Abstract
Renal transplant recipients are predisposed to urinary tract infections caused by both common uropathogens and opportunistic bacteria resulting frequently in significant polymicrobial infections. In this study, a culture-independent 16S rRNA-based approach was established to identify unusual, fastidious, or anaerobic bacteria and to investigate bacterial diversity in urinary tract specimens. Similarly sized amplicons encompassing the V6 to V8 region of the 16S rRNA were analyzed with denaturing high-performance liquid chromatography (DHPLC) (WAVE System). Artificial mixtures of single amplicons from commonly encountered uropathogenic bacteria produced distinct peak profiles whose identities were confirmed by sequencing individually collected peak products. We evaluated the application of the method on 109 urinary tract specimens from renal transplant recipients; 100% correlation was found for culture-positive specimens, and DHPLC generated peak profiles. However, for culture-negative specimens, DHPLC facilitated the detection of novel peak profiles. DNA sequencing of these individual peaks was used to identify the bacteria involved. Thus, in PCR-positive but culture-negative samples the method allowed detection of previously known uropathogens such as Corynebacterium urealyticum and Gardnerella vaginalis, but also unusual agents including Anaerococcus lactolyticus, Bacteroides vulgatus, Dialister invisus, Fusobacterium nucleatum, Lactobacillus iners, Leptotrichia amnionii, Prevotella buccalis, Prevotella ruminicola, Rahnella aquatilis, and Streptococcus intermedius were detected as single pathogens or as constituents of polymicrobial infections. The method described is reproducible and rapidly and enables both DHPLC-based profiling and sequence-based investigation of microbial communities and polymicrobial infections. A detailed understanding of infections found in recipients of renal transplants will guide antibiotic therapy regimens and provide new perspectives for decreasing the risk of graft rejection.
Currently, one of the major problems for successful kidney transplantation is the reaction of the immune system of the recipient against the donor organ, which in the case of unsuccessful immunosuppressive treatment can result in the loss of the transplant. In order to prevent or treat such rejection episodes and to maintain graft function the application of immunosuppressive agents is standard practice. The incidence of bacterial infection in renal transplant recipients is directly related to the net immunosuppressive effect achieved and the duration of time over which this therapy is administered. Bacterial urinary tract infections (UTIs) are frequently associated with the early onset of chronic rejection and may also lead to reduced transplant survival (15, 18). Studies have shown that in 40 to 60% of transplant recipients the urinary tract is the source of septicemia and that in patients with urosepsis the recurrence rate was approximately 40% (1, 13). The bacterial species causing UTIs in renal transplant recipients are reported to be similar to those causing UTIs in the general population (15). Because of the treatment with immunosuppressive drugs the patients not only suffer from common uropathogens but are also prone to opportunistic infections with unusual uropathogens (15).
The most frequently isolated causative agents of UTIs are members of the family Enterobacteriaceae, in particular Escherichia coli, and gram-positive cocci, such as Enterococcus spp. and Staphylococcus spp (20). Conventional microbial urine diagnostics employ a standard combination of nonselective and selective agar plates for enumeration and identification of bacteria, respectively. Agar plates are usually incubated at 37°C under aerobic conditions for 18 to 24 h. In the case of positive leukocyte esterase or nitrite tests, which often correlate with uropathogenic infections, a prolonged incubation period up to 48 or 72 h and a Giemsa or Gram stain are recommended (3).
Phenotypic characteristics can not be used for the identification of noncultivable organisms or for organisms with unusual biochemical profiles. An identification based on phenotypic characteristics may further be complicated in the case of fastidious organisms, whose isolation requires specific nutrients and specific temperatures or atmospheric conditions, including anaerobic conditions for the isolation of obligate anaerobes. The last decade has shown considerable development in the diagnostic application of molecular techniques, especially those based on the 16S rRNA encoding genomic region, to study microbial diversity in ecosystems and the human microbial flora (2, 5). However, as the rRNA approach uses broad-range primers, the amplification of complex bacterial communities yields mixed amplicons of the same size, which hamper sequence-based identification of the constituent species (5, 21). Methods such as denaturing gradient gel electrophoresis (DGGE) (4) or temperature gradient gel electrophoresis (TGGE) (14) have therefore been developed to analyze complex microbial communities, as they facilitate sequence-specific separation of 16S rRNA amplicons. Denaturing high-performance liquid chromatography (DHPLC) is a new technology that has found extensive use in the discovery of genetic variations in diploid genomes. The principle underlying scanning for mutations by DHPLC can clearly also be applied to the detection of sequence variations in genes such as rRNA that are highly conserved in bacteria.
A significant proportion of urinary tract infections are caused by polymicrobial agents, comprising sometimes as many as four to six species, each at ≥105 CFU/ml (20). The aim of this study was to use a culture-independent, molecular approach to examine the bacterial diversity of urinary tract specimens from renal transplant recipients. We used the DHPLC-based approach involving the WAVE System to achieve rapid, reliable and reproducible analysis of individual or mixed amplicons deriving from bacteria in clinical samples. In particular, we aimed at generating distinct peak profiles which could be used for the identification of infectious agents by sequence analysis and the establishment of a routine approach facilitating the later identification of pathogens simply on the basis of the DHPLC profile obtained. We analyzed mixed amplicons derived from the V6 to V8 region of the 16S rRNA (11, 22) with an empirically determined temperature gradient to separate mixed amplicons of polymicrobial infections for sequence-based identification. The analytical gradient “mixed species” was evaluated with artificially mixed PCR products of single species and used to identify polymicrobial UTIs and infections with fastidious or anaerobic uropathogenic bacteria.
MATERIALS AND METHODS
Enumeration and identification of bacteria in urinary tract specimens by culture.
In order to enumerate bacteria in urinary tract specimens, 0.01 and 0.001 ml from midstream collections were streaked on CLED agar plates (Merck, Darmstadt, Germany) with calibrated plastic loops. CLED agar plates are commonly used for the isolation and enumeration of microorganisms from urine. For identification, bacteria were selectively cultivated on MacConkey and on 5% sheep blood agar plates and Candida spp. on Sabouraud agar (Merck, Darmstadt, Germany). Agar plates were incubated at 37°C for 24 h under aerobic conditions. In the case of no bacterial growth but a positive leukocyte esterase test for the urinary tract specimen (Combur test, Roche, Mannheim, Germany), incubation was continued for 48 h. The isolated bacteria were identified because of their biochemical characteristics or by sequencing of the 16S rRNA and database analysis (10; National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov). Bacteriuria was defined as 5 × 104 to ≥1 × 105 CFU/ml for midstream urine.
The presence of anti-infectiva in urinary tract specimens was routinely examined with the disk diffusion method with Bacillus stearothermophilus spores in the agar plates (10).
Microscopic examination of urinary tract specimens.
For microscopic analysis centrifuged urinary tract specimens were stained with Giemsa (Merck, Darmstadt, Germany). Briefly, 50 μl of the sample was air dried on a slide and fixed with methanol for 10 min. After rinsing the slide with water the sample was stained with Giemsa solution (75 ml of water and 25 ml of Giemsa) for 10 min and again rinsed with water. The samples were examined microscopically.
DNA extraction from pure bacterial colonies and urinary tract specimens.
Chromosomal DNA of uropathogenic bacteria from plate-cultured urinary tract specimens was isolated with an alkaline extraction method. From 2 × 109 to 3 × 109 bacteria were carefully resuspended in 50 μl of sterile water and 50 μl of 0.1 N NaOH was added. The solution was thoroughly mixed and boiled at 95°C for 15 min. After chilling on ice the solution was neutralized by adding 8 μl of Tris-HCl, pH 7.0. Undisrupted bacteria and cell debris were pelleted by centrifugation (15,000 × g, 2 min) and the clear supernatant was transferred into a new reaction tube.
Bacterial DNA of urinary tract specimens was isolated with the Qiagen Kit (Qiagen, Hilden, Germany) as recommended by the vendor. Briefly, a urine sample was carefully mixed and then 500 μl was centrifuged at 7,500 × g for 10 min. The pellet was treated with proteinase K, boiled, and the DNA was purified and extracted on DNA-binding columns with AL, AW, and AE buffers (Qiagen). The final volume of the bacterial DNA extract was 100 μl. Five microliters of DNA extract from both the bacterial colonies and urinary tract specimens was used for PCR amplification.
Amplification of the 16S rRNA.
For DHPLC/WAVE System-based analysis, 5 μl of template containing 50 to 100 ng of extracted chromosomal DNA and 95 μl of a master mix containing 20 pmol of the forward primer 0933F and the reverse primer 1407R (Table 1), 30 mM KCl, 10 mM Tris-HCl, pH 8.3, 2.5 μg bovine serum albumin, 1.5 mM MgCl2, 0.4 mM deoxynucleoside triphosphates, and 1.5 units of AmpliTaq DNA polymerase were mixed for the partial amplification of the 16S rRNA and downstream WAVE System analysis.
TABLE 1.
Primers and their potential modifications for amplification of 16S rRNA
| 5′-3′ sequence | Primera | Reference |
|---|---|---|
| GCA CAA GCG GTG GAG CAT GTG G | 0933F | This study |
| GAC GGG CGG TGT GTA CAA G | 1407R | This study |
| CAG GCC TAA CAC ATG CAA GTC | 0063F | 9 |
| GGG CGG WGT GTA CAA GGC | 1387R | 9 |
| GCCCCCGCCG | GC(10F) | This study |
| CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG | GC(40F) | 16 |
F, forward primer; R, reverse primer. 10F and 40F, GC clamp of 10 or 40 bp, respectively, as attachment at the 5′ end of forward primer 0933F.
For sequence-based identification of plate-cultured bacteria with GenBank (National Center for Bio/Technology Information, http://www.ncbi.nlm.nih.gov), the 16S rRNA genes of the isolates were amplified with primers 0063F and 1387R (Table 1). Thermocycling was performed in a PCR-Express cycler with tube control (Hybaid Limited, Ashford, United Kingdom) with initial denaturation at 94°C for 7 min, followed by 25 cycles of denaturation at 94°C for 40 s, annealing at 56°C for 40 s, and extension at 72°C for 1 min, with a final extension step at 72°C for 5 min. The amplification products were analyzed by gel electrophoresis with a horizontal 1.5% agarose gel for 1 h at 150 V at room temperature in TBE running buffer. The gel was stained with SYBR-Gold (Molecular Probes, Goettingen, Germany) and recorded under UV light with the Image Master VDS System (Amersham Biosciences, Freiburg, Germany).
HPLC gradients for the separation of mixed amplicons with the WAVE 3500 DNA fragment analysis system.
The amplicons of single and mixed bacterial species were analyzed on the WAVE System (Transgenomic, Omaha, Neb.). We employed the DNASep HT cartridge which uses alkylated nonporous polystyrene-divinylbenzene copolymer microspheres for high-performance nucleic acid separations. For the particular analysis and separation of mixed bacterial species we generated the gradient “mixed species” and used it at an oven temperature of 62.0°C (Table 2). The gradient was formed by buffer A, consisting of 0.1 M triethylammonium acetate (TEAA), pH 7.0, and buffer B, consisting of 0.1 M TEAA and 25% acetonitrile, pH 7.0. Buffer C, consisting of 25% water and 75% acetonitrile, was used for washing the column. The buffers used were obtained from Transgenomic Inc. at analytical grade. The analysis was accomplished with Wavemaker software version 4.1.44.
TABLE 2.
Analytical gradient mixed species for the separation of mixed 16S rRNA amplicons C (0933F/1407R) at 62.0°C with the WAVE 3500 DNA fragment analysis systema
| Step | Time (mins) | Buffer A (%) | Buffer B (%) | Buffer C (%) |
|---|---|---|---|---|
| Loading | 0.0 | 46 | 54 | 0 |
| Step 1 (5.6 min) | 0.5 | 40 | 60 | 0 |
| Step 2 (5.6 min) | 2.5 | 39 | 61 | 0 |
| Step 3 (5.6 min) | 4.5 | 38 | 62 | 0 |
| Step 4 (5.6 min) | 8.5 | 37 | 63 | 0 |
| Step 5 (5.6 min) | 10.5 | 36 | 64 | 0 |
| Start clean | 10.6 | 46 | 54 | 0 |
| Stop clean | 10.7 | 46 | 54 | 0 |
| Start equilibrate | 10.8 | 46 | 54 | 0 |
| Stop equilibrate | 11.6 | 46 | 54 | 0 |
The flow rate was 0.35 mH/min.
Reamplification and determination of the DNA sequence of separated and collected amplicons.
The WAVE System was provided with the fragment collector FCW 200 which enabled fully automated collection of the peak samples of interest for reamplification and sequencing. Two hundred microliters of each peak sample was collected with a 96-well microtiter plate, and the solution was rapidly dried with a Savant Speedvac Plus SC210A (ThermoQuest, Egelsbach, Germany). The pellet was carefully resuspended in 5 μl of water at 37°C in a heating block for 1 h. In order to reamplify the collected amplicons, 2.5 μl was mixed with 47.5 μl of the same master mix and AmpliTaq DNA polymerase and amplified according to the scheme described above. The sequencing reactions were accomplished with primers 0933F and 1407R and the DYEnamic ET dye terminator kit (Amersham Biosciences, Freiburg, Germany), and the nucleic acid sequences were determined on the DNA sequencing system MegaBACE 1000 (Molecular Dynamics-Amersham Biosciences, Uppsala, Sweden) as recommended by the vendor.
Calculation of the CFU of bacteria in urinary tract specimens with a standard curve.
In order to calculate the CFU of bacteria in urinary tract specimens that were culture negative but PCR positive, we generated a standard curve. Pseudomonas aeruginosa was grown in brain heart infusion (BHI) broth at 37°C to an optical density at 600 nm of ≈1.0. Tenfold serial dilutions of the culture from 10−1 to 10−6 were prepared, plated on BHI agar plates, and the CFU were determined; 500 μl of each diluted culture was centrifuged at 7,500 × g for 10 min, the bacterial DNA was isolated with the Qiagen kit, and the 16S rRNA was amplified as described above. Then 20 μl of each sample was analyzed on the WAVE System, and the corresponding values of the absorbance were used to generate a regression line.
Computational analysis.
DNA sequences of the 16S rRNA were aligned with the Clustal method from MegAlign (DNAStar Inc., Madison, Wis.). The obtained nucleic acid sequences were analyzed with the algorithm Blastn at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and the GOLD genomes online database (http://wit.integratedgenomics.com/GOLD/) from Integrated Genomics.
Nucleotide sequence accession numbers.
The sequences used for compilation were AF016390, AB043971, L38654, AB065370, AB016246, BAU42284, AF091367, BHU42292, BPU91839, BSU91838, AE001347, AB045282, CJU87815, AB036835, KPU33121, LFU62624, AL592022, AL591824, AF023664, Z83862, U02968, AE000008, AE002551, AF037105, AF037106, NCU92799, PSEIAM12, PSEIAM19, PSU22427, SEU90318, SPU88546, STU88545, AP003359, AF004220, AE014210, AF003929, AF003932, AF003933, AF003930, NC_003485, AF003928, AF009475, AE000520, AF222894, AE003852, AB013297, NC_004088, NC_000907, AE004439, and AE008922.
RESULTS
Primer selection and broad-range amplification of the 16S rRNA.
We targeted the V6 to V8 region of the 16S rRNA for our study (11), which has been used previously for TGGE analysis of bacteria from complex microbial communities (12, 22). We reanalyzed these regions by aligning additional sequences from gram-negative and gram-positive bacteria pathogenic for humans, animals, or plants that were not included in the previous compilation (11). Our analysis revealed that the primer pair 0933F/1407R, matching sequences flanking the V6 to V8 region, showed a very high degree of conservation (data not shown). We used this primer pair to amplify the ≈470-bp V6 to V8 region of the 16S rRNA from several pure bacterial cultures and from urinary tract specimens (Fig. 1A and 1B). All of the selected uropathogenic bacteria showed positive PCRs, and in every case the amount of amplified DNA was at least 1 μg/μl.
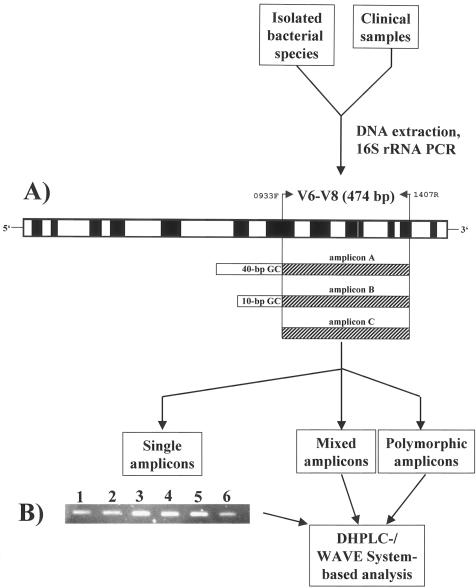
FIG. 1.
Flow chart for DHPLC/WAVE System-based analysis of the 16S rRNA from isolated bacteria and clinical samples. (A) Schematic representation of the compiled 16S rRNA from the 5′ to the 3′ end. Hypervariable regions are depicted in white boxes, and highly conserved regions in black (11; this study). Primers 0933F and 1407R encompass the V6 to V8 region (474 bp). Amplicon A harbors a 40-bp GC clamp at the 5′ end and amplicon B a 10-bp GC clamp. Amplicon C is devoid of a GC clamp. F, forward; R, reverse; V, variable. (B) SYBR-Gold-stained agarose gel electrophoresis of amplicon C from the 16S rRNA of selected uropathogens. Lane 1, E. faecalis; lane 2, P. vulgaris; lane 3, E. coli; lane 4, K. pneumoniae; lane 5, P. aeruginosa; lane 6, S. saprophyticus.
Comparison of culture-dependent and -independent detection of bacteria in urinary tract specimens.
We examined 109 urinary tract specimens from renal transplant recipients by culture and by 16S rRNA PCR (Fig. 1A). Thirty-two (29.4%) showed bacterial and four (3.7%) showed fungal growth, and 73 (66.9%) revealed no growth on agar plates. Urinary tract specimens which were culture negative but tested leukocyte esterase positive, an indicator of possible bacterial infection, were incubated for an additional 48 h. However, in no case was any bacterial growth detected. Urinary tract specimens that contained only fungi (Candida species) but were both PCR negative and culture negative for bacteria were not examined further. Each of the 32 culture-positive urinary tract specimens also exhibited a positive PCR signal. Inhibition of the PCR by inhibitors possibly present in urinary tract specimens was not observed. Moreover, we identified 10 samples out of the 109 which showed no bacterial growth on agar plates but were nevertheless PCR positive (9.2%), indicating that primer pair 0933F/1407R was capable of detecting both cultivable and noncultivable bacteria. Two of these samples also contained Candida species (Table 3).
TABLE 3.
Culture-dependent and -independent detection of prokaryotes and Candida species in 109 urinary tract specimens
| Object | Growth on agar platesa | 16S rRNA PCR |
|---|---|---|
| Bacteria | 32 | 40 |
| Candida spp. | 2 | NDb |
| Bacteria/Candida spp. | 0/2 | 2/ND |
CLED, MacConkey, 5% sheep blood, and Sabouraud agar plates.
ND, not determined.
The biochemical identification and microsequencing of the amplified 16S rRNAs with primers 0063F and 1387R of cultivated bacteria revealed 18 Enterococcus faecalis/faecium, nine Proteus mirabilis/vulgaris, eight E. coli, five Klebsiella oxytoca/pneumoniae, two Staphylococcus epidermidis/saprophyticus, and one each of Achromobacter xylosoxidans, Enterobacter cloacae, P. aeruginosa, Serratia liquefaciens, Streptococcus agalactiae, and S. mitis in significant amounts (5 × 104 to ≥105 bacteria per ml). The most prominent bacteria were E. faecalis, followed by Enterobacteriaceae such as Proteus spp., E. coli, and Klebsiella spp. The remaining species were individual cases. Additionally, we observed urinary tract infections caused not only by a single species but also by a mixture of bacteria of two and even three different bacterial genera. From the 32 culture-positive samples, 20 (65.6%) urinary tract infections were caused by only one, seven (21.9%) by two, and four (12.5%) by three etiologic agents.
DHPLC analysis performed with the WAVE System generates reproducible and reliable distinct peak profiles of single species.
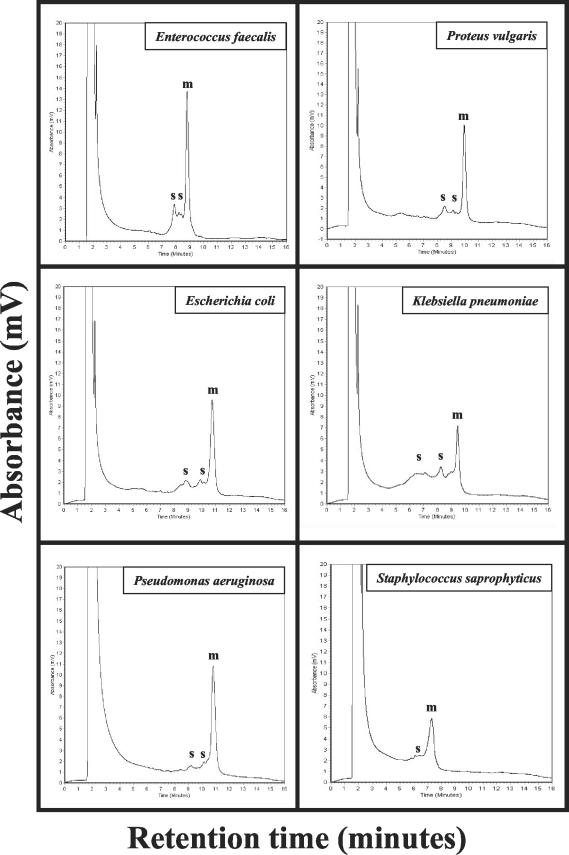
The V6 to V8 region of the 16S rRNA chosen for amplification and identification comprises of a region of 54% highly conserved and 46% hypervariable nucleic acid sequences (Fig. 1A). In order to separate mixed amplicons with respect to their nucleotide composition, we used the WAVE System. The 16S rRNA amplicons that were derived from pure cultures of uropathogenic bacteria E. faecalis, E. faecium, P. vulgaris, E. coli, K. pneumoniae, P. aeruginosa, S. liquefaciens, S. agalactiae, S. mitis, S. aureus, and S. saprophyticus were independently analyzed in triplicate on the WAVE System with the gradient “mixed species” at 62.0°C (Fig. 2 and Tables 2 and 4). Almost all of the examined species exhibited distinct peak profiles consisting of one major peak and in some cases of one to two small additional peaks with characteristic retention times. Each peak was represented by a particular retention time that showed reliable and reproducible mean values and significant small standard deviations. Two species, P. vulgaris and E. faecium, showed identical retention times (Table 4). The height of each particular peak was specified as the absorbance in mV and depended on the volume and the DNA concentration injected. We used a total volume of 10 μl for the analysis of the single species at a DNA concentration of 200 ng/μl ± 20 ng/μl.
FIG. 2.
Distinct peak profiles of selected uropathogens. Amplicon C (0933F/1407R) was analyzed with the analytical gradient mixed species at 62.0°C. Ten microliters of each PCR-product was used for injection. The retention times are the time points when the DNA fragments eluted from the column of the WAVE System and are indicated in minutes. The amount of DNA eluted is indicated as the absorbance in millivolts. The experiments were done in triplicate. The major and the smaller peaks are indicated by m and s, respectively. The duration of one analysis was 16 min.
TABLE 4.
Retention times of 16S rRNA amplicons C from selected uropathogenic bacteria in analytical gradient mixed species at 62.0°C
| Species | Retention time (mV) ± SDa
|
||
|---|---|---|---|
| Peak 1b | Peak 2b | Peak 3c | |
| Enterococcus faecalis | 8.20 ± 0.29 | 8.63 ± 0.31 | 9.27 ± 0.32 |
| Enterococcus faecium | 8.44 ± 0.07 | 9.00 ± 0.12 | 9.80 ± 0.19 |
| Proteus vulgaris | 8.49 ± 0.02 | 9.10 ± 0.04 | 9.86 ± 0.06 |
| Escherichia coli | 8.75 ± 0.04 | 9.72 ± 0.11 | 10.55 ± 0.16 |
| Klebsiella pneumoniae | 6.38 ± 0.02 | 8.19 ± 0.07 | 9.32 ± 0.18 |
| Pseudomonas aeruginosa | 9.36 ± 0.11 | 10.39 ± 0.16 | 11.09 ± 0.20 |
| Serratia liquefaciens | 7.67 ± 0.23 | 8.18 ± 0.05 | 9.21 ± 0.16 |
| Streptococcus agalactiae | 9.50 ± 0.12 | 10.70 ± 0.13 | 11.57 ± 0.13 |
| Enterobacter aerogenes | 7.51 ± 0.06 | 8.78 ± 0.11 | 10.82 ± 0.04 |
| Streptococcus mitis | 7.82 ± 0.05 | 9.46 ± 0.35 | 9.94 ± 0.36 |
| Staphylococcus aureus | 6.03 ± 0.14 | 6.70 ± 0.18 | 7.51 ± 0.12 |
| Staphylococcus saprophyticus | -d | 6.02 ± 0.25 | 7.12 ± 0.31 |
Mean value of three independent experiments ± standard deviation.
Small peak (s).
Major peak (m).
Peak absent.
Separation and sequence-based identification of artificially mixed PCR products of uropathogenic bacteria.
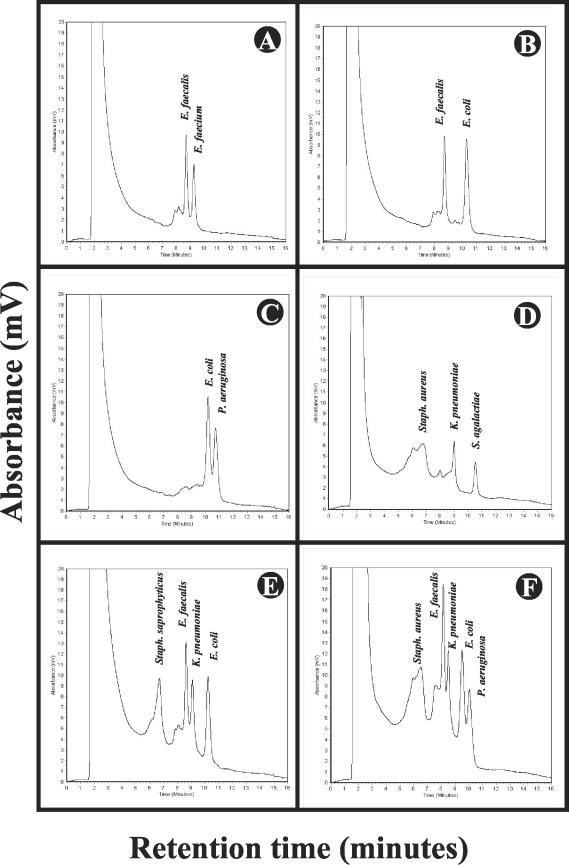
Our analysis showed that the major peaks of the examined species were eluted from the column with highly reproducible retention times (Fig. 2, Table 4). This permitted the separation of a mixture of amplicons or PCR products obtained from a DNA extraction of mixed species. To address this further, we artificially mixed equimolar amounts of PCR products from single species and analyzed the mixture on the WAVE System with the analytical gradient “mixed species” at 62.0°C. The gradient was capable of separating two species (Fig. 3A to C), three species (Fig. 3D), four species (Fig. 3E), and even five species (Fig. 3F). Even highly related species such as E. faecalis and E. faecium (Fig. 3A) or unrelated species such as E. coli and P. aeruginosa (Fig. 3C and 3F) and E. faecalis and K. pneumoniae (Fig. 3E and 3F) with close retention times (Table 4) were unambiguously separated. The identities of the species correlated with their characteristic retention times and were confirmed by identification based on sequence analysis (Fig. 3). An artificial mixture of P. vulgaris and E. faecium which showed identical retention times (Table 4) could not be separated (data not shown).
FIG.3.
Analysis of artificially mixed PCR products of selected uropathogens. Several equimolar mixtures of amplicon C (0933F/1407R) from selected uropathogens were analyzed with the analytical gradient mixed species at 62.0°C. Twenty microliters of each mixture was used for injection. The retention times are the time points when the DNA fragments are eluted from the column of the WAVE System and are indicated in minutes. The amount of DNA eluted is indicated as the absorbance (in millivolts). The experiments were done in triplicate. (A) mixture of the species E. faecalis and E. faecium; (B) E. faecalis and E. coli; (C) E. coli and P. aeruginosa; (D) mixture of the three species S. saprophyticus, K. pneumoniae, and S. agalactiae; (E) mixture of the four species S. saprophyticus, E. faecalis, K. pneumoniae, and E. coli. Panel F: mixture of the five species S. aureus, E. faecalis, K. pneumoniae, E. coli, and P. aeruginosa. The duration of one analysis was 16 min.
Culture-independent, sequence-based identification of bacteria from urinary tract specimens of renal transplant recipients.
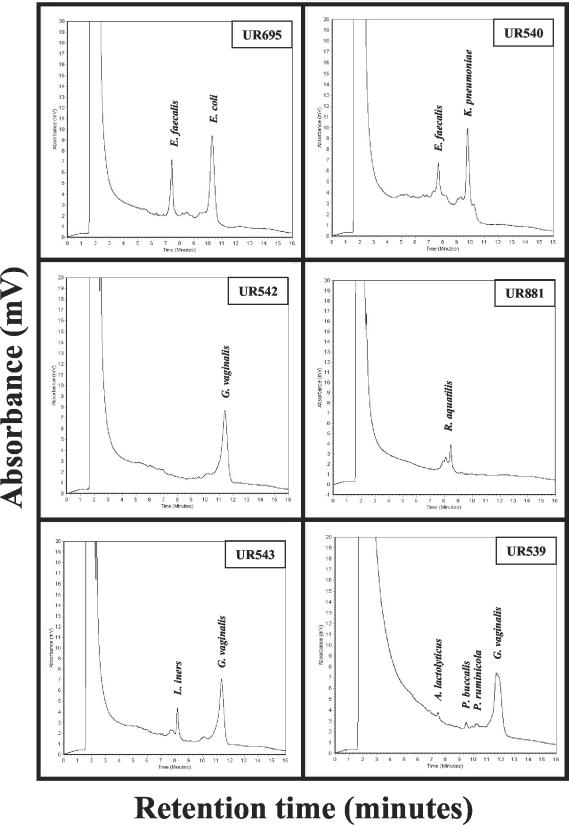
The analysis of 109 urinary tract specimens from renal transplant recipients revealed 42 PCR-positive samples, but only 32 samples showed visible bacterial growth on agar plates (Table 3). The PCR products of the 32 culture-positive samples were analyzed on the WAVE System and were subjected to identification by sequence analysis. Computational analysis of the DNA sequences with the GenBank environment at the NCBI (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov) confirmed the biochemical identification. Two representative examples of the analyses are shown in Fig. 4. Both the biochemical and the nucleotide sequence analysis of the urinary tract specimen UR695 and UR540 identified E. faecalis/E. coli and E. faecalis/K. pneumoniae, respectively (Fig. 4).
FIG. 4.
Culture-independent analysis of urinary tract specimen and comparison with the culture on agar plates. Total DNA of urinary tract specimens was extracted, amplicon C (0933F/1407R) was generated and analyzed with the analytical gradient mixed species at 62.0°C. Twenty microliters of each PCR product was used for injection. The retention times are the time points when the DNA fragments are eluted from the column of the WAVE System and are indicated in minutes. The amount of DNA eluted is indicated as the absorbance (in millivolts). The .experiments were done in triplicate. UR659: E. faecalis and E. coli were detected by both culture and PCR. UR540: E. faecalis and K. pneumoniae were detected by both culture and PCR. UR542: G. vaginalis was detected only by PCR. Coinfection with C. albicans was detected on Sabouraud agar. UR881: R. aquatilis was detected only by PCR. Coinfection with C. albicans was detected on Sabouraud agar. UR543: L. iners and G. vaginalis were detected only by PCR. UR539: A. lactolyticus, P. buccalis, P. ruminicola, and G. vaginalis were detected only by PCR. The duration of one analysis was 16 min.
The culture-independent examination of the 10 culture-negative but PCR-positive specimens (9.2%), suggested that these samples contained either noncultivated bacteria, dead bacteria or residual bacterial DNA. Two of these samples, UR542 and UR881, showed a coinfection with Candida species (Fig. 4, Tables 3 and 5). Sequence analysis of the PCR products of urinary tract specimens UR539 (Fig. 4), UR542 (Fig. 4), UR543 (Fig. 4), UR555, UR656, UR711, UR768, UR772, UR794, and UR881 (Fig. 4) revealed the bacteria A. lactolyticus, B. vulgatus, C. urealyticum, D. invisus, F. nucleatum, G. vaginalis, L. iners, L. amnionii, P. buccalis, P. ruminicola, S. intermedius, and R. aquatilis (Table 5). One urinary tract specimen (UR543) contained two bacteria, G. vaginalis and L. iners, and a second sample (UR539) four bacteria, A. lactolyticus, P. buccalis, P. ruminicola, and G. vaginalis (Fig. 4; Table 5).
TABLE 5.
Identification of bacteria in urinary tract specimens that did not grow on agar plates under standard conditions
| Sample | Gender | Drugsa | Leukocytesb | Species |
|---|---|---|---|---|
| UR539 | F | No | Positive | Anaerococcus lactolyticus, Prevotella buccalis, Prevotella ruminicola, Gardnerella vaginalis |
| UR542 | F | No | Positive | Gardnerella vaginalis, Candida albicans (coinfection) |
| UR543 | F | No | Positive | Lactobacillus iners, Gardnerella vaginalis |
| UR555 | F | No | Positive | Streptococcus intermedius, Fusobacterium nucleatum |
| UR656 | F | No | Positive | Leptotrichia amnionii |
| UR711 | M | No | Negative | Corynebacterium urealyticum |
| UR768 | M | No | Negative | Bacteroides vulgatus, Dialister invisus, Prevotella buccalis |
| UR772 | F | No | Negative | Prevotella buccalis |
| UR794 | M | No | Positive | Corynebacterium urealyticum |
| UR881 | F | No | Positive | Rahnella aquatilis, Candida albicans (coinfection) |
Treatment with anti-infectiva as determined with the disk diffusion test and from clinical records.
Leukocyte esterase was detected with the Combur test (Roche, Mannheim, Germany).
DISCUSSION
In this study we established a culture-independent, molecular approach to identify polymicrobial infections and unusual bacteria from the genitourinary tract of renal transplant recipients without fastidious cultivation. Although more than 90% of UTIs in the general population are caused by a single bacterial species, true polymicrobial infections are very likely in patients with underlying structural abnormalities, chronic indwelling catheters, or prolonged immunosuppressive therapy. Approximately 95% of such patients have infections with two to four species at ≥105 CFU/ml (3).
The 16S rRNA approach has been primarily used to identify unknown, fastidious or slow-growing bacteria (5). In the case of polymicrobial diseases, mixed PCR-products of essentially the same size are expected. Denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) are techniques that make use of the sequence variations in the hypervariable regions to distinguish the amplicons (4, 14). However, DGGE and TGGE are labor-intensive techniques that are time-consuming and require dedicated efforts to maintain reproducibility. Therefore, these techniques are inappropriate to be used routinely in a diagnostic laboratory (6, 22).
A recent study described the use of the WAVE System for the identification of bacteria (7). Although the unique peak profiles of several bacterial species could be used for their identification, the discriminatory power of the profiles to distinguish mixed populations was not sufficient in the approach reported. In our preliminary studies, with newly designed oligonucleotide primers (0933F/1407R) to amplify the V6 to V8 region (11) of the 16S rRNA (Fig. 1) we obtained reliable amplification results with a large number and variety of clinical isolates (data not shown). To establish a cost-effective, quick and reliable method for the analysis of mixed populations we empirically examined the use of an analytical gradient, with partially denaturing temperatures, in combination with a 40-bp and a 10-bp GC-clamp in amplification and subsequent analysis.
For TGGE analysis the attachment of a 40-bp G/C-rich sequence (GC-clamp) to the 5′-end of the forward primer has been recommended to improve detection of base-pair changes (16, 22). The analysis of the sequences of the V6 to V8 region with Wavemaker software advised the attachment of a GC clamp to the forward primer 0933F, but in a truncated form comprising only a 10-bp GC-rich sequence (Table 1). Therefore, we generated three different PCR-products of the 16S rRNA for DHPLC/WAVE System-based analysis which harbored either a 40-bp GC clamp (amplicon A) or a 10-bp GC clamp (amplicon B) at their 5′ ends, or which were devoid of a GC clamp (amplicon C) (Fig. 1 and Table 1) and examined several different gradients at different temperatures. We investigated the influence of 56.0°C, 58.0°C, 60.0°C, 61.0°C, 62.0°C, and 63.0°C and a narrower range from 61.2°C up to 62.8°C in steps of 0.2°C. In order to ascertain the optimal gradient for the separation of heterologous PCR-products of the same size, we artificially mixed equimolar amounts of the amplicons from different species and analyzed the mixtures as well as the single PCR products. Amplicon C (0933F/1407R) most consistently showed distinct peak profiles at characteristic retention times for all species examined when the analytical gradient “mixed species” at 62.0°C was employed (Fig. 1 and 2, Table 4). The artificial mixtures were unambiguously separated, and each species-specific peak could be confirmed by sequencing. With this method we were able to separate up to five species in one sample (Fig. 3). No discrepancy was observed when the experiments were repeated with PCR products generated from DNA extracts of mixed species. The other amplicons and gradients examined showed poorer resolution and were not suitable for the analysis of polymicrobial infections.
Albeit the mean values of the retention times generated were reliable and reproducible with significantly small standard deviations (Fig. 3), we found species with very close retention times, such as E. coli/P. aeruginosa and E. faecalis/K. pneumoniae, and species with essentially the same retention time, such as E. faecium/P. vulgaris (Table 4). Despite their close retention times, E. coli/P. aeruginosa and E. faecalis/K. pneumoniae were unambiguously separated (Fig. 3C and 3F). Interestingly, mixed amplicon populations exhibited slightly accelerated elution of the respective peaks, in the range of 0.5 to 1.0 min, compared to the single amplicon profiles (Table 4, Fig. 3). We have currently no reasonable explanation for this observation. The inability of DHPLC to distinguish between amplicons generated by bacteria such as E. faecium and P. vulgaris with identical retention times (Table 4) is not a limitation of DHPLC per se and could probably be overcome by the use of different buffer compositions, temperature gradients, column chemistries, etc. The use of additional targets suitable for phylogenetic differentiation such as the rpoB gene, encoding a subunit of the RNA polymerase also offers an alternative solution to the problem.
Although not observed in this study, possible column variations should be addressed appropriately in view of previously reported results (7). We used two new columns during this study, and the specificity of each species' retention time examined between the two columns was 100% (Fig. 3, Table 4). However, to ensure the reproducibility of peak profiles, we recommend an equilibration step with artificially mixed amplicons to calibrate the column and the WAVE System.
Although the analysis of the amplicons C on agarose gels showed only the product of the expected size (Fig. 1), DHPLC analysis revealed small additional peaks with shorter retention times than the major peak (Fig. 2, Table 4). These additional peaks were collected and subjected to sequence analysis. This revealed that the DNA of the collected peaks belonged to the species examined, but contained sequence polymorphisms. Repetition of the experiments showed that the variations were significant and were not the result of spurious nucleotide incorporation by the AmpliTaq DNA polymerase used (data not shown). A comparison of the sequences obtained to known 16S rRNAs of a single species represented by more than one allele (Fig. 2) in public databases (Integrated Genomics, NCBI) indicated that they indeed correspond to individual 16S rRNA alleles of a single bacterial species. Hence, the analytical gradient “mixed species” could be employed for both the generation of distinct peak profiles with characteristic retention times and for the analysis of the sequence heterogeneities in 16S rRNA alleles of a single species.
The comparison of the culture-dependent and -independent analysis of urinary tract specimens revealed 10 PCR-positive samples that exhibited no bacterial growth on agar plates (Fig. 4, Table 5). One explanation for this observation was that the bacteria were dead due to the treatment of the patients with anti-infectiva. However, clinical records showed that these patients were not treated with antibiotics or with antimycotics during the active urine sampling period. Alternative explanations include that residual DNA from dead and lysed bacteria were attached to the sedimented cells in urinary tract specimen or that the bacteria could not be cultivated under the culture conditions employed. Since microscopic analysis of these specimens exhibited intact typical rods or cocci we suspected infections with fastidious bacteria or agents that required a microaerophilic or strictly anaerobic environment. Indeed, the 16S rRNA approach revealed anaerobic bacteria such as A. lactolyticus, B. vulgatus, D. invisus, F. nucleatum, L. amnionii, P. buccalis and P. ruminicola. Other bacteria required microaerophilic conditions, e.g., 5% CO2, and a prolonged incubation time of 48 to 72 h, such as L. iners. Different S. intermedius strains have been reported to grow under either microaerophilic or anaerobic conditions. Some of them are fastidious bacteria and required selective media for cultivation such as G. vaginalis and C. urealyticum (10). The latter are well-known uropathogens: G. vaginalis is remotely related to the genus Bifidobacterium and is strongly associated with bacterial vaginosis. C. urealyticum belongs to the common Corynebacteria but is strongly associated with urinary tract infections and has been reported to be a frequent causative agent of chronic cystitis in women (10). L. amnionii and L. iners are novel, recently described bacteria. L. amnionii was isolated from the amniotic fluid of a woman who experienced intrauterine fetal demise (17) and L. iners is a member of the dominating vaginal flora (19). But even Lactobacillus spp. can cause severe infections in immunocompromised hosts (8). One exception was sample UR881, which contained C. albicans and a gram-negative bacterium, R. aquatilis (Fig. 4, Table 5). R. aquatilis belongs to the Enterobacteriaceae, is often misidentified as E. agglomerans, and grows generally well under standard conditions (10). However, we detected it only with the 16S rRNA approach, indicating that the bacteria were not viable or belonged to a fastidious strain.
With a standard curve, we calculated the numbers of G. vaginalis, L. amnionii, L. iners, and C. urealyticum in the examined urine samples to be in the significant range of 5 × 104 to ≥1 × 105 CFU/ml (data not shown). Thus, they have to be considered as the etiologic agents of UTIs. The numbers of the remaining bacteria were calculated to be in the range of between 1 × 104 and 5 × 104 CFU/ml. Consequently, these bacteria were assumed to be opportunistic agents in the cases analyzed. No members of the indigenous microbial flora such as viridans streptococci or coagulase-negative staphylococci from the skin were detected.
The small peaks in the complex sample UR539 were identified by sequencing as A. lactolyticus, P. buccalis, and P. ruminicola (Fig. 4). We did not detect interferences with small peaks characteristic for sequence heterogeneities in the 16S rRNA alleles observed for single species (Fig. 2). Amplicons from a single bacteria species show dominance of an allele (major peak) with minor peaks reflecting sequence variations among alleles. Such ratios would also be expected to be maintained when amplifying complex bacterial mixtures comprised of other minor peaks which represent the presence of other species. Thus, minor peaks in such samples would rather be indicative of the relative contribution of the respective bacterial population and further detailed analysis would be required to decipher the additional minor alleles deriving from these bacteria. At this time we are unable to comment as to whether the bacteria detected in lower titers could indeed play a relevant pathogenic role in renal transplant recipients receiving immunosuppressive treatment. However, most of the patients examined were positive for leukocyte esterase, an indicator of potential bacterial infection (Table 5). To decrease the risk of graft rejection, it is therefore advisable to examine both opportunistic and unusual infections since this may be highly relevant for the consideration of the antibiotic treatment administered.
In conclusion, our study strongly recommends examination of renal transplant recipients for unusual, fastidious, and anaerobic bacteria with the approach described here, especially as it also facilitates the analysis of polymicrobial infections. An examination based on this approach, and the specific procedures described in this study can be accomplished within a 24-h period after receipt of the sample. Major elements of the procedure include the primers to generate 16S rRNA derived amplicons C by consensus PCR and the analytical gradient developed in this study. The approach reported here can be used to establish a specialized database of bacterial pathogens and to detect bacteria by the combination of distinct retention profiles of amplicons without sequencing. It is easy to envisage the use of this technique to elucidate viral, fungal and parasitic pathogens in clinical settings. The versatility of the separation method provides not only for the generation of distinct profiles but also for the detection of polymorphic sites in genes and enables direct sequence-based evaluation of human microbial communities and polymicrobial infections (gastrointestinal, genital, and periodontal diseases, abscesses, and mixed-species biofilms). Even the elucidation of complex environmental microbial communities may arise from the use of this technique.
Acknowledgments
We thank Kirsten-Susann Bommersheim for technical assistance and Leigh Marsh for critical review of the manuscript.
REFERENCES
- 1.Abbott, K. C., J. D. Oliver 3rd, I. Hypolite, L. L. Lepler, A. D. Kirk, C. W. Ko, C. A. Hawkes, C. A. Jones, and L. Y. Agodoa. 2001. Hospitalization for bacterial septicemia after renal transplantation in the United States. Am. J. Nephrol. 21:120-127. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisenstadt, J., and J. A. Washington. 1996. Diagnostic microbiology for bacteria and yeasts causing urinary tract infection, p. 29-66. In H. L. T. Mobley and J. W. Warren (ed.), Urinary tract infections — molecular pathogenesis and clinical management. American Society for Microbiology Press, Washington, D.C.
- 4.Fischer, S. G., and L. S. Lerman. 1979. Length-independent separation of DNA restriction fragments in two-dimensional gel electrophoresis. Cell 16:191-200. [DOI] [PubMed] [Google Scholar]
- 5.Fredricks, D. N., and D. A. Relman. 1996. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin. Microbiol. Rev. 9:18-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fromin, N., J. Hamelin, S. Tarnawski, D. Roesti, K. Jourdain-Miserez, N. Forestier, S. Teyssier-Cuvelle, F. Gillet, M. Aragno, and P. Rossi. 2002. Statistical analysis of denaturing gel electrophoresis fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 7.Hurtle, W., D. Shoemaker, E. Henchal, and D. Norwood. 2002. Denaturing HPLC for identifying bacteria. BioTechniques 33:386-391. [DOI] [PubMed] [Google Scholar]
- 8.Jones, R., M. Ellbogen, and R. Dunlay. 2000. Lactobacillus allograft pyelonephritis and bacteremia. Nephron 86:502. [DOI] [PubMed] [Google Scholar]
- 9.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray P. R., E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.). 2003. Manual of clinical microbiology, 8th ed. American Society for Microbiology Press, Washington, D.C.
- 11.Neefs, J.-M., Y. Van de Peer, L. Hendriks, and R. De Wachter. 1990. Compilation of small subunit RNA sequences. Nucleic Acids Res. 18(Suppl.):2237-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nübel, U., B. Engelen, A. Felske, J. Snaidr, A. Wieshuber, R. I. Amman, W. Ludwig, and H. Backhaus. 1996. Sequence heterogeneities of genes encoding 16S rRNA in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J. Bacteriol. 178:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Renoult, E., F. Aouragh, D. Mayeux, D. Hestin, A. Lataste, J. Hubert, J. L'Hermite, M. Weber, and M. Kessler. 1994. Factors influencing early urinary tract infections in kidney transplant recipients. Transplant Proc. 26:2056-2058. [PubMed] [Google Scholar]
- 14.Rosenbaum, V., and D. Riesner. 1987. Temperature-gradient gel electrophoresis-thermodynamic analysis of nucleic acids and proteins in purified form and in cellular extracts. Biophys. Chem. 26:235-246. [DOI] [PubMed] [Google Scholar]
- 15.Schmaldienst, S., E. Dittrich, and W. H. Hörl. 2002. Urinary tract infections after renal transplantation. Curr. Opin. Urol. 12:125-130. [DOI] [PubMed] [Google Scholar]
- 16.Sheffield, V. C., D. R. Cox, L. S. Lerman, and R. M. Myers. 1989. Attachment of a 40-base-pair G+C-rich sequence (GC clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl. Acad. Sci. 86:232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla, S. K., P. R. Meier, P. D. Mitchell, D. N. Frank, and K. D. Reed. 2002. Leptotrichia amnionii sp. nov., a novel bacterium isolated from the amniotic fluid of a woman after intrauterine fetal demise. J. Clin. Microbiol. 40:3346-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takai, K., J. Tollemar, H. E. Wilczek, and C. G. Groth. 1998. Urinary tract infections following renal transplantation. Clin. Transplant. 12:19-23. [PubMed] [Google Scholar]
- 19.Vasquez, A., T. Jakobsson, S. Ahrné, U. Forsum, and G. Molin. 2002. Vaginal Lactobacillus flora of healthy Swedish women. J. Clin. Microbiol. 40:2746-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren, J. W. 1996. Clinical presentations and epidemiology of urinary tract infection, p. 3-27. In H. L. T. Mobley and J. W. Warren (ed.), Urinary tract infections — molecular pathogenesis and clinical management. American Society for Microbiology Press, Washington, D.C.
- 21.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zoetendal, E. G., A. D. L. Akkermans, and W. M. de Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]