better on-target signature compared with ApoB:(9514), but both sequences had a common overall signature. The profiling data is available in GEO data base under accession number 23088 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE23088).
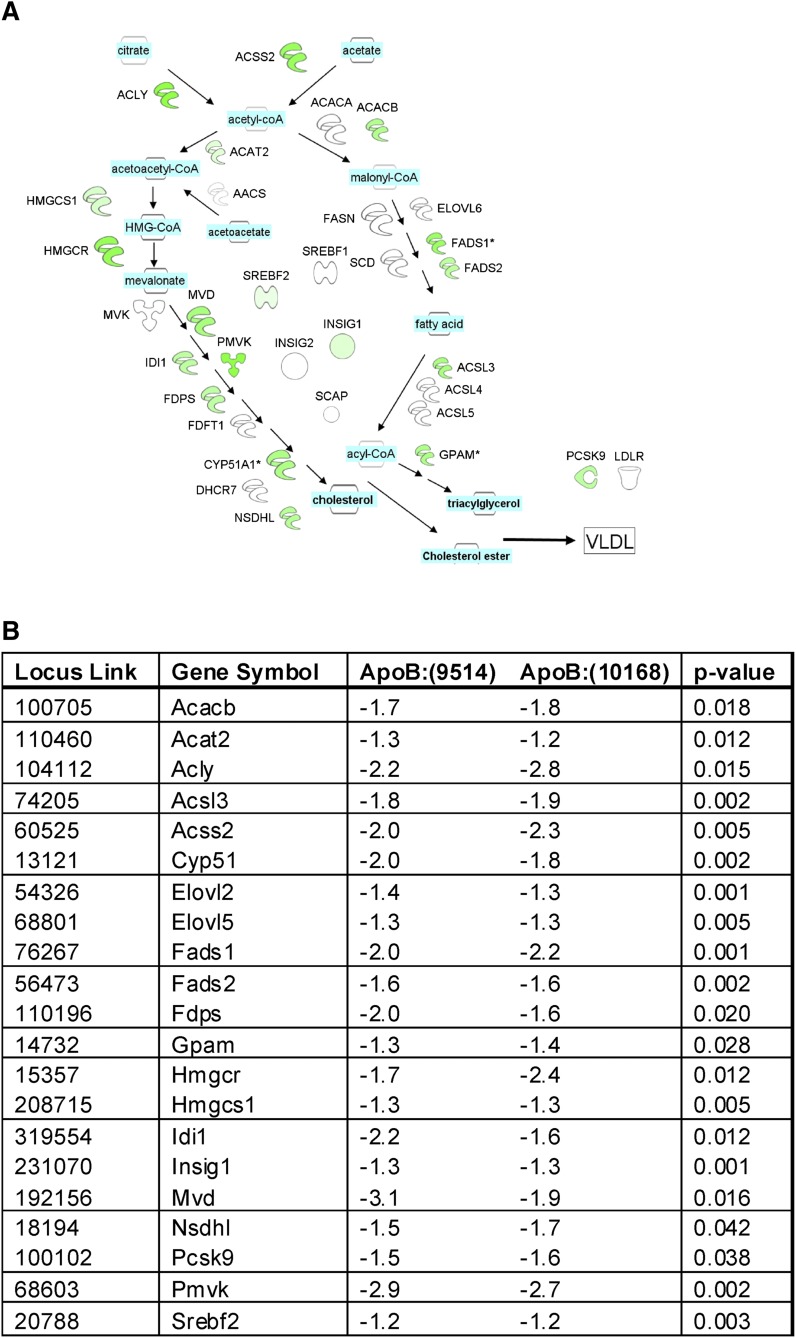
Analyses of key expression nodes affecting hepatic lipid synthesis after ApoB siRNA treatment were mapped using the Ingenuity Path Designer (Fig. 7). When ApoB siRNA was compared with PBS, network analysis revealed that cholesterol and fatty acid synthesis pathways were downregulated, as shown by a significant reduction in the mRNA of sterol regulatory element binding protein-2 (SREBP2), a key lipid regulator. In addition, insulin-induced gene 1 protein (INSIG1), PCSK9, and the cholesterol transporter STAR-related lipid transfer domain containing 4 (STARD4) were also significantly downregulated with ApoB KD independent of the tested ApoB siRNA. In addition, several enzymes involved in cholesterol biosynthesis, including 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), lanosterol synthase (LSS), mevalonate decarboxylase (MVD), and sterol-C5-desaturase (SC5D), were downregulated. Although ApoB KD resulted in hepatic steatosis (Fig. 4), fatty acid biosynthesis was downregulated through reduction in fatty acid desaturases 1 and 2 (FADS1 and FADS2). Surprisingly, ApoB KD did not reduce LDLr expression, but it did significantly reduce PCSK9 mRNA, suggesting that LDLr and PCSK9 may be decoupled in this model and supporting the idea that ApoB siRNA therapeutics may be additive to statins. Most of the gene changes were small, but they were coordinated in several components of the same molecular pathway. These data are consistent with the findings of reduced plasma cholesterol and triglycerides with ApoB KD.
Fig. 7.
A: Changes in Srebp1 and 2 pathways in mouse livers after ApoB siRNA treatment. Liver mRNAs from PBS or OCD-ApoB:(10168) siRNA groups were profiled three days post treatment by microarray as described in Materials and Methods, then mapped onto the pathways shown here using the Ingenuity Path Designer (Ingenuity®Systems). The intensity of the node color indicates the degree of upregulation (red) or downregulation (green) of mRNA expression. Shown is the OCD-ApoB:(10168) treatment profile that was normalized to the PBS treatment. B: List of Srebp1 and Srebp2 genes regulated by ApoB KD. Values represent the transcriptional changes in the liver relative to PBS-treated animals. Significance is reported as the P value for both ApoB siRNA sequences [ApoB:(9514) and ApoB:(10168)] compared with PBS.
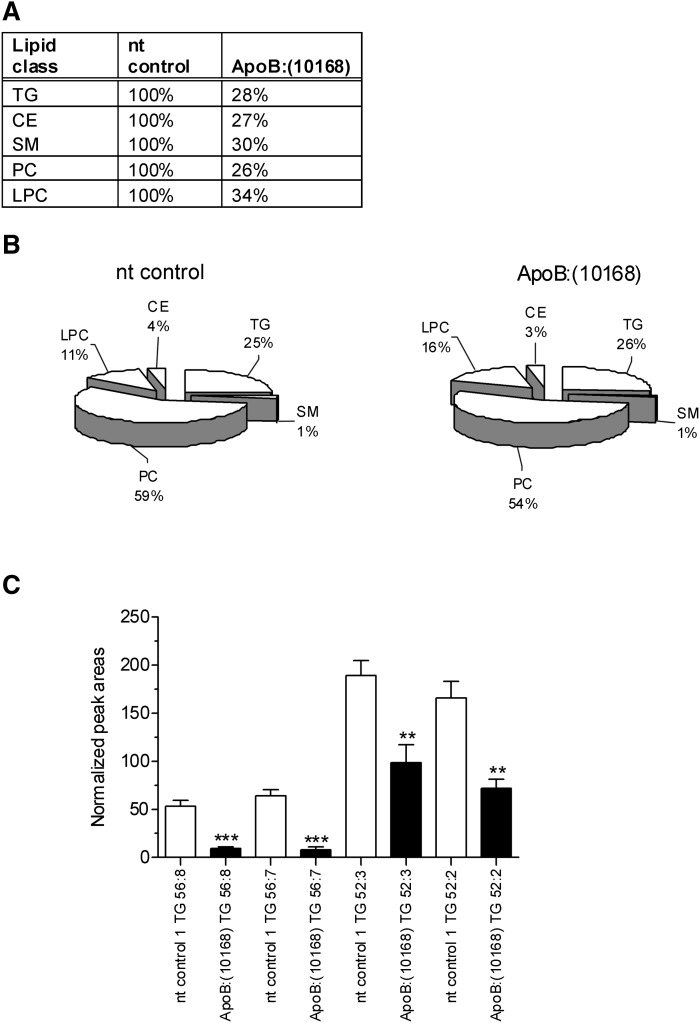
ApoB KD reduces total lipid concentrations but does not affect overall lipid distribution in plasma
To determine if ApoB KD reduces a specific lipid class in plasma, LC/MS analysis of lipids from OCD-ApoB:(10168)-treated mice were compared with the lipids from nontargeting (nt) siRNA control-treated mice. By the use of supervised MVA, it was possible to determine the major lipid classes that were downregulated by ApoB KD. A total of five lipid classes were statistically relevant: LPC, SM, PC, TG, and CE. Fig. 8A demonstrates that ApoB KD resulted in approximately 70% decrease in each of these lipids in the plasma compared with nt control siRNA treatment. Although there was a significant decrease in total lipid, Fig. 8B shows that the relative distribution of lipids did not change with ApoB KD. Following ApoB KD, the levels of circulating triglycerides in the plasma decreased dramatically (∼70%) compared with nt control siRNA treatment. When total plasma TGs were profiled, a number of triglycerides were significantly downregulated after ApoB KD. We characterized different serum triglycerides by LC/MS using lipid nomenclature as outlined by LIPID MAPS (http://lipidmaps.org) (27, 28). In the triglyceride nomenclature, the first number stands for the total number of hydrocarbons in the FFA in the TG moiety, while the second number after the colon represents the number of double bonds in the fatty acyl substituents attached to the glycerol backbone. Specifically, triglycerides TG 56:8, TG 56:7, TG 52:3, and TG 52:2 were all reduced after ApoB KD (Fig. 8C). These metabolite data, along with our microarray analysis (Fig. 7), illustrate that ApoB KD specifically decreases hepatic de novo lipogenesis.
Fig. 1.
A: Hepatic ApoB KD results in approximately 70% decrease in serum lyso-phosphatidylcholines (LPC), sphingomyelins (SM), phosphatidylcholines (PC), triglycerides (TG), and cholesteryl esters (CE). B: ApoB KD reduces total lipid concentrations but does not affect overall lipid distribution in plasma. Values represent the percentage of each general plasma lipid class in mice treated with either nontargeting control 1 (nt control 1) siRNA (left pie graph) or ApoB:(10168) siRNA (right pie graph). Animals were dosed intravenously with a single 3 mg/kg dose of OCD-siRNAs and then euthanized at day 5 postdosing. Lipids were analyzed by LC/MS as described in the Materials and Methods. C: Differences in plasma TG profile after a single 3 mg/kg dose of OCD-ApoB:(10168) siRNA at day 5 postdosing. TG composition and analysis was performed by LC/MS as outlined in Materials and Methods. Triglycerides that were downregulated by treatment of OCD-ApoB:(10168), as compared with nt control 1, are shown (TG 56:8, TG 56:7, TG 52:3, TG 52:2). The differences in all TGs shown were statistically significant (P < 0.05, 1-way ANOVA with Bonferroni posttest; nt control versus ApoB). Triglyceride nomenclature used here follows guidance as outlined by LIPID MAPS (http://lipidmaps.org). In the TG nomenclature, the first number stands for the total number of hydrocarbons in the FFA in the TG moiety, while the second number after the colon represents the number of double bonds in the fatty acyl substituents attached to the glycerol backbone.
DISCUSSION
Despite the fact that current treatments for coronary atherosclerosis and hyperlipidemia are adequate for a large number of patients, there is still a need for development of additional therapeutic approaches for patients who are unresponsive to existing medications. As ApoB is necessary for the assembly of the LDL precursor (VLDL) in the liver and its secretion into circulation, it is hypothesized that decreasing ApoB production would be an effective LDL-lowering therapy in addition to statins. ApoB has already been investigated as a potential target for development of RNAi therapeutics using both cholesterol-conjugated and LNP-formulated siRNAs and antisense oligonucelotides (ASO) (33–35). However, none of the earlier reports demonstrated liver ApoB mRNA knockdown and accompanying serum lipid changes in mice of comparable magnitude to those reported here. ASO treatments typically require 4-6 weeks of treatment to observe maximum effect and require high doses (up to 50 mg/kg twice/week). Similarly, targeting liver ApoB with cholesterol-conjugated siRNAs also requires a high dose (up to 50 mg/kg) and multiple treatments to observe maximum effect (34). Alnylam Pharmaceuticals reported LNP-siRNA targeting of liver ApoB in mice and cynomolgus monkeys (35). Reductions in ApoB protein, serum cholesterol, and LDL lasted for 11 days at a maximum siRNA dose of 5 mg/kg in C57Bl/6 mice. Data presented here showed a significant and sustained lowering of the same endpoints out to our last time point tested (21 days; Fig. 3). Therefore, it is likely that the Ldlr CETP+/– mouse model is more sensitive to lipid-lowering effects of ApoB siRNA than wild-type mice due to its unique lipid profile, which is not HDL-dominated.
ApoB liver targeting has also been shown to affect LDL and TC levels in several mouse models, including C57Bl/6 mice fed either normal or high-fat diet, Apoe-deficient mice, and Ldlr-deficient mice. Interestingly, HDL reductions were reported in all of these mouse strains, except Apoe-deficient mice, which have the highest levels of apoB-48 and the lowest levels of apoB-100 (33). Similar effects on HDL were reported for atorvastatin. In the same report, Crooke reported that Apoe-deficient mice were the least sensitive to the lipid-lowering effects of ApoB ASO, with serum TC being reduced by 25% and LDL by 40%. It has been suggested that mature HDL particles require components of apoB-containing lipoproteins, so that a significant loss of VLDL and LDL results in a reduction in the number of HDL particles (36). Alternatively, reduction in the secretion of hepatic ApoB may allow secreted ApoE to associate more readily with HDL particles in circulation and thus redirect HDL particles to a clearance via LDL receptors. This suggestion is consistent with less HDL lowering in ApoE–/– and LDLR–/– mice by ApoB KD.
We showed that siRNA-mediated targeting of ApoB mRNA in a mouse model with human-like lipid profiles can trigger significant reduction in serum LDL, TC, and total TG levels. This effect was sustained over a period of three weeks following a single 3 mg/kg dose of ApoB siRNA. Silencing of ApoB mRNA in the liver is specific and reversible. We observed similar effects on liver ApoB mRNA knockdown with multiple, independent siRNA sequences and in several mouse strains. A nontargeting control siRNA did not show ApoB mRNA reduction or serum lipid lowering. While 70% of total plasma lipids were significantly reduced with ApoB KD (Figs. 3 and 8A), the distribution of lipids was identical to nontargeting control siRNA-treated mice (Fig. 8B). However, some triglycerides in plasma (TG 52:2, TG 52:3, TG 56:7, and TG 56:8) were shown to be specifically lowered by ApoB KD (Fig. 8C). The detailed plasma lipid metabolite analysis correlated well with the hepatic microarray analysis, showing that ApoB KD directly affects de novo lipogenesis by reducing the genes involved in cholesterol and triglyceride synthesis. This action is accomplished by reducing several genes in the Srebp1 and 2 pathways. This response may be expected, as ApoB is critical for secretion of triglycerides from the liver and the liver might respond to reduced ApoB secretion by reducing the synthesis of total lipids. Although expression of TC and TG synthetic genes was reduced, the reduction was not capable of overcoming the ApoB-KD induced hepatic steatosis.
In Ldlr CETP+/– mice, we observed a fatty liver phenotype associated with ApoB KD that was both reversible and dose-dependent. Liver triglyceride levels returned to normal by week 5 following a single 3 mg/kg dose of ApoB siRNA (data not shown). Accumulation of triglycerides in the liver was not surprising, as reduction of liver ApoB protein inhibited formation of VLDL particles, and triglycerides that would normally be secreted by the liver into the circulation were now trapped in that organ. It has been reported that ApoB lowering with a microsomal triglyceride transfer protein (MTTP) inhibitor in patients homozygous for familial hypercholesterolemia almost always results in accumulation of hepatic fat (37). However, liver steatosis has not been associated with bad clinical outcomes (38). This observation is consistent with very mild increases in ALT and AST liver enzymes in spite of increased liver lipidosis in Ldlr CETP+/– mice following ApoB knockdown (Fig. 4).
Results from our molecular profiling studies are consistent with an accumulation of fatty acids and cholesterol that is concomitant with a reduction in VLDL secretion produced by ApoB KD. When less VLDL is secreted, there are more intracellular fatty acids and cholesterol available, so the cell may respond by downregulating synthesis of cholesterol and fatty acids and upregulating fatty acid catabolism. The coordinated response we observed, using two independent siRNA sequences to ApoB, was striking and suggested a reduction in total cholesterol and fatty acid levels upon ApoB KD. Furthermore, ApoB KD significantly reduced PCSK9 mRNA without significantly affecting LDLr expression. This was surprising because these two genes are usually regulated in the same direction by a SREBP-mechanism, and it suggests that siRNA therapies directed to reduce ApoB mRNA may be additive to statins.
In summary, we report the generation of a mouse model with a human-like lipid profile. Using Ldlr CETP+/– mice, we demonstrated that LNP-formulated siRNAs can be successfully used for targeting liver ApoB, resulting in significant reduction in serum total cholesterol and LDL cholesterol. These findings further support the potential of developing RNAi technology for targeting ApoB as a therapeutic approach for treatment of hyperlipidemia.
Note added in proof
The author Zhu Chen was inadvertently left out of the author list of the accepted version of this article. All other authors and the Journal's Editors-in-Chief approved the addition after the article was in proof stage. Dr. Chen will appear as an author in all forms of the article except in the originally accepted Paper in Press.
Supplementary Material
Acknowledgments
The authors thank Dipali Ruhela for providing the description of OCD-LNP and the Sirna Formulation team for providing LNP formulations used in these experiments. The authors thank Duncan Brown for siRNA design and the Process Research team for providing siRNA oligos. The authors thank Natalya Dubinina and Sirna In Vivo Biology team for generation of in vivo samples. The authors thank Harry R. Davis, Jr., and Glen Tetzloff for providing support for HPLC analysis of triglycerides in one study. The authors also thank Premier Laboratory for providing histological services and pathology review. The authors thank Julja Burchard for productive discussion, and Yong Ma and Michael Robinson for critical reading and comments on the manuscript. The authors thank Mike Lassman for providing ApoB UPLC-MS/MS protocol.
Footnotes
Abbreviations:
- ALT
- alanine aminotransferase
- apo
- apolipoprotein
- ASO
- antisense oligonucelotide
- AST
- aspartate aminotransferase
- CE
- cholesteryl ester
- CETP
- cholesteryl ester transfer protein
- Ct
- cycle threshold
- FADS
- fatty acid desaturase
- FH
- familial hypercholesterolemia
- FHBL
- familial hypobetalipoproteinemia
- FPLC
- fast-performance liquid chromatography
- HMGCR
- 3-hydroxy-3-methylglutaryl-CoA reductase
- INSIG1
- insulin-induced gene 1 protein
- KD
- knockdown
- LDH
- lactate dehydrogenase
- LDLr
- low density lipoprotein receptor
- LNP
- lipid nanoparticle
- LPC
- lyso-phosphatidylcholine
- LSS
- lanosterol synthase
- MTTP
- microsomal triglyceride transfer protein
- MVA
- multivariate analysis
- MVD
- mevalonate decarboxylase
- nt
- nontarget
- OPLS-DA
- orthogonal partial least squares-discrimination analysis
- PC
- phosphatidylcholine
- PCA
- principal component analysis
- PCSK9
- proprotein convertase subtilisin-kexin type 9
- RISC
- RNA-induced silencing complex
- dsRNA
- double-stranded RNA
- siRNA
- short-interfering RNA
- RNAi
- RNA interference
- SC5D
- sterol-C5-desaturase
- SM
- sphingomyelin
- SREBP2
- sterol regulatory element binding protein-2
- STARD4
- STAR-related lipid transfer domain containing 4
- TC
- total cholesterol
- TG
- triglyceride
- UPLC
- ultra-performance liquid chromatography
Sirna Therapeutics, Inc., is a wholly owned subsidiary of Merck & Co., Inc.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures and two tables.
REFERENCES
- 1.Wouters K., Shiri-Sverdlov R., van Gorp P. J., van Bilsen M., Hofker M. H. 2005. Understanding hyperlipidemia and atherosclerosis: lessons from genetically modified apoe and ldlr mice. Clin. Chem. Lab. Med. 43: 470–479. [DOI] [PubMed] [Google Scholar]
- 2.Maron D. J., Fazio S., Linton M. F. 2000. Current perspectives on statins. Circulation. 101: 207–213. [DOI] [PubMed] [Google Scholar]
- 3.Hobbs H. H., Brown M. S., Goldstein J. L. 1992. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum. Mutat. 1: 445–466. [DOI] [PubMed] [Google Scholar]
- 4.Schaefer E. J., Gregg R. E., Ghiselli G., Forte T. M., Ordavas J. M., Zech L. A., Brewer H. B., Jr 1986. Familial apolipoprotein E deficiency. J. Clin. Invest. 78: 1206–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Havekes L., de Wit E., Leuven J. G., Klasen E., Utermann G., Weber W., Beisiegel U. 1986. Apolipoprotein E3-Leiden. A new variant of human apolipoprotein E associated with familial type III hyperlipoproteinemia. Hum. Genet. 73: 157–163. [DOI] [PubMed] [Google Scholar]
- 6.Ishibashi S., Brown M. S., Goldstein J. L., Gerard R. D., Hammer R. E., Herz J. 1993. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J. Clin. Invest. 92: 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plump A. S., Smith J. D., Hayek T., Aalto-Setälä K., Walsh A., Verstuyft J. G., Rubin E. M., Breslow J. L. 1992. Severe hypercholesterolemia and atherosclerosis in apolipoprotein E-deficient mice created by homologous recombination in ES cells. Cell. 71: 343–353. [DOI] [PubMed] [Google Scholar]
- 8.Zhang S. H., Reddick R. L., Piedrahita J. A., Maeda N. 1992. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 258: 468–471. [DOI] [PubMed] [Google Scholar]
- 9.van den Maagdenberg A. M., Hofker M. H., Krimpenfort P. J., de Bruijn I., van Vlijmen B., van der Boom H., Havekes L. M., Frants R. R. 1993. Transgenic mice carrying the apolipoprotein E3-Leiden gene exhibit hyperlipoproteinemia. J. Biol. Chem. 268: 10540–10545. [PubMed] [Google Scholar]
- 10.Westerterp M., van der Hoogt C. C., de Haan W., Offerman E. H., Dallinga-Thie G. M., Jukema J. W., Havekes L. M., Rensen P. C. 2006. Cholesteryl ester transfer protein decreases high-density lipoprotein and severely aggravates atherosclerosis in APOE*3-Leiden mice. Arterioscler. Thromb. Vasc. Biol. 26: 2552–2559. [DOI] [PubMed] [Google Scholar]
- 11.de Vries-van der Weij J., Zadelaar S., Toet K., Havekes L. M., Kooistra T., Rensen P. C. 2009. Human CETP aggravates atherosclerosis by increasing VLDL-cholesterol rather than by decreasing HDL-cholesterol in APOE*3-Leiden mice. Atherosclerosis. 206: 153–158. [DOI] [PubMed] [Google Scholar]
- 12.Elovson J., Chatterton J. E., Bell G. T., Schumaker V. N., Reuben M. A., Puppione D. L., Reeve J. R., Jr, Young N. L. 1988. Plasma very low density lipoproteins contain a single molecule of apolipoprotein B. J. Lipid Res. 29: 1461–1473. [PubMed] [Google Scholar]
- 13.Brown M. S., Goldstein J. L. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science. 232: 34–47. [DOI] [PubMed] [Google Scholar]
- 14.Davis R. A., Hui T. Y. 2001. 2000 George Lyman Duff Memorial Lecture: atherosclerosis is a liver disease of the heart. Arterioscler. Thromb. Vasc. Biol. 21: 887–898. [DOI] [PubMed] [Google Scholar]
- 15.Glueck C. J., Kelley W., Gupta A., Fontaine R. N., Wang P., Gartside P. S. 1997. Prospective 10-year evaluation of hypobetalipoproteinemia in a cohort of 772 firefighters and cross-sectional evaluation of hypocholesterolemia in 1,479 men in the National Health and Nutrition Examination Survey I. Metabolism. 46: 625–633. [DOI] [PubMed] [Google Scholar]
- 16.Tomari Y., Zamore P. D. 2005. Perspective: machines for RNAi. Genes Dev. 19: 517–529. [DOI] [PubMed] [Google Scholar]
- 17.Aagaard L., Rossi J. J. 2007. RNAi therapeutics: principles, prospects and challenges. Adv. Drug Deliv. Rev. 59: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrissey D. V., Blanchard K., Shaw L., Jensen K., Lockridge J. A., Dickinson B., McSwiggen J. A., Vargeese C., Bowman K., Shaffer C. S., et al. 2005. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 41: 1349–1356. [DOI] [PubMed] [Google Scholar]
- 19.Morrissey D. V., Lockridge J. A., Shaw L., Blanchard K., Jensen K., Breen W., Hartsough K., Machemer L., Radka S., Jadhav V., et al. 2005. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 23: 1002–1007. [DOI] [PubMed] [Google Scholar]
- 20.Strapps W. R., Pickering V., Muiru G. T., Rice J., Orsborn S., Polisky B. A., Sachs A. B., Bartz S. R. 2010. The siRNA sequence and guide strand overhangs are determinants of in vivo duration of silencing. Nucleic Acids Res. 38: 4788–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burchard J., Jackson A. L., Malkov V., Needham R. H., Tan Y., Bartz S. R., Dai H., Sachs A. B., Linsley P. S. 2009. MicroRNA-like off-target transcript regulation by siRNAs is species specific. RNA. 15: 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrams M. T., Koser M. L., Seitzer J., Williams S. C., DiPietro M. A., Wang W., Shaw A. W., Mao X., Jadhav V., Davide J. P., et al. 2010. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment. Mol. Ther. 18: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 24.Folch J., Lees M., Sloane Stanley G. H. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 25.Burrier R. E., Smith A. A., McGregor D. G., Hoos L. M., Zilli D. L., Jr. Davis H. R. 1995. The effect of acyl CoA: cholesterol acyltransferase inhibition on the uptake, esterification and secretion of cholesterol by the hamster small intestine. J. Pharmacol. Exp. Ther. 272: 156–163. [PubMed] [Google Scholar]
- 26.Wiklund S., Johansson E., Sjöström L., Mellerowicz E. J., Edlund U., Shockcor J. P., Gottfries J., Moritz T., Trygg J. 2008. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 80: 115–122. [DOI] [PubMed] [Google Scholar]
- 27.Fahy E., Subramaniam S., Brown H. A., Glass C. K., Merrill A. H., Jr, Murphy R. C., Raetz C. R., Russell D. W., Seyama Y., Shaw W., et al. 2005. A comprehensive classification system for lipids. J. Lipid Res. 46: 839–861. [DOI] [PubMed] [Google Scholar]
- 28.Castro-Perez J. M., Kamphorst J., DeGroot J., Lafeber F., Goshawk J., Yu K., Shockcor J. P., Vreeken R. J., Hankemeier T. 2010. Comprehensive LC-MS E lipidomic analysis using a shotgun approach and its application to biomarker detection and identification in osteoarthritis patients. J. Proteome Res. 9: 2377–2389. [DOI] [PubMed] [Google Scholar]
- 29.Dai M., Wang P., Boyd A. D., Kostov G., Athey B., Jones E. G., Bunney W. E., Myers R. M., Speed T. P., Akil H., et al. 2005. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greco D., Leo D., di Porzio U., Perrone Capano C., Auvinen P. 2008. Pre-filtering improves reliability of Affymetrix GeneChips results when used to analyze gene expression in complex tissues. Mol. Cell. Probes. 22: 115–121. [DOI] [PubMed] [Google Scholar]
- 32.Harada L. M., Carrilho A. J., Oliveira H. C., Nakandakare E. P., Quintão E. C. 2006. Regulation of hepatic cholesterol metabolism in CETP/LDLr mice by cholesterol feeding and by drugs (cholestyramine and lovastatin) that lower plasma cholesterol. Clin. Exp. Pharmacol. Physiol. 33: 1209–1215. [DOI] [PubMed] [Google Scholar]
- 33.Crooke R. M., Graham M. J., Lemonidis K. M., Whipple C. P., Koo S., Perera R. J. 2005. An apolipoprotein B antisense oligonucleotide lowers LDL cholesterol in hyperlipidemic mice without causing hepatic steatosis. J. Lipid Res. 46: 872–884. [DOI] [PubMed] [Google Scholar]
- 34.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J., et al. 2004. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 432: 173–178. [DOI] [PubMed] [Google Scholar]
- 35.Zimmermann T. S., Lee A. C., Akinc A., Bramlage B., Bumcrot D., Fedoruk M. N., Harborth J., Heyes J. A., Jeffs L. B., John M., et al. 2006. RNAi-mediated gene silencing in non-human primates. Nature. 441: 111–114. [DOI] [PubMed] [Google Scholar]
- 36.Lieu H. D., Withycombe S. K., Walker Q., Rong J. X., Walzem R. L., Wong J. S., Hamilton R. L., Fisher E. A., Young S. G. 2003. Eliminating atherogenesis in mice by switching off hepatic lipoprotein secretion. Circulation. 107: 1315–1321. [DOI] [PubMed] [Google Scholar]
- 37.Cuchel M., Bloedon L. T., Szapary P. O., Kolansky D. M., Wolfe M. L., Sarkis A., Millar J. S., Ikewaki K., Siegelman E. S., Gregg R. E., et al. 2007. Inhibition of microsomal triglyceride transfer protein in familial hypercholesterolemia. N. Engl. J. Med. 356: 148–156. [DOI] [PubMed] [Google Scholar]
- 38.Schonfeld G., Lin X., Yue P. 2005. Familial hypobetalipoproteinemia: genetics and metabolism. Cell. Mol. Life Sci. 62: 1372–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.