Abstract
Genome-wide association (GWA) studies represent a powerful strategy for identifying susceptibility genes for complex diseases in human populations but results must be confirmed and replicated. Because of the close homology between mouse and human genomes, the mouse can be used to add evidence to genes suggested by human studies. We used the mouse quantitative trait loci (QTL) map to interpret results from a GWA study for genes associated with plasma HDL cholesterol levels. We first positioned single nucleotide polymorphisms (SNPs) from a human GWA study on the genomic map for mouse HDL QTL. We then used mouse bioinformatics, sequencing, and expression studies to add evidence for one well-known HDL gene (Abca1) and three newly identified genes (Galnt2, Wwox, and Cdh13), thus supporting the results of the human study. For GWA peaks that occur in human haplotype blocks with multiple genes, we examined the homologous regions in the mouse to prioritize the genes using expression, sequencing, and bioinformatics from the mouse model, showing that some genes were unlikely candidates and adding evidence for candidate genes Mvk and Mmab in one haplotype block and Fads1 and Fads2 in the second haplotype block. Our study highlights the value of mouse genetics for evaluating genes found in human GWA studies.
Keywords: genomics, high density lipoprotein, comparative genomics, quantitative trait loci, mouse model
Coronary artery disease (CAD) is the leading cause of death in the United States. Clinical and mouse studies show that a high plasma cholesterol level is a major predictor of CAD, whereas a high level of HDL cholesterol protects against CAD (1). Complex traits such as HDL-cholesterol levels are the result of a combination of multiple genetic and environmental factors. Quantitative trait loci (QTL) analysis has been used to study the genetics of HDL levels in humans and mice. By 2005, approximately 40 HDL-regulating QTL had been identified in each species (2). In the past 2 years, numerous human genome-wide association (GWA) studies have confirmed known genes and identified new genomic loci associated with HDL levels (3–11). Human GWA studies typically identify only a few significant genes because of the stringent statistical threshold, but they also highlight a number of genes with suggestive P values that are not investigated further due to the potential of identifying a high number of false positives. With the mouse, we can independently evaluate both significant and suggestive loci from GWA studies. One method is to test in vivo mouse models such as knockout or transgenic mice. Another method is to use the mouse model for sequencing, expression studies, or breeding when a QTL has been detected in the mouse homologous region. Even when GWA peaks are replicated in additional human studies, an experimental model such as the mouse is useful for elucidating gene function.
This approach is based on the hypothesis that a QTL for a trait in the mouse that maps to the homologous location as a QTL or GWA peak for the same trait in humans is most likely caused by the same gene. Although it may not always be true, this is a reasonable hypothesis that has been used to find the causative genes for many traits. In addition, if two genes are closely linked to each other and have the same or similar functions, then a mutation in one could affect HDL in humans and a mutation in the other could affect HDL in mice.
Mouse QTL are detected by crossing two strains [as reviewed in Peters et al. (12)], phenotyping all offspring, and genotyping polymorphisms at regular intervals in the genome. We use the mouse “toolbox” to investigate candidate genes within a locus and ultimately to identify the QTL gene (13, 14). This toolbox is based on genomic comparison of the parental strains of the QTL cross. The list of evidence includes databases and bioinformatic tools to compare haplotype blocks, differences in gene expression, and nonsynonymous polymorphisms predicted to change function.
In this study, we demonstrate how the published mouse QTL map helps interpret human GWA studies for HDL. For the genes identified in human GWA studies, we use the mouse model to provide bioinformatics, sequencing, and expression studies to test whether the gene is likely to be an HDL candidate gene in the mouse, thus supporting the human GWA findings. Then, for GWA peaks that point to multiple genes, we evaluate each gene, pinpoint the most credible HDL genes, and judge which genes are unlikely candidates.
MATERIALS AND METHODS
Human dataset
The GWA meta-analysis (5, 6) comprised three studies: the Diabetes Genetics Initiative (DGI), the Finland-United States Investigation of NIDDM Genetics (FUSION) and SardiNIA Study of Aging (5, 6). The dataset included the results from the association analysis between the genotypes of approximately 2.5 million SNPs (genotyped and inferred) and the phenotype (HDL levels) for 8,816 individuals. SNPs with P < 10−3 were sorted into bins by linkage disequilibrium using LDSelect with a cut-off value of r2 = 0.5 (15).
Mouse datasets
We used the published mouse HDL QTL map as a reference (2); we updated it with recent publications. The updated mouse HDL QTL list is provided in supplementary Tables I and II. The PERA/EiJxDBA/2J (PERAxD2), PERA/EiJxI/LnJ (PERAxI), CAST/EiJx129S1/SvImJ (CASTx129), and DBA/2JxCAST/EiJ (D2xCAST) crosses had previously been published by our group (16–18) and the raw data was available to us to perform combined QTL analysis as described in (18, 19).
Mapping approach
We used the human genome map from NCBI (build 36) and the refSNP position to place the GWA SNPs onto the human genome. To identify the mouse orthologs of the human GWA genes, we downloaded the human-mouse orthology gene list from the Biomart database, which integrates genetic resources including the Ensembl homology (20).
Mouse bioinformatics toolbox
The mouse bioinformatic toolbox has previously been described in Burgess-Herbert et al. (14) and DiPetrillo et al. (13). In summary, a mouse HDL QTL gene should present bioinformatics and molecular evidence including: 1) haplotype differences between the parental QTL strains, and 2) either a gene expression difference or the presence of a nonsynonymous coding polymorphism between the QTL parental strains. For haplotype analysis, we used the Center for Genome Dynamics (CGD) new mouse diversity array genotype data (21) and the CGD strain comparison tool, which allow the determination of genomic region identical between two or more strains. Genes located within these identical regions can be excluded as candidate genes for the QTL, as they are not likely to carry the causative polymorphism. Amino acid changes were assessed by examining the CGD imputed database available on their website (22), by the Mouse Phenome Database (MPD), or by resequencing of the parental strains. Additionally, we used the Sanger Institute database when the strains of a QTL were among the fully resequenced 17 strains. Expression differences were assessed using the results of the 12-strain microarray survey from Shockley et al. (23), available at the CGD website, or by performing real-time PCR in the parental strains using mice of the same sex and fed the same diet as used to detect the QTL. We assessed the potential functionality of the single nucleotide polymorphisms (SNPs) located in the promoter region by using Alibaba v2.1, which uses the Transfac database as a reference for transcription factor binding sites.
Expression analysis
For real-time PCR, we generated cDNA from liver RNA that was collected for male and female mice from 12 strains fed a chow or atherogenic diet as described previously for microarray survey experiments (23). We also collected tissues from additional strains [C57BL/6J (B6), CAST, NZB/BINJ (NZB), and NZW/LacJ (NZW)] in an identical manner. Gene-specific primers were designed by Primerdesign Ltd. (Southampton, UK) (supplementary Table III). Real-time PCR was performed using the ABI Prism 7000 sequence detection system (Applied Biosystems, Inc., Foster City, CA) using SYBR green. Beta-2 microglobulin or Gapdh were used as controls. Data were analyzed by LinRegPCR (v11.0) (24). Expression ratios between the two strains were calculated using the Relative Expression Software Tool (REST©) (25).
Sequencing of candidate genes
We obtained strain-specific genomic DNA from the Mouse DNA Resource at The Jackson Laboratory. Using custom primers, we resequenced the exons and splice sites of Abca1 (NM_013454.3) in CAST, PERA, I, D2, and 129, and Wwox (NM_019573.3) and Galnt2 (NC_000074.5) in B6, CAST, and CASA/RkJ (CASA). We also resequenced 713 bp upstream of Abca1 and the 3′UTR of Galnt2 in the relevant strains. Exon1 of Galnt2 was not resequenced due to technical difficulties. For Cdh13, we searched the CGD imputed SNP database for polymorphisms that contain CAST and B6 but not CASA. Direct sequencing was performed on the PCR products using Big Dye Terminator Cycle Sequencing Chemistry and the ABI 3700 Sequence Detection System (Applied Biosystems, Inc.). Results were analyzed using Sequencher software (version 4.2). All SNPs and genotypes have been reported to NCBI dbSNP. Each polymorphism was evaluated for functionality using the MatchTM Algorithm (BIOBASE Biological Databases GmbH, Wolfenbüttel, Germany) for polymorphisms in the promoter region and PolyPhen and SIFT for nonsynonymous coding polymorphisms (26, 27). We used the University of California Santa Cruz (UCSC) database to assess if the amino acid was conserved in mammals.
Abca1 promoter activity assay
We amplified 713 bp upstream of the transcription start site of the mouse Abca1 gene from strains CAST and D2 and ligated each amplicon to the PGL3-enhancer vector (Promega, Madison, WI). Each promoter construct was cotransfected to HEK293 cells (American Type Culture Collection, Manassas, VA) with plasmids CMX-BGAL, CMX-mlXRa, and CMX-mRXRa. Transfection was accomplished using FuGENE 6 according to the manufacturer's instructions (Roche Diagnostics, Indianapolis, IN). Transfected cells were incubated for 24 h with vehicle alone (ethanol), 22OH (10 μM), 9cRA (10 μM), or with both ligands. After washes, cells were collected and lysed; supernatant was collected after centrifugation. Luciferase activity was measured using a luminescence counter (Victor 2, Perkin Elmer, Boston, MA) immediately after addition of luciferase assay reagent to an aliquot of cell lysate.
RESULTS
Mapping of GWA study genes onto the mouse QTL map for HDL levels
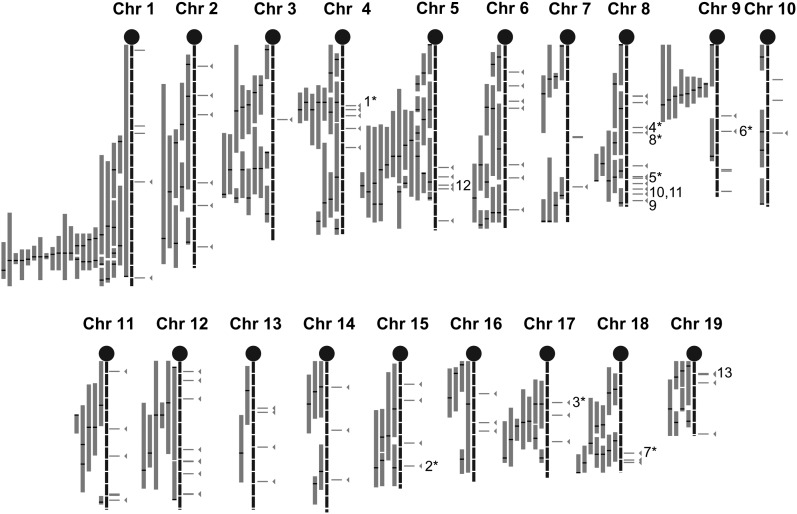
We mapped results of the human GWA study from Kathiresan et al. (5) onto homologous locations on the mouse genome, investigating the seven significant loci (104 SNPs with P < 5 × 10−8 as well as suggestive SNPs, defined as P < 10−3. These 3,882 SNPs were first binned by linkage disequilibrium into 1,121 bins using r2 = 0.5. To reduce the number of potential false positive results from the human GWA study, we only selected the most significant bins carrying at least one SNP with P < 10−4 (171 bins or 1,324 SNPs in total indicated in supplementary Table IV). Each bin contained between one and 116 SNPs (supplementary Table IV). The SNPs within each bin were mapped on the human genome (build 36). If the SNP fell within a gene, we assigned the gene to that bin; if the SNP fell in an intergenic region, we assigned it the closest gene. In the latter case, the SNPs were within 138 Kb of the closest gene on average. For 62 bins, we also pooled adjacent bins into a single locus (supplementary Table V) because they mapped to the same or adjacent neighboring genes, leaving 109 unique loci. For each human gene, we identified the mouse homolog using the Biomart database. A total of 17 loci were removed from the list because the mouse does not have a homolog for the human gene or the locus did not map to a mouse location accurately; i.e., the human SNP fell into an intergenic region and its proximal and distal genes mapped to different mouse chromosomes (supplementary Table VI). This left the 92 unique loci listed in supplementary Table VII and represented in Fig. 1. Seventy-two loci contained one gene, 18 loci contained between two and six genes, and two loci, both on mouse chromosome (Chr) 8, contained 18 and 30 genes, respectively. The largest locus was an intergenic region of 753 Kb on mouse Chr 10.
Fig. 1.
Mouse HDL QTL map overlapped with the homologous genes identified in a human GWA study. HDL QTL information was gathered from Wang and Paigen (2). We updated this list with recent HDL QTL results. A QTL is defined as a main effect QTL with a suggestive or significant LOD score as reported in the study. Each QTL peak positions, CIs, strains, sex, diet, and reference study are indicated in supplementary data (supplementary Tables I, II). Gray bars represent the 95% CI of each mouse QTL, black bars indicate QTL peaks. SNPs from the GWA study with a P < 10−3 were selected and binned by linkage disequilibrium using r2 = 0.5. Horizontal gray lines to the right of the chromosomes represent the 92 selected GWA study hits (bins), lines with arrows represent genes falling within the 95% CI of the HDL QTL. Genes discussed in this report are also indicated in this map: Abca1 (1), Ppara (2), Angptl4 (3), Cpe (4), Lcat (5), Lipc (6), Lipg (7), Lpl (8), Galnt2 (9), Cdh13 (10), Wwox (11), Mvk/Mmab (12), Fads1/Fads2 (13). Numbers with * indicate a gene with a reported in vivo mouse model (knock-out, transgenic or mutant mouse) showing a variation in HDL level compared with controls.
Mapping these 92 loci onto the mouse map showed that 49 (53%) of the GWA loci were within 5 Mb of a mouse QTL peak. Eight human GWA loci mapped to genes known to be involved in HDL metabolism (Abca1, Ppara, Angptl4, Cpe, Lcat, Lipc, Lipg, and Lpl) for which an in vivo mouse model (knockout, transgenic or spontaneous mutation) shows a difference in HDL level (Fig. 1). We then used the mouse model to obtain supporting evidence for some of the GWA peaks starting with a known HDL gene (Abca1) as “proof of principle” and continuing with three unknown HDL genes (Galnt2, Wwox, and Cdh13).
Evidence for a known mouse HDL gene, Abca1
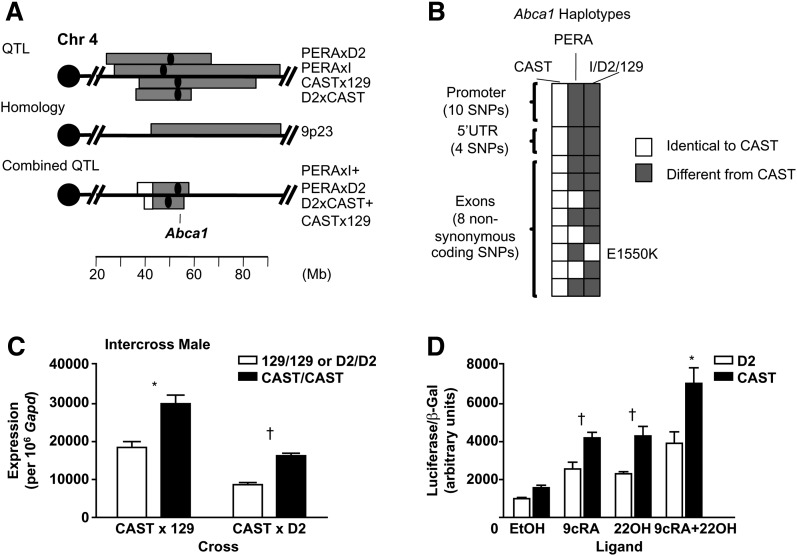
Abca1, identified in the GWA with P = 7.36 × 10−8 (5), encodes the ATP-binding cassette, sub-family A member 1. HDL QTL have been identified at that location in humans (Chr 9p23) and in mouse crosses (129xCAST, CASTxD2, PERAxI, PERAxD2, NODxB6.129apoE−/−, NZBxNZW, and B6xC3H) at the Abca1 location at 53 Mb (Fig. 2A) (16–18, 28, 29). The homologous region between mouse and human QTL contained 496 genes (Fig. 2A). We combined the data from four QTL crosses for which overlapping wild-derived strains were used: 129xCAST with CASTxD2 and PERAxI with PERAxD2 (supplementary ). The combined QTL exhibits a reduced confidence interval (CI) and a higher logarithm of the odds (LOD) score, two consistent characteristics indicating that the same gene causes a QTL in two crosses (Fig. 2A). The QTL peak is located at 28 cM (53 Mb) for the PERA crosses and 22-26 cM for the CAST crosses; Abca1 is at 28.6 cM. This combined approach reduced the region from 495 genes to 171 genes for the PERA crosses, and to 208 genes for the CAST crosses; 157 genes, including the known HDL gene Abca1, were found in both PERA and CAST crosses (supplementary Table VIII).
Fig. 2.
Bioinformatic tools and molecular evidence for Abca1 in the mouse on Chr 4. The 95% CI of the QTL involving CAST and PERA are indicated as the horizontal gray bar with the black dot as the QTL peak (A). A combined QTL was performed using R/qtl (supplementary ); the reduced 95% CI overlapping with the homologous region is indicated as the gray bar (A). The GWA gene is indicated at 53 Mb (Abca1). Sequencing results and haplotypes of Abca1 are indicated in B. Additional information regarding the resequencing is available in supplementary Table IX. E1550K was predicted to be damaging. Expression difference for Abca1 was evaluated between CAST, D2, and 129 by quantitative PCR (C). RNA was extracted from liver tissue from 16-week-old males on a high-fat diet. Promoter activity difference was evaluated in CAST and D2 (D). Both constructs (CAST and D2) were cotransfected with Beta-galactosidase (bGAL), liver X receptor alpha (LXRa), and retinoid X receptor alpha (RXRa) plasmids into HEK 293 cells. LXRa and RXRa ensured transcriptional activation of Abca1 and were activated with ligands 22-hydroxycholesterol (22OH, 10 µM) or 9-cis-retinoic acid (9cRA, 10 µM), respectively, or with both ligands. The results summarize four replicates for each construct. The CAST promoter exhibited higher transcriptional activity compared with that of D2. *P < 0.001, †P < 0.05 (C, D).
We resequenced the exons and about 1Kb upstream of the gene in the parental strains (PERA, CAST, I, D2, and 129) to encompass the proximal promoter region of Abca1 (supplementary Table IX). Through this resequencing, we identified three haplotypes within the gene region specific to CAST, PERA, and I/D2/129 (Fig. 2B). This finding is consistent with the results of the combined QTL cross; I and D2 are identical but different from PERA, D2 and 129 are identical but different from CAST (Fig. 2B).
We then examined Abca1 for a causal polymorphism: either a nonsynonymous coding polymorphism that differs between the parental strains and is likely to change function or a difference in expression between the parental strains, implying a regulatory polymorphism. For PERA crosses, we identified eight polymorphisms that changed amino acids between PERA and the strains I and D2; one of these, E1550K, was damaging according to Polyphen (Table 1 and supplementary Table IX). Therefore, we suggest that this may be the causal polymorphism that results in an HDL QTL between PERA and the strains I and D2.
TABLE 1.
Mouse bioinformatic evidence for human GWA genes
| HDL QTL Crossa | Gene Expression Differencesb | Amino Acid Substitutionc | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Strains | Diet | Sex | Fold Change(P) | Amino Acid | Charge/Polarity | Functional? | Conserved? |
| Known HDL gene | ||||||||
| Abca1 | D2xCAST | HF | M | +1.67(<0.001) | T251I | Polarity | No | No |
| V343M | No | No | No | |||||
| S658G | No | No | No | |||||
| I691V | No | No | Yes | |||||
| N1889T | No | No | Yes | |||||
| CASTx129 | HF | M | +1.56 (<0.001) | |||||
| PERAxI | HF | F | +1.15 (0.165) | S658G | No | No | No | |
| PERAxD2 | HF | F | +1.05 (0.542) | V1291I | No | No | No | |
| E1550K | Polarity and charge | Yes | Yes | |||||
| T1565N | No | No | No | |||||
| Unknown HDL gene | ||||||||
| Galnt2 | B6xCASAa | C | F | -1.27 (0.010) | None | — | — | — |
| Wwox | B6xCAST | C | F | −1.16 (0.342) | V186M | No | Yes (SIFT) | No |
| Cdh13 | B6xCAST | C | F | +1.16 (0.052) | N56S | No | No | No |
| Y214F | Polarity | Yes (SIFT) | Yes | |||||
| I660M | No | No | Yes | |||||
HF, high-fat diet; C, chow diet; M, male; F, female.
Bold indicates the high allele strain. CAST was used as a surrogate for CASA.
Gene expression differences between the QTL parental strains. All quantitative PCR performed regarding both loci are reported here. Quantitative PCR was performed on cDNA from liver samples from at least three mice for the most relevant strain/diet/sex. Quantitative PCR was performed for Abca1, Galnt2, and Wwox using β2 microglobulin as the endogenous control. The microarray result is indicated for Cdh13 (probe 1431824_at). The fold change is indicated as the gene expression of the high allele strain (bold) compared with the low allele strain.
For Cdh13, amino acid substitutions were determined using the high-density SNP component of the MPD database and the imputed SNPs from the CGD with a minimum confidence level of 0.9. For Galnt2 and Wwox, amino acid substitutions were determined by resequencing of B6 and CAST. Functionality was determined by using Polyphen and SIFT. Conservation was determined by comparison with mammals at the UCSC website. For Abca1, the amino acid substitutions are arranged in two groups: the crosses involving CAST and the crosses involving PERA. Additional information on the sequencing results is available in supplementary Table IX and XII.
Nevertheless, this polymorphism cannot explain the QTL in the CAST crosses because CAST, D2, and 129 did not have any polymorphisms that changed amino acids. However, the sequence of the promoter region differs by 10 polymorphisms in CAST (Fig. 2B and supplementary Table IX) compared with D2 and 129. Therefore, we hypothesized that a difference in Abca1 expression may explain the CAST crosses. Using our 12-strain microarray survey, we identified an expression difference between CAST and (D2 and 129) males on a high-fat diet but no expression difference among PERA, I, and D2 females (Table 1 and supplementary Table X). This result was confirmed by real-time PCR (Fig. 2C). By luciferase assay, we extended this observation by showing that a cis-acting element in the promoter region controls the Abca1 mRNA expression (Fig. 2D); the CAST promoter construct displayed greater promoter activity compared with the D2 promoter. The reduced expression in the D2 strain, leading to a decrease in HDL level, is consistent with the Abca1 knockout mouse model that shows lower HDL (30).
Therefore, in the four crosses involving a wild-derived mouse strain, we conclude that Abca1 is the causal QTL gene but that in the two CAST crosses, the causative polymorphism is located in the promoter region and causes an expression difference whereas in the two PERA crosses, an amino acid change causes a functional change in protein activity. Thus, the Abca1 gene has two different mutations, providing an allelic series and strengthening the evidence that Abca1 is the causal QTL gene.
Additionally, we investigated if any Abca1 polymorphisms could also explain the three additional QTL located in the region (B6xC3H, NZBxNZW, and NODxB6.129apoE−/−). A derived sub-strain of B6 (C57BL/6NJ), C3H, and NOD have been fully resequenced by the Sanger Institute, and the sequences of the B6, C3H, and NOD strains are identical at the Abca1 locus (Abca1 gene and promoter). In addition, as indicated by the CDG database, NZB and NZW have identical haplotypes. These results indicates that Abca1 is not the QTL gene for the B6xC3H, NZBxNZW, and NODxB6.129apoE−/− crosses and further investigation will determine which gene is responsible for these QTL.
Mouse evidence for novel genes found in the GWA study: Galnt2, Wwox, and Cdh13
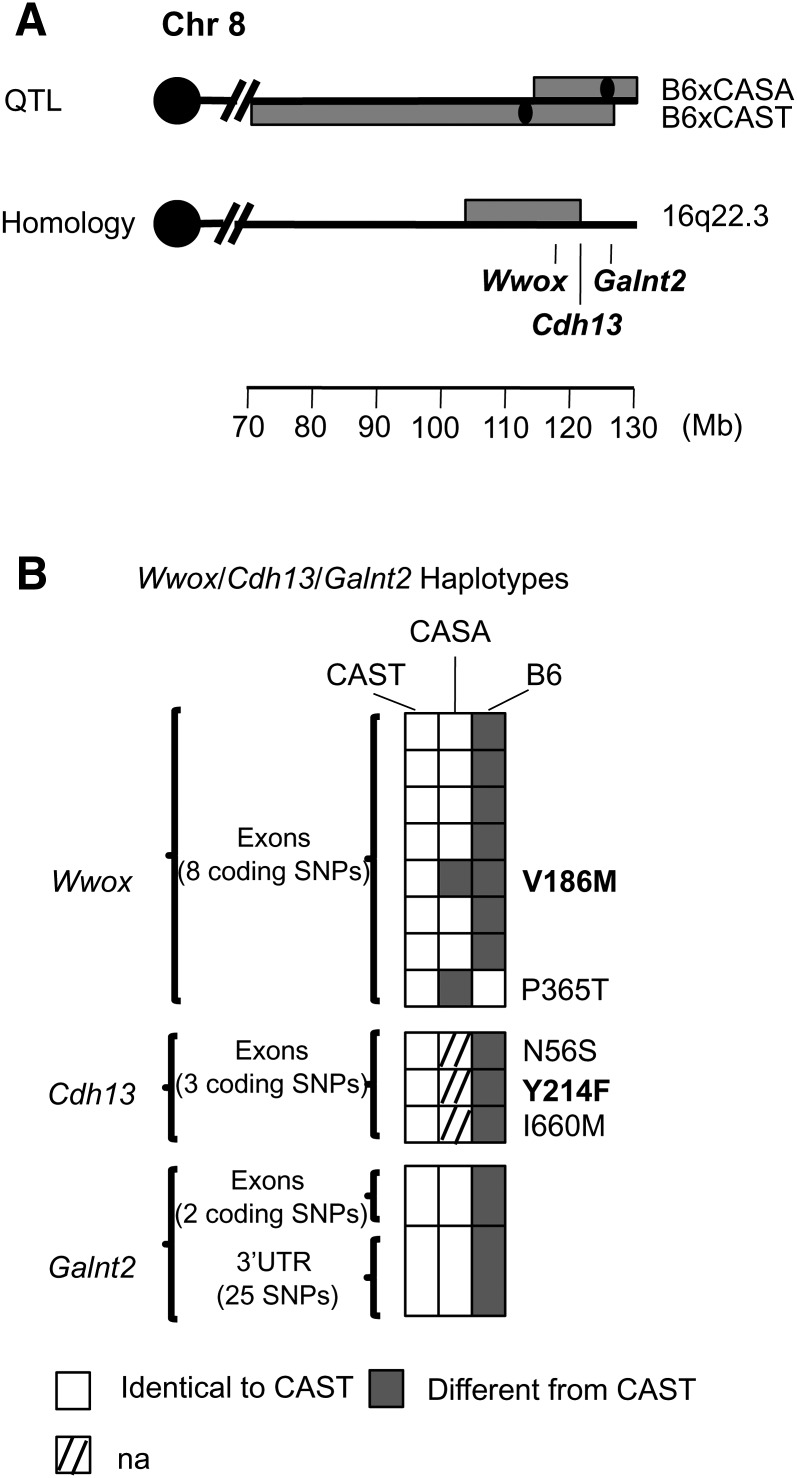
Three potential HDL genes, whose role in regulating HDL was previously unknown, were identified by the GWA study. These candidates mapped to distal mouse Chr 8: at 126 Mb for Galnt2 (P = 2.86 × 10−7 in the GWA), at 117 Mb for Wwox (P = 2.25 × 10−5 in the GWA), and at 121 Mb for Cdh13 (P = 2.46 × 10−5 in the GWA). Two mouse crosses previously identified a QTL in this region: B6xCAST and B6xCASA in males and females on a chow diet with peaks at 111 Mb (LOD = 3.6) and 129 Mb (LOD = 4.7), respectively (Fig. 3A) (31, 32). CAST and CASA are closely related; they were inbred from the same group of wild-derived Castaneus mice. B6 was the high HDL strain, CAST and CASA were the low HDL strains. Because the peaks of the two crosses were 18 Mb apart, we suggest that both distinct QTL and colocalizing QTL are present. The distal region of mouse Chr 8 containing Galnt2 is homologous to human chromosome 1, whereas the homologous region containing Cdh13, Wwox, and the peak of the B6xCAST QTL is located on human chromosome 16 (16q23.3), which carries a human HDL QTL (Fig. 3A). Human homology reduced the genomic locus to 15 Mb for the B6xCAST cross (106 to 121 Mb) (Fig. 3A). In addition, haplotype analysis reduced the number of candidate genes to 184 for the B6xCAST cross (including Cdh13 and Wwox) and 154 for the B6xCASA cross (including Galnt2); the 4 Mb overlap between both QTL contained 23 genes (supplementary Table XI). Because the GWA study genes were among the mouse HDL QTL candidate genes identified by haplotype analysis, we searched for additional molecular evidence for all three genes in the mouse.
Fig. 3.
Bioinformatic tools for the mouse Chr 8 locus. The 95% CI of the B6xCAST and B6xCASA QTL are indicated as the horizontal gray bar with the black dot as the QTL peak (A). Haplotypes based on resequencing of the genes in B6, CAST, and CASA are indicated (B). Additional resequencing information is available in supplementary Table XII. Amino acid substitutions predicted to be deleterious are indicated in bold.
For Galnt2, resequencing did not identify any nonsynonymous polymorphisms but did establish that the haplotypes within the gene are identical between CAST and CASA but different from B6 (Fig. 3B and supplementary Table XII). CASA was not in our 12-strain microarray survey, and CASA mice were not available for this study, but both CASA and CAST have the same haplotype in regions surrounding and within the gene (Fig. 3B). Therefore, we used CAST as a surrogate for CASA in our expression analysis. In our microarray study, Galnt2 was differentially expressed between B6 and CAST in liver of females and males on a chow diet (supplementary Table X). Real-time PCR verified these results in females (1.27-fold decrease in expression in B6 compared with CAST, P = 0.01) (Table 1). This result is consistent with recent in vitro results showing that inhibiting Galnt2 expression increases HDL (11); B6 is the high HDL strain at this locus and showed lower expression of Galnt2 compared with CAST/CASA.
For Wwox, we performed real-time PCR between B6 and CAST on chow in males and females but did not find any difference in expression (P > 0.05) (Table 1). However, by resequencing, we identified a nonsynonymous coding polymorphism predicted to change function (V186M) in an exon present in four out of the five transcripts reported in Ensembl (Fig. 3B and supplementary Table XII). The polymorphism segregated between B6 and CAST but not between B6 and CASA. We also identified another nonsynonymous coding polymorphism (P365T) that segregated between B6 and CASA (supplementary Table XII). However, this polymorphism was specific to only one transcript and its functionality could not be assessed due to the lack of sequence alignment at that position within SIFT. A QTL gene may have an expression difference or an amino acid difference that changes function; because V186M may affect the function or structure of WWOX, we think that Wwox is a QTL gene for the B6xCAST cross. These results support this gene being associated with HDL metabolism.
For Cdh13, no difference in expression was observed in our 12-strain microarray survey between B6 and CAST fed chow in males and females (P > 0.05) (Table 1 and supplementary Table X). However, use of SIFT showed that one out of the three amino acid changes reported in MPD was characterized as damaging (Y214F).
Therefore, based on the evidence outlined above for these three genes, the B6xCAST cross has polymorphisms in Wwox and Cdh13, and differs in expression at Galnt2 (consistent with the broad QTL and large 95% CI). The B6xCASA cross differs in expression at Galnt2, whereas the effect of P365T in Wwox is yet to be determined for this cross.
Using mouse genetics as a tool to resolve regions with multiple genes
MVK/MMAB locus.
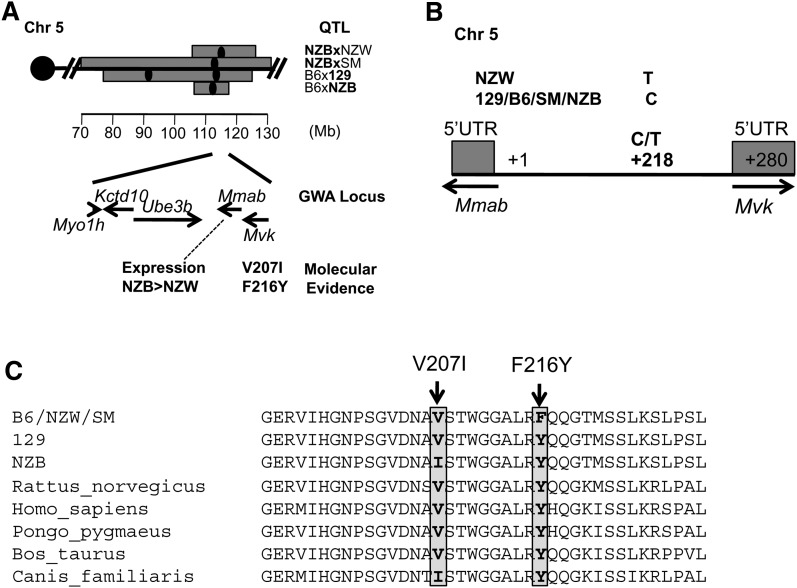
The human GWA identified a locus on human chromosome 12 at 108 Mb (12q23-24) encompassing five genes: MYO1H, KCTD10, UBE3B, MVK, and MMAB (P = 1.88 × 10−6). These genes cannot be dissociated through GWA studies or replication in additional human populations because of the underlying high linkage disequilibrium in the region. The issue is which gene is the actual HDL gene? Homologous genes in the mouse are located in the same sequential order on Chr 5 at 114 Mb. This region in the mouse has multiple QTL, some with multiple peaks. The most relevant crosses, which we selected based on peak location (within 5 Mb of the gene locus), are B6xNZB (peak, 113 Mb, LOD = 3.6), NZBxSM/J (SM) (peak, 113 Mb, LOD = 5.6), NZBxNZW (peak, 115 Mb, LOD = 12.7), and B6 × 129 (peak, 114 Mb, LOD = 3.3) (29, 33–35) (Fig. 4A). NZB and 129 carry the high HDL allele, strains B6, NZW, and SM carry the low HDL allele. Overlapping QTL could be due to different genes. For this particular locus, however, we assumed that the same gene was responsible for all four crosses because three of these crosses had one mouse strain in common (NZB), suggesting that a polymorphism specific to this strain was responsible for the QTL.
Fig. 4.
Polymorphisms in the promoter and coding regions of Mmab and Mvk. A: Mouse HDL QTL located on Chr 5. The five genes identified by the GWA are indicated in their sequential order in the mouse with their molecular evidence. B: One polymorphism was identified in the promoter region at 218 bp downstream of the 5 ′ UTR of Mmab or 62 bp upstream of the 5 ′ UTR of Mvk that is specific to NZW. C: The exons and promoter regions of NZB, NZW, B6, and SM were resequenced. Two nonsynonymous coding polymorphims, V207I and F216Y, were identified in Mvk.
We first examined the gene expression differences for these five genes between the QTL parental strains. Myo1 h and Ube3b had no expression differences among any QTL strains in our 12-strain microarray survey (P > 0.01) (supplementary Table X). Real-time PCR on Mvk, Mmab, and Kctd10 showed no significant expression differences between the QTL strains (Table 2) except for Mmab in a single cross, NZBxNZW. Mmab and Mvk are positioned in the opposite direction with only 280 bp between their respective 5′ UTR. By resequencing, we identified a G/A polymorphism 62 bp upstream of Mvk that was specific to NZW (Fig. 4B). However, we could not identify a transcription factor binding site at this location. Because we found no difference in expression for four of these genes and the expression difference for Mmab could only explain one cross (NZBxNZW), we investigated the possibility of a nonsynonymous coding polymorphism that could explain the QTL.
TABLE 2.
Mouse bioinformatic evidence for human GWA genes in LD block
| HDL QTL Crossa | Gene Expression Differencesb | Amino Acid Substitutionc | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Strains | Diet | Sex | Fold Change(P) | Amino Acid | Charge/Polarity | Functional? | Conserved? |
| Multiple genes locus on Chr 5 | ||||||||
| Kctd10 | B6xNZB | HF | F | +1.13 (0.828) | None | — | — | — |
| NZBxNZW | HF | F | +1.28 (0.767) | None | — | — | — | |
| NZBxSM | HF | F | +1.23 (0.733) | None | — | — | — | |
| C | F | +1.18 (0.883) | ||||||
| B6x129 | HF | F | +1.05 (0.912) | None | — | — | — | |
| C | F | +1.29 (0.453) | ||||||
| Mmab | B6xNZB | HF | F | −1.22 (0.736) | None | — | — | — |
| NZBxNZW | HF | F | −2.08 (0.001) | None | — | — | — | |
| NZBxSM | HF | F | +1.21 (0.690) | None | — | — | — | |
| C | F | +1.05 (0.998) | ||||||
| B6x129 | HF | F | −1.24 (0.426) | None | — | — | — | |
| C | F | +1.14 (0.769) | ||||||
| Mvk | B6xNZB | HF | F | −1.04 (0.955) | V207IF216Y | NoNo | NoNo | YesYes |
| NZBxNZW | HF | F | −1.35 (0.854) | V207IF216Y | NoNo | NoNo | YesYes | |
| NZBxSM | HF | F | +1.03 (0.912) | V207IF216Y | NoNo | NoNo | YesYes | |
| C | F | +1.27 (0.669) | ||||||
| B6x129 | HF | F | −1.00 (0.989) | F216Y | No | No | Yes | |
| C | F | +1.06 (0.873) | ||||||
| Multiple genes locus on Chr 19 | ||||||||
| Fads1 | BALBxB6 | C | M | +1.66 (0.044) | V163I | No | No | Yes |
| C | F | +1.40 (0.129) | ||||||
| Fads2 | BALBxB6 | C | M | +2.32 (0.025) | ||||
HF, high-fat diet; C, chow diet; M, male; F, female.
Bold indicates the high allele strain.
Gene expression differences between the QTL parental strains. All quantitative PCR performed regarding both loci are reported here. Experiments were performed in liver samples from at least three mice for the most relevant strain/sex/diet combination using β2 microglobulin as the endogenous control. The fold change is indicated as the gene expression of the high allele strain (bold) compared with the low allele strain. Only females were used to verify the microarray results provided in supplementary Table X.
Sequencing results are available in supplementary Tables XIII and XIV. Functionality was determined by using Polyphen and SIFT. Conservation was determined by comparison with mammals at the UCSC website.
We resequenced the coding regions of all five genes in NZB, 129, B6, SM, and NZW (supplementary Table XIII). We did not identify any nonsynonymous coding polymorphisms in Myo1 h, Kctd10, Ube3b, and Mmab. Mvk contained two nonsynonymous polymorphisms, I207V and F216Y (Fig. 4C). Neither polymorphism is predicted to change the function of the protein (Polyphen/SIFT) but both are located in a highly conserved region. F216Y changes the polarity of the amino acid and segregates between the parental strains of all crosses. Based on this evidence, we think Mvk is the most likely causal gene for the crosses, due to the presence of a conserved amino acid change (F216Y) that segregates between high and low HDL strains. Although we have no proof that this amino acid changes the protein function, we suggest that further testing may show a causal functional change. This result illustrates how merging crosses can help narrow QTL and pinpoint potential causal genes and polymorphisms.
FADS1/FADS2 locus.
The GWA study also identified a region on human chromosome 11 at 61.3 Mb that encompasses six genes in high linkage disequilibrium: C11orf9, C11orf10, FEN1, FADS1, FADS2, and FADS3. The mouse orthologous genes are located on Chr 19 at 10.1 Mb, Gm98 and 1810006K21Rik are the orthologous genes of C11orf9 and C11orf10, respectively. Two mouse crosses have identified an HDL QTL within 5 Mb of this location: C57BL/6J x A/J (B6xA) and C57BL/6J x BALB/cJ (B6xBALB) in both males and females fed chow (36, 37). B6 was the high allele strain in the B6xA cross and the low allele strain in the B6xBALB cross, so the QTL in these two crosses are most likely not caused by the same gene. Other unpublished work from our laboratory has tentatively identified the gene for the B6xA QTL and it is not located in this cluster; therefore, we focused on the B6xBALB cross. By haplotype analysis, we eliminated Fen1 as a likely candidate gene because the haplotypes within the gene were identical in both B6 and BALB. We judged Gm98, 1810006K21Rik, and Fads3 to be unlikely candidates because none of these three genes had nonsynonymous polymorphisms between B6 and BALB (based on SNP databases) and none showed a difference in liver expression between B6 and BALB, based on our 12-strain microarray database (supplementary Table X). Therefore, we focused our attention on Fads1 and Fads2. We identified a nonsynonymous SNP in Fads2 between B6 and BALB by resequencing, V163I (supplementary Table XIV). However, we do not think it is responsible for the QTL because the amino acid change does not change polarity or charge, it is not located in a conserved region, and it was considered nonfunctional (Polyphen/SIFT). Using the 12-strain microarray database, we found a difference in expression between B6 and BALB for Fads1 and Fads2 (supplementary Table XIV), which we confirmed by real-time PCR (Table 2). Expression of both Fads1 and Fads2 was higher in BALB compared with B6 and both genes are involved in the same biological pathway. Further work in humans and mice is required to determine whether Fads1 or Fads2 (or both) is the QTL gene, but using the mouse model did help to narrow the focus of future work from six to two genes.
DISCUSSION
Recent human GWA studies have identified well-known HDL genes such as ABCA1, LIPG, LPL, and LIPC (3–10), as well as new genes such as GALNT2. However, GWA studies may not detect HDL genes because some GWA peaks are only suggestive and others map to haplotype blocks with multiple genes. Because 90% of the HDL QTL overlap between humans and mice (2), we suggest an innovative strategy based on mouse genetics to help decipher and add support to the results of human GWA studies. We first showed a strong similarity between results of a human GWA study from Kathiresan et al. (5) and the mouse HDL QTL map (2). When a gene identified by a human GWA study mapped within a mouse HDL QTL at a homologous location, we compared parental strains of the mouse QTL cross for additional bioinformatic and molecular evidence. We demonstrated this strategy as a proof of principle to a well-known HDL gene (ABCA1) and a newly identified HDL gene (GALNT2) (11). We then applied this strategy to candidate genes for two other categories of GWA study results: 1) when the new gene reaches only a suggestive P-value (WWOX and CDH13), and 2) when the identified locus contains more than one gene (MVK/MMAB and FADS loci).
In this report, we explain and provide examples of our strategy to add evidence to human loci identified at the significant and suggestive level by GWA studies by using a mouse bioinformatics toolbox(13, 14). Our analyses are based on recommendations by the Complex Trait Consortium, which in 2003, published criteria for determining when a gene is a QTL gene (38). They recommended using several independent lines of evidence for each gene. Within each mouse QTL, we use haplotype analysis between parental strains of QTL crosses. We then use the high-density SNP databases and expression profile databases among multiple inbred mouse strains to add evidence to the genes located within each QTL (13, 14). To consider an HDL gene as a viable candidate, we require it to have either a molecular difference, such as a nonsynonymous coding difference that affects either the structure/function of the protein (Abca1, Wwox, Cdh13, and Mvk), or a difference in gene expression level (Abca1, Galnt2, Mmab). A current limitation of our bioinformatics toolbox is the requirement to further investigate alternative splicing and posttranslational modification differences between the parental strains in mice. Both of these are possible causes of QTL genes, but databases today do not have sufficient information for a bioinformatic approach. Until such information is available, these possibilities must be examined with sequencing or with direct measurement of protein levels with antibodies.
Among the candidate genes investigated in this report, in vivo mouse models for some of the genes are available. For instance, the Wwox knockout mouse has previously been shown to exhibit hypercholesterolemia (39); HDL was not measured, but hypercholesterolemia in a chow-fed mouse would indicate increased HDL because 80% to 90% of cholesterol in a chow-fed mouse is HDL. The Mvk−/− mouse does not have elevated cholesterol when fed chow (40), a fact that does not support Mvk as the candidate gene. However, three of the four crosses that detected this QTL were found in mice fed a high-fat, atherogenic diet; the Mvk−/− mouse needs to be tested on such a diet. Mouse models for Cdh13, Fads1, and Fads2 have not been tested for HDL cholesterol levels, but based on the GWA study and our bioinformatics results in the mouse, the genes should be prioritized for further study. CDH13 encodes T-cadherin and binds to LDLs (41). The mechanism and the physiological consequences of this interaction are not completely understood but the binding of CDH13 to LDL could play a role in HDL metabolism. Fads1 encodes the delta 5 desaturase in fatty acid metabolism, Fads2 encodes the delta 6 desaturase. These two genes are part of the same pathway that produces arachidonic acid from linoleic acid. In the mouse, both genes have a similar expression difference between the strains that detected the QTL. As a result, no choice can be made between these two genes without further work. In fact, it may be that a single regulatory polymorphism causes the expression difference for both genes. Because there are two possible QTL genes and because they are so closely related in function, one or both could play a role in HDL metabolism.
Molecular evidence in human populations that suggests cis-regulation of WWOX (42) and FADS1 (7) is confirmed by our results in the mouse. The direction of the difference in expression matches what we found in the mouse for both genes. For instance, in humans, FADS1 was positively correlated with HDL levels whereas in the mouse, a higher expression of the gene was observed in the high HDL strain (7). Additionally, at the MVK/MMAB locus, cis-expression of MMAB has been observed in some (7, 43, 44) but not all (45–47) human populations, and thus has been implied as responsible for HDL differences in some of these studies. We did identify an Mmab expression difference consistent with observations in humans in one mouse cross (NZBxNZW) (7) but not in the other crosses. Instead, the mouse shows stronger evidence for Mvk (as opposed to Mmab) based on a nonsynonymous coding polymorphism that may affect the function or structure of the protein. Perhaps both genes are involved in HDL regulation, but during evolution, one gene mutated in mice and the other mutated in humans. Our approach is based on the hypothesis that a QTL for a trait in the mouse that maps to the homologous location as a QTL or GWA peak for the same trait in humans is most likely caused by the same gene. This is a reasonable hypothesis and has been used to find the causative genes for many traits. However, if two genes are closely linked to each other and have the same or similar functions, then a mutation in one could affect HDL in humans and a mutation in the other could affect HDL in mice. To our knowledge, neither MMAB not MVK has been screened for coding mutations in humans. Therefore, although there is expression evidence for MMAB in human populations, the possibility of deleterious mutations affecting the structure and function of the MVK protein should be investigated in humans by resequencing before rejecting the gene as an HDL gene. In addition, we have evidence for Acads as a candidate gene for this locus in the mouse (48). Acads is located just 2 Mb away from Mvk/Mmab in the mouse but more than 10 Mb from these genes in humans. The human GWA study did not point to Acads. Therefore, it is possible that both genes (Acads and Mvk) are involved in HDL regulation in mice but that only the MVK/MMAB locus is involved in humans.
Although we expect the genes involved in HDL cholesterol to be the same among species, the molecular mutation that causes the QTL may differ within and among species. For instance, in this report, we found that a nonsynonymous coding polymorphism in Abca1 was responsible for the QTL from the CAST crosses on Chr 4 whereas a difference in expression of Abca1 was responsible for the QTL from the PERA crosses. In addition, Lipg is accepted as a known HDL gene, but the molecular basis of observed association and linkage with HDL vary from a nonsynonymous coding polymorphism in humans (49) to a difference in expression of the gene in mice and baboons (29, 50).
A limitation of our strategy is the existence of important species differences in HDL metabolism, such as the absence of the CETP gene in the mouse. CETP produces the exchange of cholesteryl ester between HDL and VLDL, which participates in HDL degradation in humans. This difference could explain the fact that some human loci were found outside a mouse HDL QTL. However, we also recently showed that the underlying genes for HDL QTL are likely to be the same between rats and mice (51). Therefore, we think that comparative genomics is a very powerful approach to identify candidate genes for complex traits such as HDL.
To conclude, by combining bioinformatics tools with limited laboratory experiments in the mouse, our human-mouse strategy can add support to human GWA results. This approach is advantageous with phenotypes for which mouse QTL results are publicly available, such as triglyceride level, bone mineral density, or other complex traits. We have provided here our current list of HDL QTL as supplementary material. When the mouse homologous gene from a GWA gene is located under a mouse QTL for the same phenotype, one can apply the bioinformatics toolbox and gather evidence before progressing to more targeted studies using animal models, such as knockout and transgenic mice, which are time consuming and expensive but offer great advantages for studying the direct effect of a candidate gene in vivo. Importantly, our human-mouse comparative strategy represents a fast pragmatic approach to help human geneticists extend and clarify GWA studies that identify candidate genes for other complex traits.
Supplementary Material
Acknowledgments
The authors thank Harry Whitmore and Fred Rumill for their help with mouse husbandry, Joanne Currer for editing the manuscript, and Jesse Hammer for graphical assistance.
Footnotes
Abbreviations:
- CAD
- coronary artery disease
- CGD
- Center for Genome Dynamics
- CI
- confidence interval
- GWA
- genome-wide association
- LOD
- logarithm of the odds
- MPD
- Mouse Phenome Database
- QTL
- quantitative trait loci
- SNP
- single nucleotide polymorphism
This study was funded by National Institutes of Health grants (HL077796 and HL081162 to B.P., HL095668 to R.K.), the American Heart Association post-doctoral Fellowship (to M.S.L.), and the National Cancer Institute Core grant to The Jackson Laboratory (CA034196). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies. S. Kathiresan has received grant support from Pfizer and Alnylam Pharmaceuticals and has served on scientific advisory boards for Pfizer, Merck, and Daiichi Sankyo.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure and 14 tables.
REFERENCES
- 1.Castelli W. P., Doyle J. T., Gordon T., Hames C. G., Hjortland M. C., Hulley S. B., Kagan A., Zukel W. J. 1977. HDL cholesterol and other lipids in coronary heart disease. The cooperative lipoprotein phenotyping study. Circulation. 55: 767–772. [DOI] [PubMed] [Google Scholar]
- 2.Wang X., Paigen B. 2005. Genetics of variation in HDL cholesterol in humans and mice. Circ. Res. 96: 27–42. [DOI] [PubMed] [Google Scholar]
- 3.Saxena R., Voight B. F., Lyssenko V., Burtt N. P., de Bakker P. I., Chen H., Roix J. J., Kathiresan S., Hirschhorn J. N., Daly M. J., et al. 2007. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 316: 1331–1336. [DOI] [PubMed] [Google Scholar]
- 4.Kathiresan S., Manning A. K., Demissie S., D'Agostino R. B., Surti A., Guiducci C., Gianniny L., Burtt N. P., Melander O., Orho-Melander M., et al. 2007. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med. Genet. 8(Suppl 1): S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., et al. 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aulchenko Y. S., Ripatti S., Lindqvist I., Boomsma D., Heid I. M., Pramstaller P. P., Penninx B. W., Janssens A. C., Wilson J. F., Spector T., et al. 2009. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 41: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabatti C., Service S. K., Hartikainen A. L., Pouta A., Ripatti S., Brodsky J., Jones C. G., Zaitlen N. A., Varilo T., Kaakinen M., et al. 2009. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 41: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chasman D. I., Pare G., Mora S., Hopewell J. C., Peloso G., Clarke R., Cupples L. A., Hamsten A., Kathiresan S., Malarstig A., et al. 2009. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 5: e1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 466: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters L. L., Robledo R. F., Bult C. J., Churchill G. A., Paigen B. J., Svenson K. L. 2007. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat. Rev. Genet. 8: 58–69. [DOI] [PubMed] [Google Scholar]
- 13.DiPetrillo K., Wang X., Stylianou I. M., Paigen B. 2005. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 21: 683–692. [DOI] [PubMed] [Google Scholar]
- 14.Burgess-Herbert S. L., Cox A., Tsaih S. W., Paigen B. 2008. Practical applications of the bioinformatics toolbox for narrowing quantitative trait loci. Genetics. 180: 2227–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlson C. S., Eberle M. A., Rieder M. J., Yi Q., Kruglyak L., Nickerson D. A. 2004. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 74: 106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyons M. A., Wittenburg H., Li R., Walsh K. A., Churchill G. A., Carey M. C., Paigen B. 2003. Quantitative trait loci that determine lipoprotein cholesterol levels in DBA/2J and CAST/Ei inbred mice. J. Lipid Res. 44: 953–967. [DOI] [PubMed] [Google Scholar]
- 17.Lyons M. A., Wittenburg H., Li R., Walsh K. A., Korstanje R., Churchill G. A., Carey M. C., Paigen B. 2004. Quantitative trait loci that determine lipoprotein cholesterol levels in an intercross of 129S1/SvImJ and CAST/Ei inbred mice. Physiol. Genomics. 17: 60–68. [DOI] [PubMed] [Google Scholar]
- 18.Wittenburg H., Lyons M. A., Li R., Kurtz U., Wang X., Mossner J., Churchill G. A., Carey M. C., Paigen B. 2006. QTL mapping for genetic determinants of lipoprotein cholesterol levels in combined crosses of inbred mouse strains. J. Lipid Res. 47: 1780–1790. [DOI] [PubMed] [Google Scholar]
- 19.Li R., Lyons M. A., Wittenburg H., Paigen B., Churchill G. A. 2005. Combining data from multiple inbred line crosses improves the power and resolution of quantitative trait loci mapping. Genetics. 169: 1699–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smedley D., Haider S., Ballester B., Holland R., London D., Thorisson G., Kasprzyk A. 2009. BioMart–biological queries made easy. BMC Genomics. 10: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H., Ding Y., Hutchins L. N., Szatkiewicz J., Bell T. A., Paigen B. J., Graber J. H., de Villena F. P., Churchill G. A. 2009. A customized and versatile high-density genotyping array for the mouse. Nat. Methods. 6: 663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szatkiewicz J. P., Beane G. L., Ding Y., Hutchins L., Pardo-Manuel de Villena F., Churchill G. A. 2008. An imputed genotype resource for the laboratory mouse. Mamm. Genome. 19: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shockley K. R., Witmer D., Burgess-Herbert S. L., Paigen B., Churchill G. A. 2009. Effects of atherogenic diet on hepatic gene expression across mouse strains. Physiol. Genomics. 39: 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruijter J. M., Ramakers C., Hoogaars W. M., Karlen Y., Bakker O., van den Hoff M. J., Moorman A. F. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfaffl M. W., Horgan G. W., Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng P. C., Henikoff S. 2001. Predicting deleterious amino acid substitutions. Genome Res. 11: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sunyaev S., Ramensky V., Koch I., Lathe W., 3rd, Kondrashov A. S., Bork P. 2001. Prediction of deleterious human alleles. Hum. Mol. Genet. 10: 591–597. [DOI] [PubMed] [Google Scholar]
- 28.Pajukanta P., Allayee H., Krass K. L., Kuraishy A., Soro A., Lilja H. E., Mar R., Taskinen M. R., Nuotio I., Laakso M., et al. 2003. Combined analysis of genome scans of Dutch and Finnish families reveals a susceptibility locus for high-density lipoprotein cholesterol on chromosome 16q. Am. J. Hum. Genet. 72: 903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su Z., Ishimori N., Chen Y., Leiter E. H., Churchill G. A., Paigen B., Stylianou I. M. 2009. Four additional mouse crosses improve the lipid QTL landscape and identify Lipg as a QTL gene. J. Lipid Res. 50: 2083–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNeish J., Aiello R. J., Guyot D., Turi T., Gabel C., Aldinger C., Hoppe K. L., Roach M. L., Royer L. J., de Wet J., et al. 2000. High density lipoprotein deficiency and foam cell accumulation in mice with targeted disruption of ATP-binding cassette transporter-1. Proc. Natl. Acad. Sci. USA. 97: 4245–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sehayek E., Duncan E. M., Yu H. J., Petukhova L., Breslow J. L. 2003. Loci controlling plasma non-HDL and HDL cholesterol levels in a C57BL /6J x CASA /Rk intercross. J. Lipid Res. 44: 1744–1750. [DOI] [PubMed] [Google Scholar]
- 32.Mehrabian M., Castellani L. W., Wen P. Z., Wong J., Rithaporn T., Hama S. Y., Hough G. P., Johnson D., Albers J. J., Mottino G. A., et al. 2000. Genetic control of HDL levels and composition in an interspecific mouse cross (CAST/Ei x C57BL/6J). J. Lipid Res. 41: 1936–1946. [PubMed] [Google Scholar]
- 33.Wang X., Le Roy I., Nicodeme E., Li R., Wagner R., Petros C., Churchill G. A., Harris S., Darvasi A., Kirilovsky J., et al. 2003. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome Res. 13: 1654–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su Z., Korstanje R., Tsaih S. W., Paigen B. 2008. Candidate genes for obesity revealed from a C57BL/6J x 129S1/SvImJ intercross. Int J Obes (Lond). 32: 1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell-Huynh D. A., Weinreb A., Castellani L. W., Mehrabian M., Doolittle M. H., Lusis A. J. 1995. Genetic factors in lipoprotein metabolism. Analysis of a genetic cross between inbred mouse strains NZB/BINJ and SM/J using a complete linkage map approach. J. Clin. Invest. 96: 1845–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stylianou I. M., Tsaih S. W., DiPetrillo K., Ishimori N., Li R., Paigen B., Churchill G. 2006. Complex genetic architecture revealed by analysis of high-density lipoprotein cholesterol in chromosome substitution strains and F2 crosses. Genetics. 174: 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welch C. L., Bretschger S., Wen P. Z., Mehrabian M., Latib N., Fruchart-Najib J., Fruchart J. C., Myrick C., Lusis A. J. 2004. Novel QTLs for HDL levels identified in mice by controlling for Apoa2 allelic effects: confirmation of a chromosome 6 locus in a congenic strain. Physiol. Genomics. 17: 48–59. [DOI] [PubMed] [Google Scholar]
- 38.Abiola O., Angel J. M., Avner P., Bachmanov A. A., Belknap J. K., Bennett B., Blankenhorn E. P., Blizard D. A., Bolivar V., Brockmann G. A., et al. 2003. The nature and identification of quantitative trait loci: a community's view. Nat. Rev. Genet. 4: 911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aqeilan R. I., Trapasso F., Hussain S., Costinean S., Marshall D., Pekarsky Y., Hagan J. P., Zanesi N., Kaou M., Stein G. S., et al. 2007. Targeted deletion of Wwox reveals a tumor suppressor function. Proc. Natl. Acad. Sci. USA. 104: 3949–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hager E. J., Tse H. M., Piganelli J. D., Gupta M., Baetscher M., Tse T. E., Pappu A. S., Steiner R. D., Hoffmann G. F., Gibson K. M. 2007. Deletion of a single mevalonate kinase (Mvk) allele yields a murine model of hyper-IgD syndrome. J. Inherit. Metab. Dis. 30: 888–895. [DOI] [PubMed] [Google Scholar]
- 41.Rubina K., Talovskaya E., Cherenkov V., Ivanov D., Stambolsky D., Storozhevykh T., Pinelis V., Shevelev A., Parfyonova Y., Resink T., et al. 2005. LDL induces intracellular signalling and cell migration via atypical LDL-binding protein T-cadherin. Mol. Cell. Biochem. 273: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J. C., Weissglas-Volkov D., Kyttala M., Dastani Z., Cantor R. M., Sobel E. M., Plaisier C. L., Engert J. C., van Greevenbroek M. M., Kane J. P., et al. 2008. WW-domain-containing oxidoreductase is associated with low plasma HDL-C levels. Am. J. Hum. Genet. 83: 180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schadt E. E., Molony C., Chudin E., Hao K., Yang X., Lum P. Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. 2008. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6: e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fogarty M. P., Xiao R., Prokunina-Olsson L., Scott L. J., Mohlke K. L. 2010. Allelic expression imbalance at high-density lipoprotein cholesterol locus MMAB-MVK. Hum. Mol. Genet. 19: 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goring H. H., Curran J. E., Johnson M. P., Dyer T. D., Charlesworth J., Cole S. A., Jowett J. B., Abraham L. J., Rainwater D. L., Comuzzie A. G., et al. 2007. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 39: 1208–1216. [DOI] [PubMed] [Google Scholar]
- 46.Dixon A. L., Liang L., Moffatt M. F., Chen W., Heath S., Wong K. C., Taylor J., Burnett E., Gut I., Farrall M., et al. 2007. A genome-wide association study of global gene expression. Nat. Genet. 39: 1202–1207. [DOI] [PubMed] [Google Scholar]
- 47.Stranger B. E., Nica A. C., Forrest M. S., Dimas A., Bird C. P., Beazley C., Ingle C. E., Dunning M., Flicek P., Koller D., et al. 2007. Population genomics of human gene expression. Nat. Genet. 39: 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su Z., Leduc M. S., Korstanje R., Paigen B. 2010. Untangling HDL quantitative trait loci on mouse chromosome 5 and identifying Scarb1 and Acads as the underlying genes. J. Lipid Res. 51: 2706–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edmondson A. C., Brown R. J., Kathiresan S., Cupples L. A., Demissie S., Manning A. K., Jensen M. K., Rimm E. B., Wang J., Rodrigues A., et al. 2009. Loss-of-function variants in endothelial lipase are a cause of elevated HDL cholesterol in humans. J. Clin. Invest. 119: 1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cox L. A., Birnbaum S., Mahaney M. C., Rainwater D. L., Williams J. T., VandeBerg J. L. 2007. Identification of promoter variants in baboon endothelial lipase that regulate high-density lipoprotein cholesterol levels. Circulation. 116: 1185–1195. [DOI] [PubMed] [Google Scholar]
- 51.Cox A., Sheehan S. M., Kloting I., Paigen B., Korstanje R. 2010. Combining QTL data for HDL cholesterol levels from two different species leads to smaller confidence intervals. Heredity 105: 426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.