Abstract
Defensins comprise a potent class of membrane disruptive antimicrobial peptides (AMPs) with well-characterized broad spectrum and selective microbicidal effects. By using high-resolution synchrotron small angle x-ray scattering to investigate interactions between heterogeneous membranes and members of the defensin subfamilies, α-defensins (Crp-4), β-defensins (HBD-2, HBD-3), and θ-defensins (RTD-1, BTD-7), we show how these peptides all permeabilize model bacterial membranes but not model eukaryotic membranes: defensins selectively generate saddle-splay (‘negative Gaussian’) membrane curvature in model membranes rich in negative curvature lipids such as those with phosphoethanolamine (PE) headgroups. These results are shown to be consistent with vesicle leakage assays. A mechanism of action based on saddle-splay membrane curvature generation is broadly enabling, since it is a necessary condition for processes such as pore formation, blebbing, budding, vesicularization, all of which destabilize the barrier function of cell membranes. Importantly, saddle-splay membrane curvature generation places constraints on the amino acid composition of membrane disruptive peptides. For example, we show that the requirement for generating saddle-splay curvature implies that a decrease in arginine content in an AMP can be offset by an increase in both lysine and hydrophobic content. This ‘design rule’ is consistent with the amino acid compositions of 1,080 known cationic AMPs.
1. Introduction
Antimicrobial peptides (AMPs) comprise an important component of the innate host defense system. Collectively, AMPs have broad spectrum antimicrobial activity.1–4 Although AMPs are diverse in sequence and structure, most share two general structural features: they are amphipathic and cationic.1,2 From in vitro studies, AMP amphipathicity is inferred to disrupt membranes through a combination of electrostatic interactions of the cationic AMP with the anionic membrane followed by insertion of hydrophobic patches into the non-polar interior of the bilayer.1–3,5–7 Membranes can be disrupted by AMPs via a diverse range of processes, including pore formation, blebbing, budding, vesicularization.1–12 This diversity of outcomes has impeded the development of a detailed, molecular understanding of how AMPs destabilize bacterial membranes specifically. Such knowledge can in principle provide ‘design rules’ that impact development of synthetic antimicrobials.
Mammals produce two main types of membrane active AMPs: defensins,8–10 and cathelicidins.11 Defensins are synthesized by varied epithelia and by phagocytes, and function as a biochemical line of defense against microbial infection.8 Vertebrate defensins share general features, including cationic charge, MW of 2–5 kDa, distinctive tridisulfide arrays, and β-strand structures constrained by the disulfide pairings.8,9 They can be separated into three subfamilies, α-defensins, β-defensins, and θ-defensins.9 Since many aspects of defensin biology are known, this is an ideal prototypical family of AMPs to investigate the detailed molecular mechanisms responsible for selective activity against bacterial but not mammalian membranes.
In this work we use synchrotron small angle x-ray scattering (SAXS) to map out the fundamental curvature deformation modes induced in model cell membranes by defensins, and elucidate causative relationships between curvature deformations and membrane disruption processes. Mammalian defensins are systematically investigated, using members from the three defensin subfamilies, including rhesus monkey θ-defensin-1 (RTD-1), baboon θ-defensin-7 (BTD-7), mouse Paneth cell α-defensin cryptdin-4 (Crp-4), human β-defensin-2 (HBD-2), and human β-defensin-3 (HBD-3). The bactericidal activity of these defensins is correlated with the induction of saddle-splay (equivalently negative Gaussian) membrane curvature, which enables membrane destabilizing processes.13 All peptides restructured vesicles into porous bicontinuous phases rich in saddle-splay curvature when lipid compositions mimic those of bacterial membranes, but not when the lipid compositions are more representative of mammalian membranes. A key parameter for activity is the concentration of negative intrinsic curvature (c0 < 0) lipids, such as those with PE headgroups, which exist at significantly higher concentrations in bacterial cytoplasmic membranes compared to eukaryotic membranes. Existence of homologous behavior in synthetic antimicrobials,14–16 suggests a common root mechanism for selective membrane permeation. In general, AMPs destabilize membranes via a diverse range of processes, many of which involve saddle-splay curvature. We deduce a criterion for amino acid compositions of AMPs based on the requirement for generating saddle-splay membrane curvature, and show that it is consistent with trends in amino acid composition of 1,080 known cationic AMPs.
2. Experimental Section
2.1. Preparation of Peptides
Recombinant Crp4 was expressed and purified as His6-tagged fusion proteins as described.17–19 Expression of recombinant fusion proteins was induced by adjusting exponentially-growing E. coli BL21-CodonPlus (DE3)-RIL cells to 0.1 mM isopropyl-β-D-1-thiogalactopyranoside and incubating at 37 °C for 6 h in Terrific Broth as described in earlier reports. Cells were lysed by sonication in 6 M guanidine-HCl, 100 mM Tris (pH 8.0), and the suspension was clarified by centrifugation. His6-tagged fusion proteins purified by nickel-nitrilotriacetic acid resin affinity chromatography (Qiagen, Valencia, CA) were cleaved with cyanogen bromide and purified by analytical C18 reverse phase high pressure liquid chromatography (RP-HPLC). Peptide homogeneity was confirmed in analytical acid-urea polyacrylamide gel electrophoresis (AU-PAGE), a highly sensitive measure of defensin foldamers. Molecular masses were verified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and peptides were quantified using extinction coefficient calculations at 280nm performed at ExPASY.20 θ-defensins RTD-1 and BTD-7 were produced by solid phase synthesis, purified and characterized as described previously.21,22 HBD-2 and HBD-3 are expressed and purified as described in23,24.
2.2. Liposome Preparation for X-ray Measurements
DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine), DOPG {1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (sodium salt)}, and DOPS {1,2-dioleoyl-sn-glycero-3-phospho-L-serine (sodium salt)}, lyophilized lipids from Avanti Polar lipids were used without further purification. For x-ray experiments we prepared Small Unilamellar Vesicles (SUVs) by sonication. DOPG, DOPS were dissolved in chloroform/methanol, and DOPC, DOPE were dissolved in chloroform for stock solutions. Mixtures of DOPG and DOPE were prepared at 1:4 mass ratio to approximate bacterial lipid compositions, while DOPS, DOPC, and DOPE at 1:4:0, and 1:2:2 mass ratios were used to approximate the membranes of eukaryotic cells. Lipid mixtures were dried under N2, dessicated overnight under vacuum, and resuspended at a final concentration of 20 mg/ml in Millipore H2O. Solutions were incubated at 37°C overnight and then sonicated until clear. SUV’s were obtained via extrusi on (0.2 µm pore Nucleopore filter).
2.3. SAXS Experiments
Defensin and lipid solutions were thoroughly mixed at specific peptide to lipid ratios (P/L) and sealed in quartz capillaries. All samples were prepared in 100–150 mM NaCl to simulate physiological salt conditions. For SAXS experiments at the Stanford Synchrotron Radiation Laboratory (BL4-2) and at the Advanced Photon Source (BESSRC-CAT BL-12IDC), monochromatic X-rays with energies of 9–11 and 12 keV, respectively, were used. Scattered radiation was collected using a MAR Research CCD area detector (pixel size, 79 µm). Samples were checked for consistency by multiple measurements with different x-ray sources. No radiation damage was observed for the incident beam intensities and the exposure times used. 2D SAXS powder patterns were integrated using the Nika 1.2 package,25 and FIT2D.26
2.4. Vesicle Dye Leakage Experiments
Giant Unilamellar Vesicles (GUVs) were prepared using the swelling method. Stock solutions of lipids DOPS, DOPE, and DOPC in chloroform were deposited (100µl at 20mg/ml) at the desired mass ratios onto roughened, cleaned Teflon, along with dye DiO (10µl at 1mg/ml). After drying under vacuum the Teflon was hydrated with moist nitrogen gas for 30min, and then 5 ml swelling solution was added (100 mM sucrose plus 20 µM Alexa 633 maleimide), and incubated for 2 days at 37°C. After swelling, the GUV suspension was diluted 40× into 200 mM glucose. 200 µl of this dilution was placed on a coverslip soaked in 1% BSA, and Crp4 (10µl at 1 mg/ml) was added. A Leica SP2 laser scanning confocal microscope was used.
2.5. Analysis of Data from the Antimicrobial Peptide Database
27 Data was obtained via the online antimicrobial peptide database.28
Comparison of helical peptides with defensins: Eligible peptides were required to be active against Gram-negative or Gram-positive bacteria. A pool of 299 peptides with helix structure is compared with a pool of 143 defensins. ‘Average % of AMP composition’ is the number of times an amino acid is found in a pool divided by the total number of amino acids in that pool.
Lysine/arginine ratio versus % hydrophobicity: Eligible peptides were required to have a net positive charge and be active against either Gram-negative or Gram-positive bacteria. Percentage of lysine, %K, and percentage of arginine, %R, were read off from the sequence information for a set hydrophobic bin. Here %K (%R) = Number of lysines (arginines) found in the bin/Total number of amino acids in the bin. Note that %K / %R = NK / NR (defined below).
Lysine/arginine ratio versus <hydrophobicity>: Eligible peptides were required to have a net positive charge and be active against either Gram-negative or Gram-positive bacteria. These criteria retrieved a pool of 1,080 peptides. For a given peptide, j, its average hydrophobicity is defined by:
Where n = number of amino acids in the peptide, wi = the hydrophobicity of the ith amino acid in the peptide. This value is set by the particular hydrophobicity scale used. To facilitate easier comparison across scales, the value of the apparent free energy of membrane insertion for an amino acid, ΔGaaapp, based upon the Wimley-White biological scale was reversed in sign, i.e. ΔGaaapp→ −ΔGaaapp. The published hydrophobicity values were used for all other scales.
To partition each peptide into a bin the hydrophobic extremes for the pool were determined and used to set the range of hydrophobicity scale. The range was divided into 100 equal bins. For the M peptides in a given bin, we define:
NK / NR versus <hydrophobicity> is plotted for each of the 100 bins.
3. Results
3.1. θ-Defensins Generate Saddle-Splay Curvature in Model Bacterial Membranes with High PE Concentrations but Not in Model Eukaryotic Membranes with Low PE Concentrations
θ-defensins are short 18 amino acid circularized peptides expressed only in old world monkeys and orangutans.8 Previous in vitro studies have shown both rhesus θ-defensin-1 (RTD-1) and baboon θ-defensin-7 (BTD-7) have activity against Gram-positive and Gram-negative bacteria, and fungi.21,29 Moreover, RTD-1 has antiviral activity against HIV-130, and the herpes simplex virus.31 Both θ-defensins are amphipathic and contain five arginine residues.
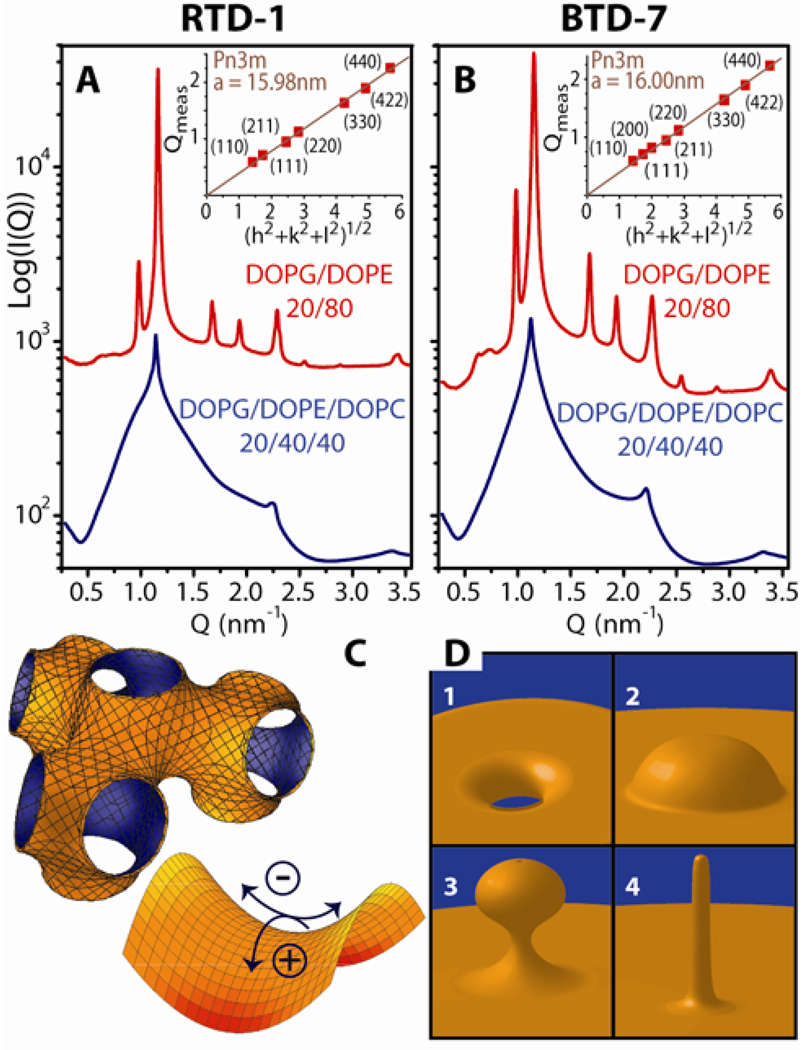
Small Unilamellar Vesicles (SUV’s) with phospholipid compositions characteristic of Gram-negative bacteria (DOPG/DOPE = 20/80) were incubated with specific defensins to study the induced structural changes. Synchrotron SAXS profiles from lipid solutions only showed a broad characteristic feature consistent with a single lipid bilayer form factor expected for unilamellar vesicles (see Supporting Information, Figure S1). When exposed to RTD-1 or BTD-7 (Figure 1A&B), the vesicles underwent a drastic structural transition and displayed characteristic correlation peaks with ratios, √2: √3: √4: √6, which indicate the formation of a cubic Pn3m ‘double-diamond’ lattice. By fitting the slopes of the measured Q-values at every peak we calculate lattice parameters, aBTD-7 = 16.00 nm, and aRTD-1 = 15.98 nm, for BTD-7 and RTD-1, respectively.32 The Pn3m (Figure 1C) is a bicontinuous cubic phase where two non-intersecting water channels are separated by a lipid bilayer.33 The center of the bilayer traces out a minimal surface with negative Gaussian curvature at every point. Although bilayer saddle-splay curvature is different than the self-connected monolayer saddle-splay curvature seen in a toroidal pore (Figure 1D), it should be noted that each constituent monolayer in a bicontinuous phase also possesses negative Gaussian curvature at every point, and is equally accessible by AMPs. On such saddle-shaped surfaces (Figure 1C), the surface curves upwards in one direction and downwards in the perpendicular direction. For example, saddle-shaped surfaces are found inside toroids, which have a single hole each. This type of curvature is not found on the surfaces of spheres, which lack holes. Furthermore, protrusions such as buds and blebs (Figure 1D), require bilayer saddle-splay curvature at their base. Saddle-splay curvature is required for membrane poration, as well as for formation of different AMP-induced destabilization mechanisms depicted in Fig 1D.
Figure 1.
Generation of negative Gaussian curvature by θ-defensins is dependent on membrane PE concentration. At peptide to lipid molar ratio, P/L = 1/30, both RTD-1 (A) and BTD-7 (B) θ-defensins, induced a Pn3m cubic phase in SUVs with lipid compositions mimicking the cell membranes of Gram-negative bacteria, DOPG/DOPE = 20/80, but not in membranes with reduced amounts of negative intrinsic curvature lipids, DOPG/DOPE/DOPC = 20/40/40. Insets in A&B show indexation of the Pn3m cubic phase by agreement between the measured peak Q positions and the Miller indices h, k, l, with relationship Qmeas = 2π√(h2+k2+l2)/a, for a cubic phase with lattice parameter, a. (C) (upper left) Illustration of the Pn3m cubic phase. The surface at the mid-plane between bilayer leaflets has zero mean curvature and negative Gaussian curvature at every point. (lower right) Negative Gaussian curvature requires positive curvature (+) in one direction and negative curvature (−) in the perpendicular direction to locally produce a saddle shape. (D) Diagrams of different manifestations of saddle-splay curvature in the interior of a pore (1) and at the base of blebs (2), buds (3), and rod-like projections (4).
To examine the role of membrane intrinsic curvature on induced saddle-splay curvature we constructed ternary lipid membranes of constant anionic charge (DOPG = 20%) but with varying ratios of DOPE (c0 < 0) and DOPC (c0 ≈ 0). In contrast to effects on PE-rich membranes, RTD-1 and BTD-7 did not restructure vesicles that mimic eukaryotic membranes with low concentrations of PE. Phases rich in saddle-splay curvature were not induced by RTD-1 or BTD-7 in membranes with DOPG:DOPE:DOPC = 20:40:40 (Figure 1A&B), as indicated by the absence of their characteristic correlation peaks. The diffraction data showed a combination of broad form factor scattering characteristic of isolated bilayers and a lamellar phase with a periodicity of dRTD-1 = 5.67 nm and dBTD-7 = 5.67 nm for RTD-1 and BTD-7, indicating that weak inter-membrane attraction is induced, although curvature is not.
It is important to discriminate between necessary and sufficient conditions for saddle-splay curvature generation. Alamethicin,34 gramicidin S,35 and protegrin-1,36 form cubic phases in pure PE lipids under specific processing conditions such as temperature cycling. The above results on defensins allow us to assess systematically the roles played by different lipid species. It has been previously proposed that anionic lipids in bacterial membranes are important for AMP function. We agree with this assessment. However, while anionic PG lipids and high curvature PE lipids in the target membrane are both necessary conditions for induction of saddle-splay curvature, neither by itself is a sufficient condition (see Supporting Information, Figure S2). Our data suggests that AMPs target distributions of lipids rather than individual lipid species.
3.2. Arginine-rich α-Defensins also Generate Saddle-Splay Curvature in Membranes Enriched with PE and Selectively Permeate PE-Rich GUVs
To investigate whether other arginine-rich defensins exhibit similar behavior we conducted similar experiments on mouse cryptdin-4 (Crp-4), a 32 amino acid α-defensin expressed in mouse Paneth cells with in vitro microbicidal activity against Gram-negative and Gram-positive bacteria.37,38 Crp-4 is amphipathic and 7 of the 10 cationic amino acids in Crp-4 are arginine residues.39
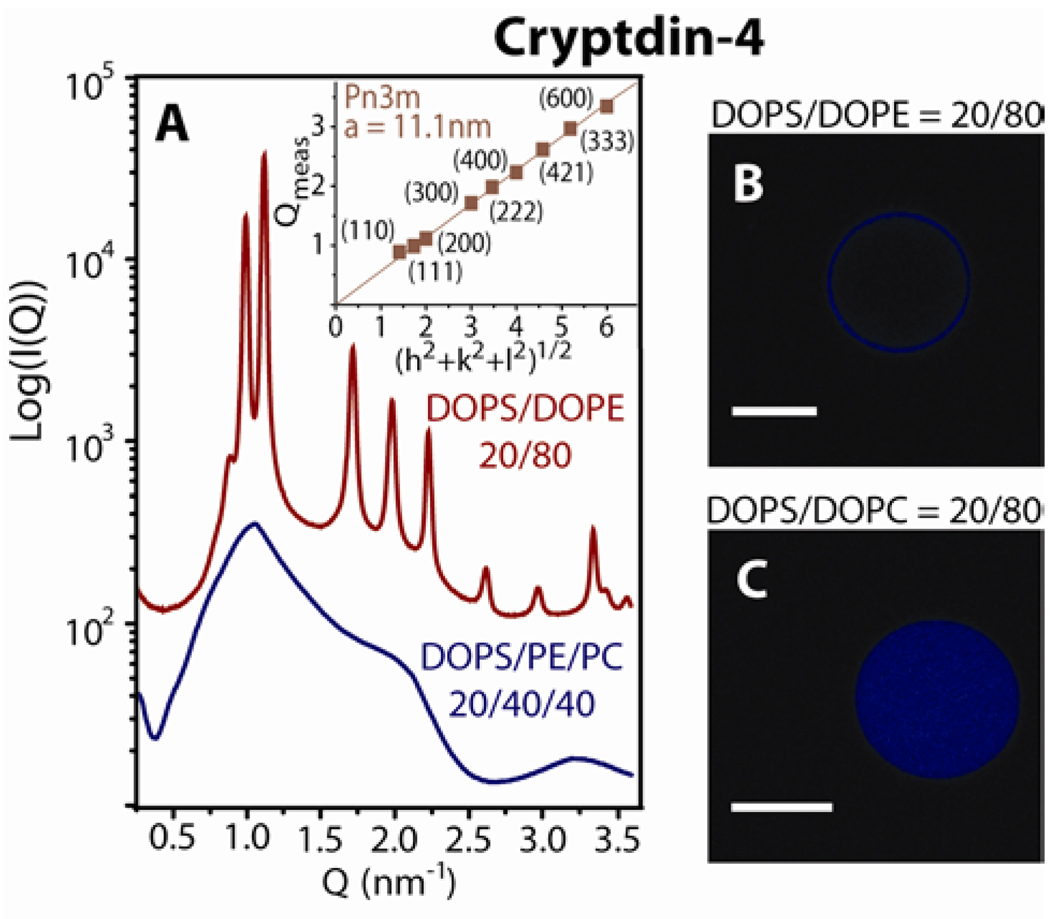
Like the θ-defensins, the α-defensin Crp-4 generates saddle-splay curvature in model bacterial membranes with high PE but not in model eukaryotic membranes with low PE. Incubation of Crp-4 with SUVs of high PE lipid composition produced a Pn3m cubic phase with lattice parameter, a = 11.1 nm (Figure 2A), while reduction of membrane DOPE content led to the disappearance of the Pn3m cubic phase. Interestingly, previous work suggests that negative intrinsic curvature lipids can promote pore formation through modification of the Gaussian modulus.40,41 Typically the Gaussian curvature modulus is negative, which exacts a free energy penalty for pore formation. However, the presence of c0 < 0 lipids (such as PE lipids) changes the bilayer Gaussian curvature modulus toward positive values.40 The decreased free energy barrier to changes in membrane topology toward porous phases is consistent with our observation that bacterial membranes rich in negative intrinsic curvature lipids are more susceptible to pore formation.
Figure 2.
The α-defensin Crp-4 showed similar phase behavior to θ-defensins which suggests a general mechansim for selectivity by defensins rich in arginine. At P/L = 1/45, Crp-4 induced a Pn3m cubic phase in DOPS/DOPE = 20/80 membranes, but induced a lamellar phase with lattice spacing, a = 6.0nm in DOPS/DOPE/DOPC = 20/40/40 membranes. Figures B and C show representative GUVs after treatment with Crp-4. Dye leakage occurred in vesicles enriched with DOPE (B), but not in vesicles enriched with DOPC (C), consistent with the relationship between membrane PE content and the creation of porous phases by defensins. White scale bars are 10µm.
Our x-ray experiments measure AMP-induced curvature deformations in a 3D lipid system, which has the freedom to connect into bulk phases such as the Pn3m. Both eukaryotic and prokaryotic cells have quasi-2D membranes, where induced saddle-splay curvature deformations can take the form of pores, blebs, or buds. To determine how peptide-induced saddle-splay curvature is manifested in 2D membranes we investigated the effect of Crp-4 on giant unilamellar vesicles (GUV) by confocal microscopy. Liposomes with lipid compositions mimicking the membranes of bacteria (PS/PE = 20/80) and of eukaryotic cells (PS/PC = 20/80) were loaded with maleimide dye and incubated with Crp-4 (molar ratio P/L=1, see Experimental Section). Dye leakage was evident in PE-rich membranes after peptide exposure (Figure 2B), consistent with Crp-4 induced pore formation in the intact vesicles. In contrast, negligible dye leakage was observed in PC-rich vesicles (Figure 2C). Selective permeation of GUVs rich in PE but not PC is consistent with the observation of pore-enabling Crp-4 induced saddle-splay curvature (Figure 2A) from SAXS experiments. Taken together, these results indicate Crp-4 preferentially creates pores in membranes with negative intrinsic curvature lipids.
3.3. Generality of Mechanism for Defensins and Relation of Curvature Generation to Antimicrobial Peptide Sequence: HBD-2 and HBD-3 as Examples
We hypothesized the selective induction of saddle-splay curvature in model bacterial membranes is a generic feature among defensins. We used human β-defensin-2 (HBD-2, DEFB4) and human β-defensin-3 (HBD-3, DEFB103A),42 to test this hypothesis. HBD-2 is a 41AA amphipathic peptide,43 which is predominately effective against Gram-negative bacteria.23 HBD-3 is 45AA, amphipathic, and is generally more potent than HBD-2.42,44 Unlike the other two defensin subfamilies which strongly favor arginine over lysine, β-defensins have intermediate arginine to lysine ratios.10 In HBD-3 the cationic residues consist of 7 arginines and 6 lysines, and HBD-2 has 2 arginines, 5 lysines, and 1 histidine.
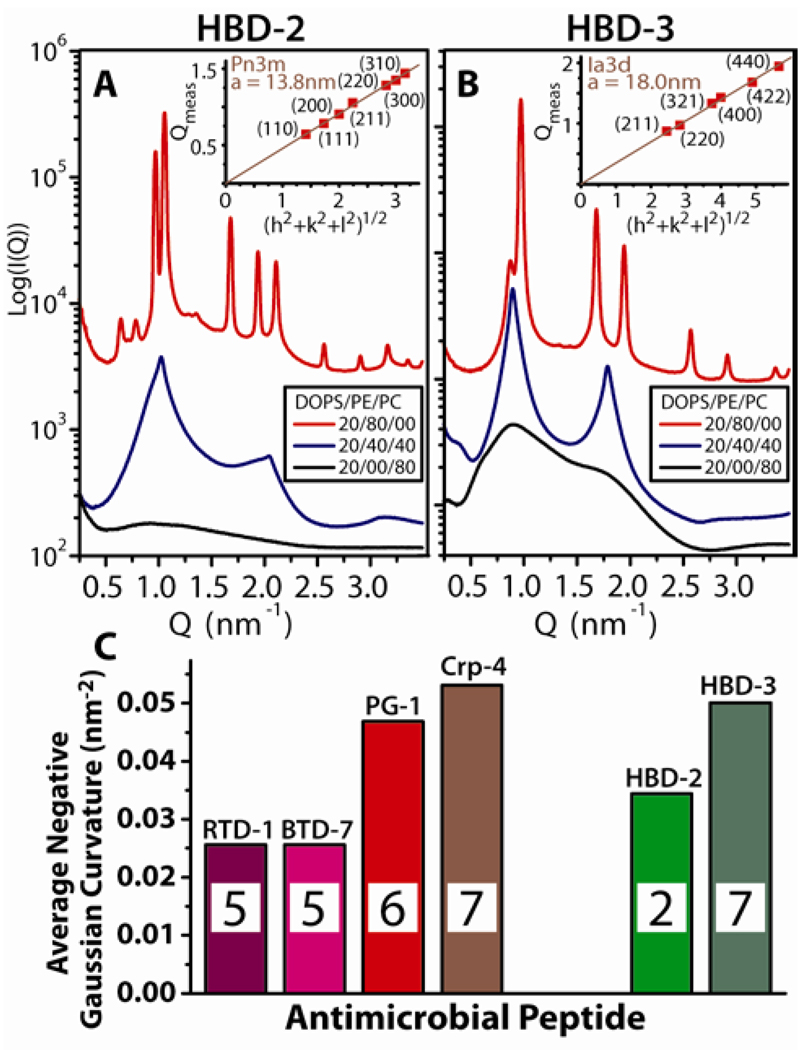
Figure 3 shows the SAXS spectra of model bacterial and model eukaryotic membranes after exposure to HBD-2 and HBD-3. The capacity for membrane–specific saddle-splay curvature generation is observed, and is consistent with behavior of θ-defensins and α-defensins. Both β-defensins showed weak interactions with membranes enriched with DOPC, and produced lamellar phases in membranes with intermediate amounts of PE/PC (Figure 3A&B). In DOPS/DOPE = 20/80 membranes, HBD-2 generated a Pn3m cubic, with lattice parameter a = 13.8 nm (Figure 3A). HBD-3 also generated saddle-splay curvature in DOPS/DOPE = 20/80 membranes, which is exhibited in reflections with ratio, √6: √8: √14: √16, characteristic of the cubic Ia3d “gyroid” phase with a = 18.0 nm (Figure 3B). Like the Pn3m, the Ia3d is a bicontinuous cubic phase consisting of two non-intersecting water channels separated by a single lipid bilayer which has saddle-splay curvature everywhere on its minimal surface.45
Figure 3.
β-defensins with intermediate arginine/lysine ratios also generated negative Gaussian curvature. Neither HBD-2 (A) or HBD-3 (B) produced cubic phases in membranes with low PE concentrations. In DOPS/DOPE/DOPC = 20/40/40 membranes, HBD-2 and HBD-3 restructured SUVs into lamellar phases with lattice spacings d = 6.2nm and 7.0nm, respectively, and DOPS/DOPC = 20/80 membranes showed only weak interaction with both β-defensins. Conversely, in DOPS/DOPE = 20/80 membranes HBD-2 induced coexisting Pn3m and inverted hexagonal (lattice spacing a = 7.5nm) phases, while HBD-3 induced an Ia3d cubic phase. P/L = 1/35 for HBD-2, and 1/55 for HBD-3. (C) Comparision of the average negative Gaussian curvature, <K>, produced in 80% PE membranes by AMPs rich in arginine (left group) and by AMPs with both lysines and arginines (right group) around the peptide to lipid isoelectric point. The inset in each bar graph corresponds to the number of arginines in the AMP.
4. Discussion
4.1. Average Induced Saddle-Splay Curvature, <K>, From SAXS Measurements Allows Quantitative Comparison of Different Antimicrobial Peptides
Our previous work on cell penetrating peptides,46,47 has shown lysine and arginine generate different types of membrane curvatures. Since lysine and arginine are cationic they both induce wrapping by anionic membranes. This is due to the entropy of counterions condensed on both the charged membrane and on the short charged polymers. Electrostatic compensation between a cationic polymer48 and an anionic membrane49 leads to the release of condensed counterions into the bulk solution and a corresponding large entropy gain, which results in strong polymer-membrane binding. This has been observed in a large number of systems50–52 Since maximal contact between the peptide and membrane leads to maximal counterion release, the membrane will tend to wrap around the peptide generating negative curvature.53,54 The guanidine group of arginine supports multi-dentate hydrogen bonding, which can organize bulky lipid head groups to generate positive curvature in the perpendicular direction, resulting in saddle-splay curvature. In contrast, the amine group of lysine can only hydrogen bond to one lipid head group at a time.55,56 Consistent with this, polylysine induces negative mean curvature on membranes to form inverted hexagonal phases, while polyarginine can also induce saddle-splay curvature to form cubic phases. Clearly more work, experimental, theoretical, and computational, is required for a full understanding of these effects. Extant theoretical studies57–59 suggest the anisotropy associated with saddle-splay curvature deformations may also provide AMPs with a way to communicate and organize cooperatively. Currently, we are directly testing effects from peptide orientation and conformation, as well as proximity effects of different amino acids.
The Pn3m, Ia3d, and Im3m cubic phases are related by a Bonnet transformation, so their average Gaussian curvatures can be quantitatively compared.33,45 The average Gaussian curvature is <K> = 2πχ/a2Ao, where a is the cubic lattice constant, χ is the Euler characteristic, and Ao is the surface area per unit cell. Figure 3C shows the <K> values extracted from the diffraction data for arginine-rich defensins (RTD-1, BTD-7, Crp-4) and protegrin-1 (PG-1),60 (see Supporting Information, Figure S3). A correlation exists between negative Gaussian curvature and arginine content in structurally similar peptides. HBD-2 and HBD-3, which have ~50% lysines and ~50% arginines, clearly deviate from this curve, although <K> is more negative for HBD-3 compared with HBD-2, consistent with the greater number of arginines in HBD-3.
4.2. The Requirement of Saddle-Splay Membrane Curvature Generation Places Strong Constraints on Amino Acid Composition of Antimicrobial Peptides: the Saddle-Splay Curvature Selection Rule
Defensins can generate pores,8–10 but AMPs have been observed to disrupt membranes via processes that do not involve pores. Generation of the required saddle-splay membrane curvature for these processes places strong constraints on the amino acid composition and composite hydrophobic and cationic components of membrane disruptive peptides. Hydrophobic amino acids generate positive curvature.5–7 Non-polar components of the amphipathic peptide are inserted into the membrane, which increases the hydrophobic volume of the perturbed lipid monolayer and thereby creates positive curvature strain. Differences in lipid headgroup chemistry will not drastically affect this mechanism.
Cationic amino acids work differently. We have demonstrated lipid head group organization by arginine and lysine generates distinct types of membrane curvature deformations. These interactions provide an AMP with complementary curvature producing mechanisms. Arginine generates negative Gaussian curvature (positive and negative curvatures along the two perpendicular principal directions), whereas lysine generates negative curvature along one direction only. Mechanisms of curvature generation based on head group re-organization are inherently sensitive to head group chemistry, therefore such mechanisms will be conducive to mediating specific peptide-lipid interactions.
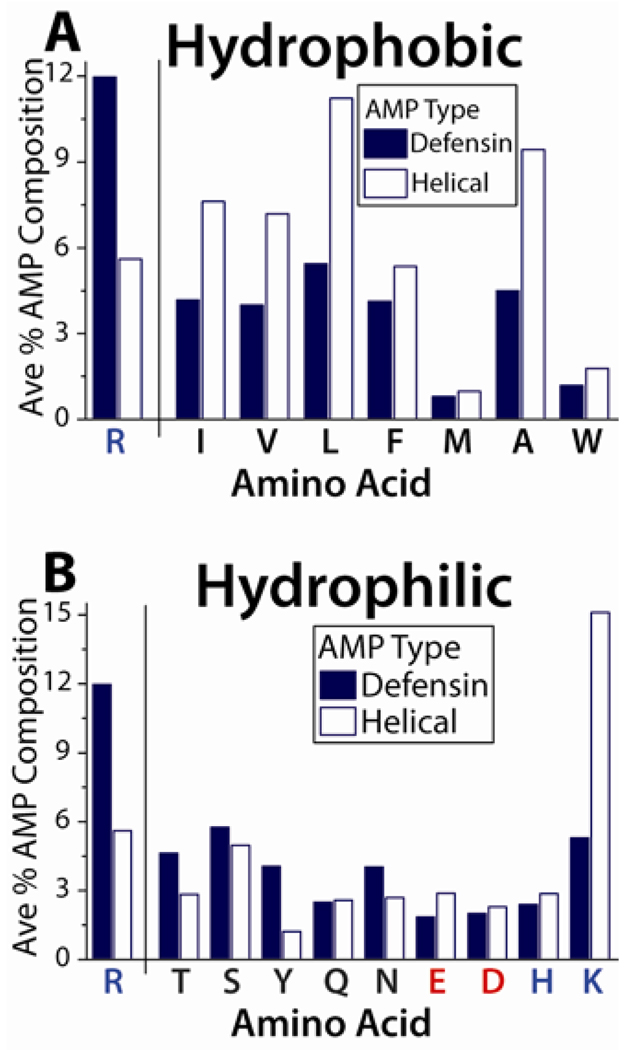
Based on the above, a series of rules governing the amino acid content of membrane disruptive antimicrobial peptides can be proposed. A decrease in arginine content in a peptide sequence will imply that less negative Gaussian curvature can be generated. This can in principle be compensated for by an increase in both lysine and hydrophobic content. This ‘exchange rate’ concept between arginines and lysines / hydrophobes is illustrated in a comparison between the amino acid content,61 of β-sheet bonded defensins and that of α-helical AMPs (Figure 4A&B). The average arginine composition of α-helical AMPs is ~50% that of defensins, but the proportion of most hydrophobic amino acids in α-helical AMPs is ~200% that found in defensins (Figure 4A). Likewise, whereas most of the hydrophilic amino acids are found in similar proportions for α-helical AMPs and defensins, the proportion of lysines in α-helical AMPs has drastically increased by ~300% relative to that in defensins (Figure 4B). Thus, there is a unifying conceptual link between α-helical AMPs and β-sheet AMPs: β-sheet AMPs generally rely more on arginines to generate curvature, while α-helical AMPs rely more on lysines and hydrophobes to do so. In both cases, the end result is the generation of saddle-splay curvature.
Figure 4.
The exchange of R with K and hydrophobicity is illustrated by comparing β-sheet defensins with α-helical AMPs. (A) On average defensins contain much greater amounts of R than α-helical AMPs, and utilize less of each type of hydrophobic amino acid. (B) Overall, the proportion of hydrophilic amino acids is comparable in defensins and α-helical AMPs. The exception is K, which is more commonly found in α-helical AMPs. Blue denotes basic amino acids, while negatively charged amino acids are colored red.
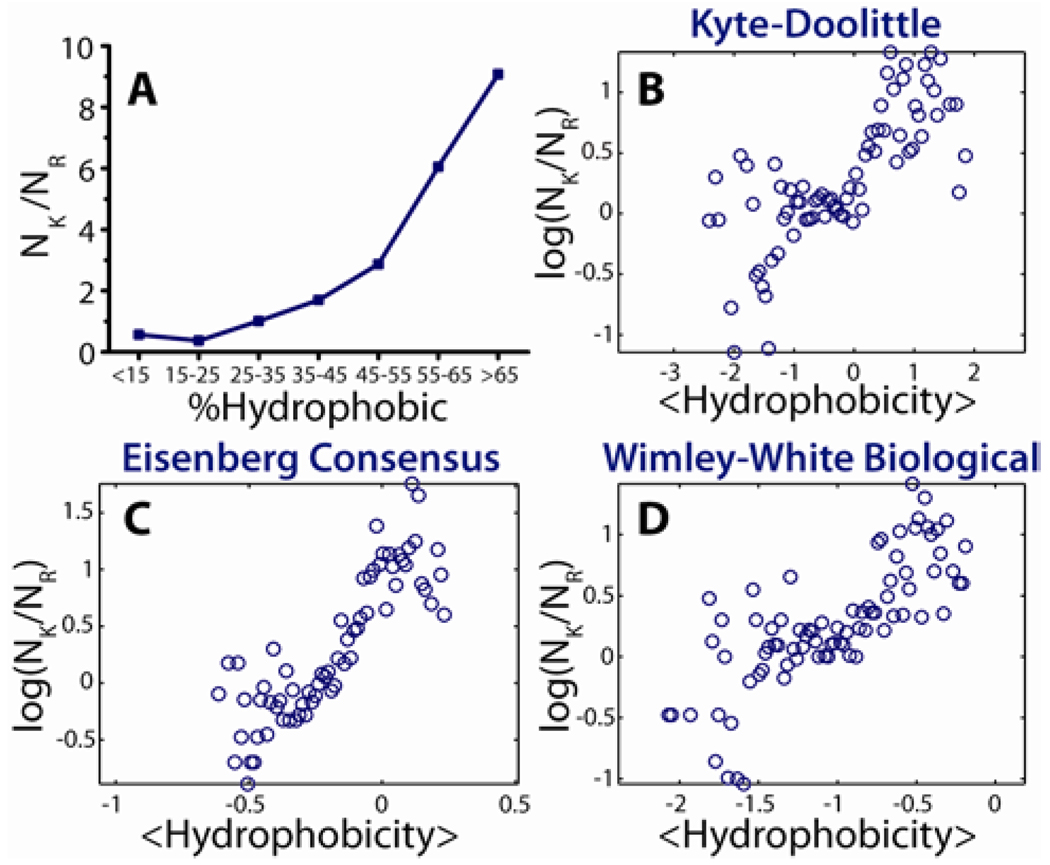
Within a more general compass, a strong manifestation of this ‘saddle-splay curvature selection rule’ for amino acid content can be observed in the compositions of AMPs. We test this selection rule against 1,080 known cationic peptides with activity against Gram positive or Gram negative bacteria.27 Figure 5A shows a plot of NK/NR (the ratio of the number of lysines to the number of arginines) vs %Hydrophobic (% of hydrophobic amino acids in peptide, see Experimental Section), in which all AMPs have been binned into 7 histograms. A strong increasing trend consistent with the saddle-splay curvature selection rule can be discerned. Figure 5B–D shows a similar analysis of AMP amino acid sequences using more sophisticated measures to quantify peptide hydrophobicity, using three established scales (Kyte-Doolittle,62, Eisenberg consensus,63, Wimley-White biological,64), in which all AMPs have been partitioned into 100 bins. A strong positive correlation is observed between hydrophobicity and log (NK/NR). Even though we do not account for effects due to proximate amino acids, sequence, and peptide conformation in our simple approach, there is strong ‘exponential-like’ dependence between hydrophobicity and NK/NR, consistent with the saddle-splay curvature selection rule.
Figure 5.
Demonstration of the exchange between R, with K and hydrophobicity based upon 1,080 cationic AMPs in the antimicrobial peptide database. (A) By organizing all 1,080 AMP sequences into 7 histogram bins, a strong positive dependence between the peptide lysine to arginine ratio and peptide hydrophobicity percentage can be observed. More robust calculations of hydrophobicity based upon the widely used Kyte-Doolittle (B), Eisenberg Consensus (C), and Wimley-White (D) hydrophobicity scales show the same relationship between the exchange of arginine with lysine and hydrophobicity consistent with the saddle-splay curvature selection rule. See Experimental Section for details and binning schemes in B–D.
5. Conclusions
The curvature selection rule is consistent with different mechanisms of barrier disruption such as budding, blebbing, pore formation, vesicularization. However, many AMPs have additional antimicrobial functions other than membrane permeation. For example, indolicidin,65 buforin,66 and tachyplesin67 are also known to bind internal targets in bacteria such as DNA. Mammalian α- and β-defensins, and cathelicidin LL-37 play other important functions in host defense including modulation of the innate immune response and wound repair.10,68 A necessary and sufficient condition for membrane disruption is not a necessary and sufficient condition for full antimicrobial activity, which may involve other mechanisms such as DNA binding. (They may share related amino acid requirements, since both membrane activity and DNA binding require high cationic charge density.) It should be noted however, that AMPs which kill bacteria by binding to interior elements must first permeabilize the cytoplasmic bacterial membrane to reach the target. Since negative Gaussian curvature is topologically required for pore formation we expect the saddle-splay curvature selection rule should also apply to non-lytic AMPs provided that membrane permeation is necessary for their function, even if it isn't a sufficient condition for full activity.
In a more general context, the saddle-splay curvature selection rule we propose does not strongly restrict the amino acid content of an AMP, since it affects only the arginine, lysine, and hydrophobic residues. Membrane destabilizing AMPs therefore inherently have significant sequence flexibility for beneficial mutations. Consistent with this observation, it is known that AMP amino acid sequences are fairly non-conserved and labile across species.1 However, when they acquire specific functions, such as Fe homeostasis activity in the hepcidins,69 their sequences tend to stabilize. That 1080 known AMPs are consistent with the saddle-splay selection rule suggests that they all have some degree of membrane curvature generating activity, but since the selection rule under-determines the full sequence, there is potential for AMPs to acquire new functions, such as optimized DNA binding.
The saddle-splay curvature selection rule ultimately originates from geometry, so the amino acid composition trends are expected to be general for a broad range of membrane-active peptides. For example, the fusion protein M2 from the influenza virus induces in vitro cell membrane budding and scission. It contains a 17 residue amphipathic helix with comparable amino acid content to AMPs and is capable of inducing budding in GUVs.70 During virion release M2 localized to the ‘neck’ of budding virions, the region of maximal negative Gaussian curvature.47,70 This suggests other membrane-active peptides and proteins utilize similar strategies to generate curvature in membranes.
In summary, we show that a broad range of defensins selectively permeate bacterial membranes by inducing saddle-splay membrane curvature in a manner that depends on target membrane lipid composition. Since the presence of saddle-splay curvature is necessary for membrane-disrupting processes, generation of saddle-splay membrane curvature by AMPs is a versatile mechanism of action. A saddle-splay curvature selection rule that places strong constraints on the amino acid composition of membrane disruptive peptides is deduced. We believe this rule can guide the rational design of antimicrobials.
Supplementary Material
Acknowledgements
We thank Michael M. Kozlov, Karl Lohner, Tom Ganz, and William DeGrado for useful discussions and suggestions. X-ray work was performed at the Stanford Synchrotron Radiation Lab (SSRL), the Advanced Photon Source (APS), and at the Fredrick Seitz Materials Research Laboratory (FS-MRL, Urbana, IL). SSRL is supported by DOE and NIH. Use of the APS is supported by DOE DE-AC02-06CH11357. This work is supported by NIH grants R01DK044632 and R01AI059346 (A.J.O.), NIH grants R37AI022931, RO1AI058129, and RO1DE15517 (MES), NIH grants P50 HL-61234 and P01 HL-091842 and the Roy J. Carver Charitable Trust (PBM), NIH grant 1UO1 AI082192-01 and NSF grants DMR-0409769 and WaterCAMPWS (GCLW).
Footnotes
Supporting Information Available: SAXS measurements on SUVs used in this study; Interaction of θ-defensin RTD-1 with pure DOPE lipids and pure DOPG lipids; SAXS experiments on the cathelicidin protegrin-1 with DOPS/DOPE/DOPC membranes. This material is available free of charge via the Internet at http://pubs.acs.org
Contributor Information
Nathan W. Schmidt, Email: nschmid2@illinois.edu.
Abhijit Mishra, Email: abhijit.mishra@uclu.edu.
Ghee Hwee Lai, Email: glai2@illinois.edu.
Matthew Davis, Email: madavis5@uiuc.edu.
Lori K. Sanders, Email: lksander@illinois.edu.
Dat Tran, Email: Dat.tran@med.usc.edu.
Angie Garcia, Email: angiegar@usc.edu.
Kenneth P. Tai, Email: ktai@uci.edu.
Paul B. McCray, Jr., Email: paulmccray@uiowa.edu.
André J. Ouellette, Email: aouellet@usc.edu;.
Michael E. Selsted, Email: selsted@usc.edu.
Gerard C. L. Wong, Email: gclwong.ucla@gmail.com.
References
- 1.Zasloff M. Nature. 2002;415:389. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Brogden KA. Nat. Rev. Microbiol. 2005;3:238. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 3.Shai Y. Biochim. Biophys. Acta, Biomembr. 1999;1462:55. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 4.Hancock REW, Sahl H-G. Nat. Biotechnol. 2006;24:1551. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 5.Matsuzaki K, Sugishita K-i, Ishibe N, Ueha M, Nakata S, Miyajima K, Epand RM. Biochemistry. 1998;37:11856. doi: 10.1021/bi980539y. [DOI] [PubMed] [Google Scholar]
- 6.Matsuzaki K. Biochim. Biophys. Acta, Biomembr. 1999;1462:1. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 7.Huang HW. Biochemistry. 2000;39:8347. doi: 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- 8.Selsted ME, Ouellette AJ. Nat. Immunol. 2005;6:551. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 9.Ganz T. Nat. Rev. Immunol. 2003;3:710. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 10.Lehrer RI. Nat. Rev. Microbiol. 2004;2:727. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- 11.Zanetti M. J. Leukocyte Biol. 2004;75:39. doi: 10.1189/jlb.0403147. [DOI] [PubMed] [Google Scholar]
- 12.Falagas ME, Kasiakou SK. Clin. Infect. Dis. 2005;40:1333. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 13.Gelbart W, Ben-Shaul A, Roux D. Micelles, Membranes, Microemulsions and Monolayers. 1st ed. New York: Springer; 1994. [Google Scholar]
- 14.Yang L, Gordon VD, Mishra A, Som A, Purdy KR, Davis MA, Tew GN, Wong GCL. J. Am. Chem. Soc. 2007;129:12141. doi: 10.1021/ja072310o. [DOI] [PubMed] [Google Scholar]
- 15.Yang L, Gordon VD, Trinkle DR, Schmidt NW, Davis MA, DeVries C, Som A, Cronan JE, Tew GN, Wong GCL. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20595. doi: 10.1073/pnas.0806456105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Som A, Yang L, Wong GCL, Tew GN. J. Am. Chem. Soc. 2009;131:15102. doi: 10.1021/ja9067063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirafuji Y, Tanabe H, Satchell DP, Henschen-Edman A, Wilson CL, Ouellette AJ. J. Biol. Chem. 2003;278:7910. doi: 10.1074/jbc.M210600200. [DOI] [PubMed] [Google Scholar]
- 18.Tanabe H, Qu X, Weeks CS, Cummings JE, Kolusheva S, Walsh KB, Jelinek R, Vanderlick TK, Selsted ME, Ouellette AJ. J. Biol. Chem. 2004;279:11976. doi: 10.1074/jbc.M310251200. [DOI] [PubMed] [Google Scholar]
- 19.Figueredo S, Mastroianni JR, Tai KP, Ouellette AJ. In: Antimicrobial Peptides. Giuliani A, Rinaldi AC, editors. Vol. 618. Humana Press; 2010. p. 47. [Google Scholar]
- 20. http://ca.expasy.org/tools. [Google Scholar]
- 21.Garcia A, Osapay G, Tran P, Yuan J, Selsted M. Infect. Immun. 2008;76:5883. doi: 10.1128/IAI.01100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran D, Tran PA, Tang Y-Q, Yuan J, Cole T, Selsted ME. J. Biol. Chem. 2002;277:3079. doi: 10.1074/jbc.M109117200. [DOI] [PubMed] [Google Scholar]
- 23.Singh PK, Jia HP, Wiles K, Hesselberth J, Liu L, Conway B-AD, Greenberg EP, Valore EV, Welsh MJ, Ganz T, Tack BF, McCray PB. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14961. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taggart CC, Greene CM, Smith SG, Levine RL, McCray PB, O’Neill S, McElvaney NG. J. Immunol. 2003;171:931. doi: 10.4049/jimmunol.171.2.931. [DOI] [PubMed] [Google Scholar]
- 25. usaxs.xor.aps.anl.gov/staff/ilavsky/nika.html.
- 26. www.esrf.eu/computing/scientific/FIT2D/ [Google Scholar]
- 27.Wang Z, Wang G. Nucleic Acids Res. 2004;32:D590. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. http://aps.unmc.edu/AP/main.php. [Google Scholar]
- 29.Tang Y-Q, Yuan J, Ösapay G, Ösapay K, Tran D, Miller C, Ouellette A, Selsted M. Science. 1999;286:498. doi: 10.1126/science.286.5439.498. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Owen S, Rudolph D, Cole A, Hong T, Waring A, Lal R, Lehrer R. J. Immunol. 2004;173:515. doi: 10.4049/jimmunol.173.1.515. [DOI] [PubMed] [Google Scholar]
- 31.Yasin B, Wang W, Pang M, Cheshenko N, Hong T, Waring A, Herold B, Wagar E, Lehrer R. J. Virol. 2004;78:5147. doi: 10.1128/JVI.78.10.5147-5156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.While coexisting phases (such as lamellar and inverted hexagonal phases) are sometimes observed at specific lipid compositions the observed reflections cannot be accounted for without the presence of a Pn3m cubic phase. The strong induced formation of cubic phases like the Pn3m which are rich in saddle-splay curvature is a generic feature in the full phase diagram of defensin-lipid structures.
- 33.Schwarz US, Gompper G. Phys. Rev. Lett. 2000;85:1472. doi: 10.1103/PhysRevLett.85.1472. [DOI] [PubMed] [Google Scholar]
- 34.Keller SL, Gruner SM, Gawrisch K. Biochim. Biophys. Acta, Biomembr. 1996;1278:241. doi: 10.1016/0005-2736(95)00229-4. [DOI] [PubMed] [Google Scholar]
- 35.Prenner EJ, Lewis RNAH, Neuman KC, Gruner SM, Kondejewski LH, Hodges RS, McElhaney RN. Biochemistry. 1997;36:7906. doi: 10.1021/bi962785k. [DOI] [PubMed] [Google Scholar]
- 36.Hickel A, Danner-Pongratz S, Amenitsch H, Degovics G, Rappolt M, Lohner K, Pabst G. Biochim. Biophys. Acta, Biomembr. 2008;1778:2325. doi: 10.1016/j.bbamem.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 37.Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Nat. Immunol. 2000;1:113. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 38.Llenado RA, Weeks CS, Cocco MJ, Ouellette AJ. Infect. Immun. 2009;77:5035. doi: 10.1128/IAI.00695-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selsted ME, Miller SI, Henschen AH, Ouellette AJ. J. Cell Biol. 1992;118:929. doi: 10.1083/jcb.118.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmerberg J, Kozlov MM. Nat. Rev. Mol. Cell Biol. 2006;7:9. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 41.Siegel DP, Kozlov MM. Biophys. J. 2004;87:366. doi: 10.1529/biophysj.104.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harder J, Bartels J, Christophers E, Schroeder J-M. J. Biol. Chem. 2001;276:5707. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 43.Harder J, Bartels J, Christophers E, Schroder JM. Nature. 1997;387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 44.Starner TD, Agerberth B, Gudmundsson GH, McCray PB. J. Immunol. 2005;174:1608. doi: 10.4049/jimmunol.174.3.1608. [DOI] [PubMed] [Google Scholar]
- 45.Shearman GC, Ces O, Templer RH, Seddon JM. J. Phys.: Condens. Matter. 2006;18:S1105. doi: 10.1088/0953-8984/18/28/S01. [DOI] [PubMed] [Google Scholar]
- 46.Mishra A, Gordon V, Yang L, Coridan R, Wong G. Angew. Chem., Int. Ed. 2008;47:2986. doi: 10.1002/anie.200704444. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt N, Mishra A, Lai GH, Wong GCL. FEBS Lett. 2010;584:1806. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 48.Manning GS. J. Chem. Phys. 1969;51:924. [Google Scholar]
- 49.Zimm B, Bret M. J. Biomol. Struct. Dyn. 1983;1:461. doi: 10.1080/07391102.1983.10507455. [DOI] [PubMed] [Google Scholar]
- 50.Rädler JO, Koltover I, Salditt T, Safinya CR. Science. 1997;275:810. doi: 10.1126/science.275.5301.810. [DOI] [PubMed] [Google Scholar]
- 51.Wong GCL, Tang JX, Lin A, Li Y, Janmey PA, Safinya CR. Science. 2000;288:2035. doi: 10.1126/science.288.5473.2035. [DOI] [PubMed] [Google Scholar]
- 52.Yang L, Liang H, Angelini TE, Butler J, Coridan R, Tang JX, Wong GCL. Nat. Mater. 2004;3:615. doi: 10.1038/nmat1195. [DOI] [PubMed] [Google Scholar]
- 53.May S, Ben-Shaul A. Biophys. J. 1997;73:2427. doi: 10.1016/S0006-3495(97)78271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koltover I, Salditt T, Rädler JO, Safinya CR. Science. 1998;281:78. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- 55.Schug KA, Lindner W. Chem. Rev. 2004;105:67. doi: 10.1021/cr040603j. [DOI] [PubMed] [Google Scholar]
- 56.Wender PA, Galliher WC, Goun EA, Jones LR, Pillow TH. Adv. Drug Delivery Rev. 2008;60:452. doi: 10.1016/j.addr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chou T, Kim KS, Oster G. Biophys. J. 2001;80:1075. doi: 10.1016/S0006-3495(01)76086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dommersnes PG, Fournier J-B. Biophys. J. 2002;83:2898. doi: 10.1016/S0006-3495(02)75299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fošnarič M, Kralj-Iglič V, Bohinc K, Iglič A, May S. J. Phys. Chem. B. 2003;107:12519. [Google Scholar]
- 60.Kokryakov VN, Harwig SSL, Panyutich EA, Shevchenko AA, Aleshina GM, Shamova OV, Korneva HA, Lehrer RI. FEBS Lett. 1993;327:231. doi: 10.1016/0014-5793(93)80175-t. [DOI] [PubMed] [Google Scholar]
- 61.In Figure 4 we exclude the amino acids cysteine, proline, and glycine which primarily influence AMP tertiary structure and are not directly involved in the cationicity or hydophobicity of AMPs. Also we consider only arginine and lysine in our exchange rate concept since they constitute the vast majority (89%) of cationic AA found in the 1,080 AMPs.
- 62.Kyte J, Doolittle RF. J. Mol. Biol. 1982;157:105. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 63.Eisenberg D, Weiss RM, Terwilliger TC, Wilcox W. Faraday Symp. Chem. Soc; 1982. p. 109. [Google Scholar]
- 64.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Nature. 2005;433:377. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 65.Hsu C-H, Chen C, Jou M-L, Lee AY-L, Lin Y-C, Yu Y-P, Huang W-T, Wu S-H. Nucleic Acids Res. 2005;33:4053. doi: 10.1093/nar/gki725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park CB, Kim HS, Kim SC. Biochem. Biophys. Res. Commun. 1998;244:253. doi: 10.1006/bbrc.1998.8159. [DOI] [PubMed] [Google Scholar]
- 67.Yonezawa A, Kuwahara J, Fujii N, Sugiura Y. Biochemistry. 1992;31:2998. doi: 10.1021/bi00126a022. [DOI] [PubMed] [Google Scholar]
- 68.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang Y-H, Homey B, Cao W, Wang Y-H, Su B, Nestle FO, Zal T, Mellman I, Schroder J-M, Liu Y-J, Gilliet M. Nature. 2007;449:564. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 69.Ganz T. Blood. 2003;102:783. doi: 10.1182/blood-2003-03-0672. [DOI] [PubMed] [Google Scholar]
- 70.Rossman JS, Jing X, Leser GP, Lamb RA. Cell. 2010;142:902. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.