Summary
A-kinase anchoring proteins (AKAPs) regulate cyclic AMP-dependent protein kinase (PKA) signaling in space and time. Dual-specific AKAP 2 (D-AKAP2) binds to the dimerization/docking (D/D) domain of both RI and RII regulatory subunits of PKA with high affinity. Here, we have determined the structures of the RIα D/D domain alone and in complex with D-AKAP2. The D/D domain presents an extensive surface for binding through a well-formed N-termina helix and this surface restricts the diversity of AKAPs that can interact. The structures also underscore the importance of a redox-sensitive disulfide in affecting AKAP binding. An unexpected shift in the helical register of D-AKAP2 compared to the RIIα:D-AKAP2 complex structure makes the mode of binding to RIα novel. Finally, the comparison allows us to deduce a molecular explanation for the sequence and spatial determinants of AKAP specificity.
Introduction
Numerous signaling pathways are regulated by phosphorylation of target proteins, a phenomenon in which cyclic AMP (cAMP)-dependent protein kinase (PKA) plays a central role. Correct spatio-temporal localization of signaling molecules such as PKA is central to the intricacies of signal transduction. A family of proteins called the A-kinase anchoring proteins (AKAPs) carries out this function for PKA and characterization of PKA:AKAP complexes is thus important for understanding the mechanisms of regulation.
PKA is a homodimer of regulatory (R) subunits bound to two catalytic (C) subunits, and based on the R subunit, the enzyme is classified as type I or II (with α and β subclasses) (Skalhegg and Tasken, 1997). While all four R subunits share a similar domain organization, they are not functionally redundant and are localized differently in the cell. For example, RIα-null mice are embryonically lethal, showing that its function is crucial for normal development (Amieux et al., 1997). Mutations in the RIα gene cause familial cardiac myxomas and Carney complex (Casey et al., 2000; Kirschner et al., 2000). Whereas RII subunits are localized to discrete parts of the cell, RI subunits are usually distributed diffusely in the cell (Wong and Scott, 2004) and then dynamically recruited to discrete sites such as the cap site of activated lymphocytes (Levy et al., 1996; Skalhegg et al., 1994).
Each R subunit is comprised of a dimerization/docking (D/D) domain at the N-terminus followed by a linker containing an inhibitor site and finally, two cAMP binding domains (Figure 1A) (reviewed in Taylor et al. (Taylor et al., 2005)). The D/D domains form an X-type, anti-parallel four-helix bundle that serves as a docking surface for AKAPs (Banky et al., 1998; Harris et al., 1994; Newlon et al., 2001), which interact with the docking surface through an amphipathic helix (ca. 16 residues) (Banky et al., 1998; Gold et al., 2006; Kinderman et al., 2006; Newlon et al., 2001). While AKAPs were initially identified by their ability to bind RII subunits, in recent years several AKAPs such as D-AKAP1 (Huang et al., 1997b), D-AKAP2 (Huang et al., 1997a), AKAP220 (Reinton et al., 2000), Merlin (Gronholm et al., 2003) and Pap7 (Li et al., 2001) have been shown to bind RI subunits as well, thereby making them dual-specific.
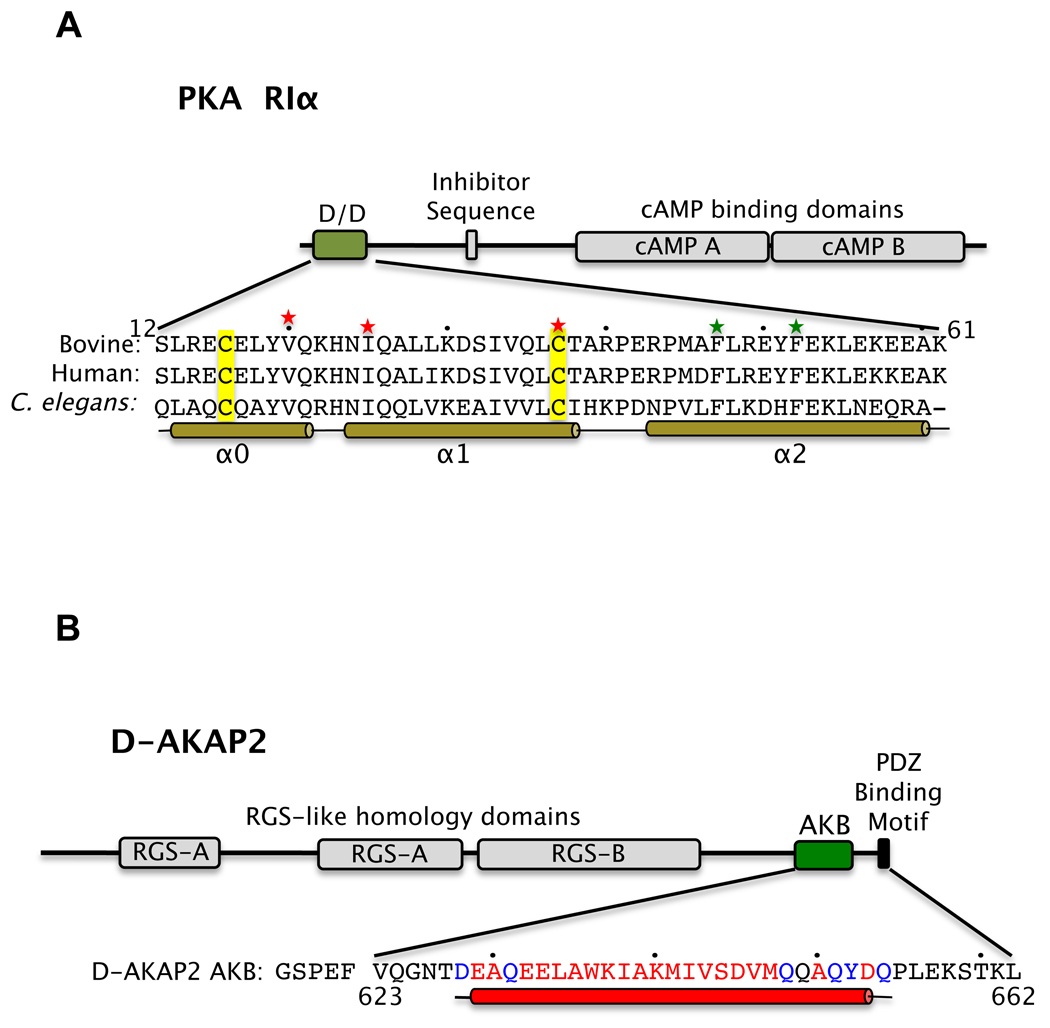
Figure 1. The functional domain structures of D-AKAP2 and RIα.
The domain structures of RIα (A) and D-AKAP2 (B) are shown. In RI, the Dimerization/Docking (D/D) domain, the inhibitor sequence and the two cAMP-binding domains are labeled. In D-AKAP2, the RGS-like domains, the A-kinase Binding (AKB) region and the C-terminal PDZ motif are indicated. The sequences of the RIα D/D domain and the D-AKAP2 AKB region used for crystallization are shown and numbered. Residues involved in α-helices are indicated by a solid bar and every tenth residue is indicated by a dot. In RIα D/D, green and red stars indicate residues crucial for dimerization and docking to AKAP, respectively. The conserved Cys residues are highlighted in yellow. In D-AKAP2, residues not modeled and modeled as Ala in the structure are indicated in black and blue respectively.
Dual-specific AKAP 2 (D-AKAP2) is a multi-subunit protein containing two putative regulator of G-protein signaling (RGS)-like homology domains, followed by a 27-residue PKA binding (AKB) domain and finally a PSD-95/DlgA/ZO-1 (PDZ)-binding motif at the C-terminus (Figure 1B) (Huang et al., 1997a). The AKB domain of D-AKAP2 can bind to the D/D domains of both RI and RII with high affinity (KD = 48 and 2 nM, respectively) (Burns et al., 2003), showing that the motif has all the information necessary and sufficient for binding both R subunits.
The binding of the AKB region to R subunits has been characterized extensively using biochemical, biophysical and structural methods. These include mutational studies of RI and RII to identify residues involved in dimerization and anchoring (Banky et al., 1998; Burns et al., 2003) and hydrogen/deuterium exchange mass spectrometry (H/D MS) of RI and RII bound to the D-AKAP2 AKB to determine the regions that are protected upon binding (Burns-Hamuro et al., 2005). Peptide arrays were used to determine RI versus RII specificity and design isoform-specific peptides to disrupt the PKA:AKAP interaction (Alto et al., 2003; Burns-Hamuro et al., 2003; Carlson et al., 2006; Gold et al., 2006). Finally, SNP analyses showed that a single amino acid change (Val to Ile) in the AKB region results in reduced binding to RIα compared to RIIα and changes in the cellular distribution of RIα (Kammerer et al., 2003). This SNP is associated with myocardial infarction (Nishihama et al., 2007; Yoshida et al., 2007), heart rate dysregulation (Tingley et al., 2007) and familial breast cancer (Wirtenberger et al., 2007).
The initial steps toward establishing a D/D domain:AKAP interaction were the NMR structure determinations of RIIα D/D (Newlon et al., 2001) and RIα D/D (Banky et al., 2003). Both structures have a similar four-helix bundle organization but unique to the RIα D/D domain is an N-terminal helix (named as α0 or N-1 helix) that is stabilized by an inter-subunit disulfide bond (Figure 1 A). While this helix is well formed, its position relative to the four-helix core is variable due to the flexibility of the α0-α1 loop (Banky et al., 2003). Recently it was also shown in cardiac myocytes that upon H2O2 treatment, there is increased disulfide bond formation in the RIα D/D and nuclear localization of PKA, an effect attributed to the enhanced anchoring by AKAP (Brennan et al., 2006). The presence of the N-terminal helix and the disulfide bonds in the RIα isoform begs the question - what roles do they play in anchoring AKAPs?
In order to further understand the molecular basis of AKAP anchoring, structures of RIIα D/D with D-AKAP2 (Kinderman et al., 2006) and with an RII-selective peptide, AKAP-IS (Gold et al., 2006) were recently determined. Through an amphipathic helix, the asymmetric sequences of the AKAPs bind diagonally across the symmetric D/D domain. Interestingly, the N-terminus of one monomer is ordered and makes critical interactions with the AKAP, while the N-terminus of the other monomer is disordered. This flexibility was suggested to give the RII isoform the ability to bind a wider variety of AKAP sequences.
The RIIα D/D:D-AKAP2 structure was used to predict isoform differences of binding. For example, it was proposed that the disulfide bonded Cys residues in RIα would be structurally equivalent to an Ile and Leu of RIIα and therefore, would play equivalent roles in AKAP binding. Further, explanations for strong preferences for certain residues in D-AKAP2 for RIIα over RIα were also made based on a predicted RIα:D-AKAP2 binding. In summary, while these structures yielded important clues, the central question still remained - what are the structural determinants that render D-AKAP2 dual-specific and AKAPs in general, RI-, RII- or dual-specific?
To answer these questions and to complete the atomic-level picture of the dual-specificity of D-AKAP2, we determined the crystal structures of the RIα D/D alone and in complex with the D-AKAP2 AKB. Extrapolating biochemical and biophysical data onto the structures allow us to gain insights into the roles of the N-terminal helix and disulfide bonds in AKAP binding. An unexpected and novel finding is the shift in the helical register of D-AKAP2 when bound to RIα D/D relative to RIIα D/D. While this result negates predictions made about AKAP binding to RIα, it now allows us to base all conclusions about isoform-specific AKAP binding on a strong structural foundation. A detailed comparison with the RIIα D/D:D-AKAP2 complex strongly suggests that this differential mode of binding to the PKA R subunits is true for all AKAPs.
Results and Discussion
Overall Apo RIα D/D Structure
Apo RIα D/D crystals contain a monomer in the asymmetric unit, with the obligatory homodimer generated by a crystallographic 2-fold axis. Sulfur single wavelength anomalous diffraction (SAD) data were used to carry out de novo phasing of the structure (Table 1). To rationally compare the structure with existing D/D domain structures, the other half of the dimer was generated by a crystallographic transformation. As with other D/D domains, the RIα D/D dimer is comprised of an X-type anti-parallel, four-helix bundle with each monomer contributing two helices, α1 and α2, to the hydrophobic dimer interface (Figure 2A) (Banky et al., 2003; Newlon et al., 2001). α0 at the N-terminus also contributes to the dimer interface through two intermolecular disulfide bonds between Cys16 and Cys37 (Figures 2A and 2B) (Banky et al., 2003). Tyr19 and Val20 stabilize the disulfide bond through packing interactions, and stacking interactions with His23, in turn, stabilize Tyr19 (Figure 2A) (Figure S1A).
Table 1.
Data collection and refinement statisticsa
| Data collection and phasing | Apo RIα D/D (S-SAD) | Apo RIα D/D | RIα D/D:D-AKAP2 | Apo RIα D/D |
|---|---|---|---|---|
| Radiation sourceb | APS; 23-ID | APS; 22-BM | ALS; 8.2.2 | In-house |
| Wavelength (Å) | 1.9 | 1.0 | 1.0 | 1.54 |
| Number of images | 804 | 360 | 200 | 36 |
| Space group | P6222 | P6222 | P212121 | P6222 |
| Unit cell (a, b, c) (Å) | 44.0, 44.0, 92.9 | 44.1, 44.1, 93.0 | 40.5, 56.2, 57.1 | 44.1, 44.1, 93.2 |
| Resolution (Å) | 100–2.40 (2.49–2.40) | 100–2.00 (2.07–2.00) | 100–2.30 (2.38–2.30) | 100–2.90 (3.00–2.90) |
| Unique reflections | 2202 (135) | 4047 (387) | 6221 (601) | 1377 (134) |
| Multiplicity | 52.0 (10.0) | 36.9 (27.6) | 7.5 (7.8) | 3.6 (3.6) |
| Completeness (%) | 91.8 (59.2) | 99.8 (100) | 99.9 (100) | 96.0 (97.8) |
| Rmeasc (%) | 8.7 (16.8) | 8.1 (36.7) | 7.2 (41.6) | 6.7 (23.3) |
| I/σ | 34.6 (7.9) | 32.4 (11.5) | 14.5 (5.5) | 17.7 (6.2) |
| FOM – Solved | 0.28 | |||
| FOM – Resolved | 0.47 | |||
| Refinement | ||||
| Resolution (Å) | 100–2.00 | 100–2.30 | ||
| Number of reflections | 3929 | 6015 | ||
| Number of amino acids | 50 | 126 | ||
| Number of solvent molecules | 29 | 30 | ||
| Number of Zn+2 ions | – | 5 | ||
| Total number of atoms | 463 | 1058 | ||
| Average B (all atoms) (Å2) | 38.6 | 53.2 | ||
| R/Rfree (%) | 18.7/24.9 | 18.6/25.4 | ||
| RMSD bonds (Å) | 0.006 | 0.007 | ||
| RMSD angles (°) | 0.835 | 0.949 | ||
| ϕ/ψ favored region (%)e | 100 | 100 | ||
Numbers in parentheses correspond to values in the highest resolution shell.
APS, Advanced Photon Source, Argonne, IL, USA and ALS, Advanced Light Source, Berkeley, CA, USA.
Rmeas is the multiplicity weighted merging R factor (Diederichs and Karplus, 1997).
Figure of Merit (FOM) both before (Solve) and after (Resolve) density modification.
Ramachandran plot quality as defined in Molprobity (Lovell et al., 2003).
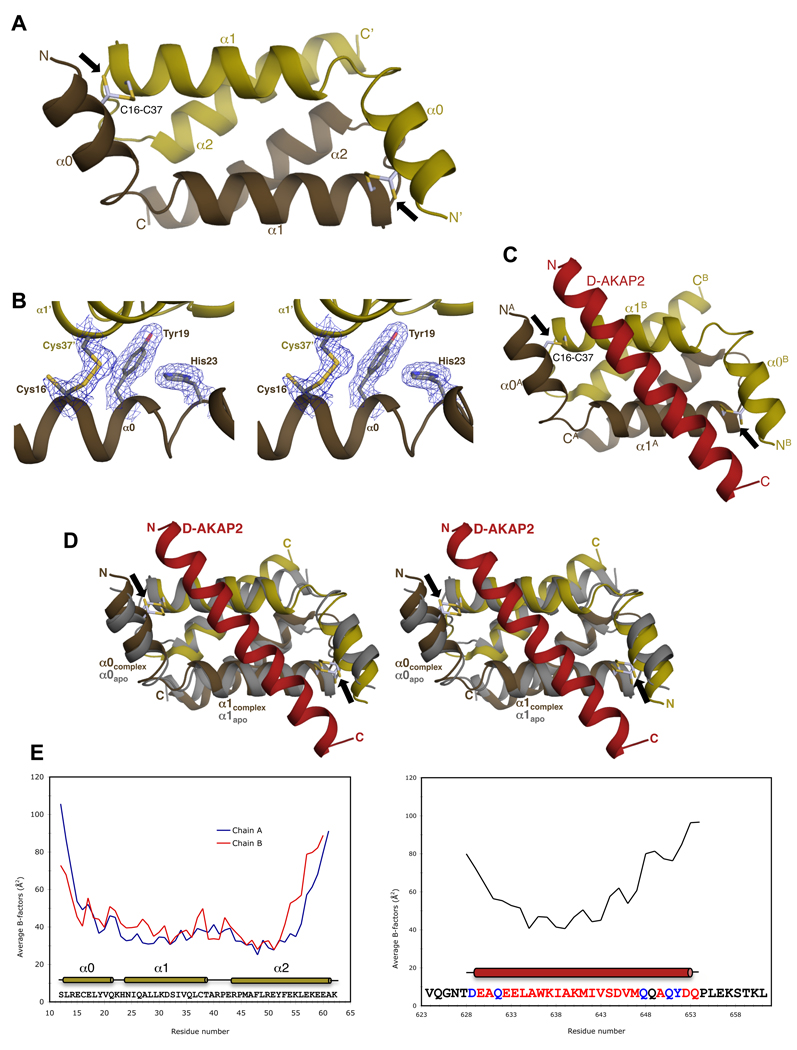
Figure 2. Overview of Apo RIα D/D and RIα D/D:D-AKAP2 structures.
(A) Overall structure of Apo RIα D/D showing the anti-parallel, four-helix bundle. The D/D domain monomer is colored gold with the other monomer, generated by the crystallographic 2-fold symmetry, colored brown. The termini and the helices are labeled. The intermolecular disulfide bonds between residues Cys16 and Cys37 are shown as ball-and-stick models and indicated by a black arrow. (B) Stereoview of the 2Fo–Fc density map contoured at 1 ρrms (root-mean-square electron density of map often reported as σ) shows clear density for the partially reduced disulfide bond and packing residues. The secondary structure elements and the residues are labeled. (C) Overall structure of the RIα D/D domain complexed with the AKB region of D-AKAP2. The monomers of the D/D domain are depicted in gold and brown and D-AKAP2 is shown in red. The locations of the disulfide bonds are indicated by black arrows. (D) Stereoview of overlay of the complex RIα D/D dimer with the apo RIα D/D. RIα D/D:D-AKAP2 structure is colored as Figure 2C and apo RIα D/D is colored gray. Secondary structural elements and the termini are labeled. Black arrows indicate the position of the disulfide bonds. (E) Correlation of crystallographic B-factors with H/D-exchange protection data. The average main chain B-factors of the two monomers of RIα D/D and the AKB region of D-AKAP2 are plotted against residue number. The sequence of the crystallized proteins and the location of the secondary structural elements are indicated below the plot. Residues not modeled or modeled as Ala in the structure are in black and blue respectively. In previous H/D MS experiments, the core of the AKB region showed increased protection from solvent upon binding to the D/D domain consistent with lower B-factors. At the termini, the lack of protection is consistent with the higher B-factors and weak electron density.
The intermolecular disulfides in the structure warrant a comment. Cys16 exists in two alternate conformations, with the major conformation being the oxidized disulfide conformation (0.7 occupancy, see Experimental Procedures) (Figure 2B). A control data set collected at the in-house X-ray source on the apo crystal shows an intact disulfide, thereby confirming that disulfide bond cleavage is an artifact of synchrotron radiation (Table 1). For this reason, we will treat it as a disulfide. In our model, therefore, one monomer forms two head-to-tail disulfides with a symmetry-generated monomer.
In general, the B-factors of the core and the C-terminus correlate well with previous backbone amide H/D MS experiments (Burns-Hamuro et al., 2005). At the N-terminus Arg14 and Glu15 form salt bridges with Glu53′ and Arg43′ respectively, via crystal packing interactions (‘prime’ symbol denotes symmetry related residues) resulting in lower than average B factors (not shown). In solution and under oxidizing conditions, the disulfide bond would primarily stabilize the N-terminal region. This interpretation of the N-terminal helix is also consistent with the apo RIα D/D NMR structures (Figure S1)(Banky et al., 2003).
RIα D/D:D-AKAP2 Complex
Overall Structure
The RIαD/D:D-AKAP2 complex crystallizes with one RIα D/D dimer bound to D-AKAP2 in the asymmetric unit (Table 1). While the majority of the residues of the RIα D/D have clear electron density, ~15 residues of D-AKAP2 at the N- and C-termini have little or no density due to disorder and, therefore, not modeled (Figure 1B).
D-AKAP2 binds diagonally across (~55°) the face of the hydrophobic groove formed by the two monomers burying ~710 Å2 of the D/D interface making extensive hydrophobic and polar interactions with the dimer. In the complex, RIα D/D is oxidized and maintains its four-helix bundle structure (Figure 2C). The N-terminal helices of both monomers are ordered and structurally similar to each other. The monomer whose N-terminus interacts with N-terminus of D-AKAP2 is labeled as ‘A’, and the other monomer is labeled ‘B’. Only residues from α0 and α1 play a role in stabilizing the D-AKAP2 binding. The root mean square deviation (RMSD) between the two RIα D/D monomers in the complex structure is ~0.5 Å (49 equivalent Cα atoms), indicating that small differences exist. On the other hand, the RMSDs between the dimer of the complex structure and apo structure are ~1.4 Å (98 equivalent Cα atoms), suggesting that the organization of the two monomers undergoes rearrangements, mainly in the α0 and α1 regions, upon binding to D-AKAP2 (Figure 2D). For example, the side chains of Leu29 and Ile33 residues undergo large conformational changes to accommodate AKAP residues. Residues Gln26 and Lys30 present a different rotamer to make hydrogen bonds with residues Glu632, Asp645 and Gln649 of D-AKAP2, thereby stabilizing the helix.
Unlike the apo structure, the N- and C-termini of the RIα D/D and D-AKAP2 are not tethered artificially by crystal packing interactions and are relatively more mobile as indicated by their higher than average B-factors (Figure 2E). Since the present structure is consistent with previous H/D MS experiments (Burns-Hamuro et al., 2005), we can conclude that the structure is an accurate representation of how AKAPs interact with the RIα D/D domain.
Stabilizing Interactions
While the same residues from each monomer are involved, high affinity binding to D-AKAP2 is achieved by local specific changes in the D/D structure (Figure 3A). We describe the interaction interface in a systematic manner for three main reasons: one, to provide a context for the structural determinants of binding; two, to set the stage for comparing other D/D:AKAP complexes with the present structure; and finally, to provide a stable foundation for modeling other AKAPs onto the structure.
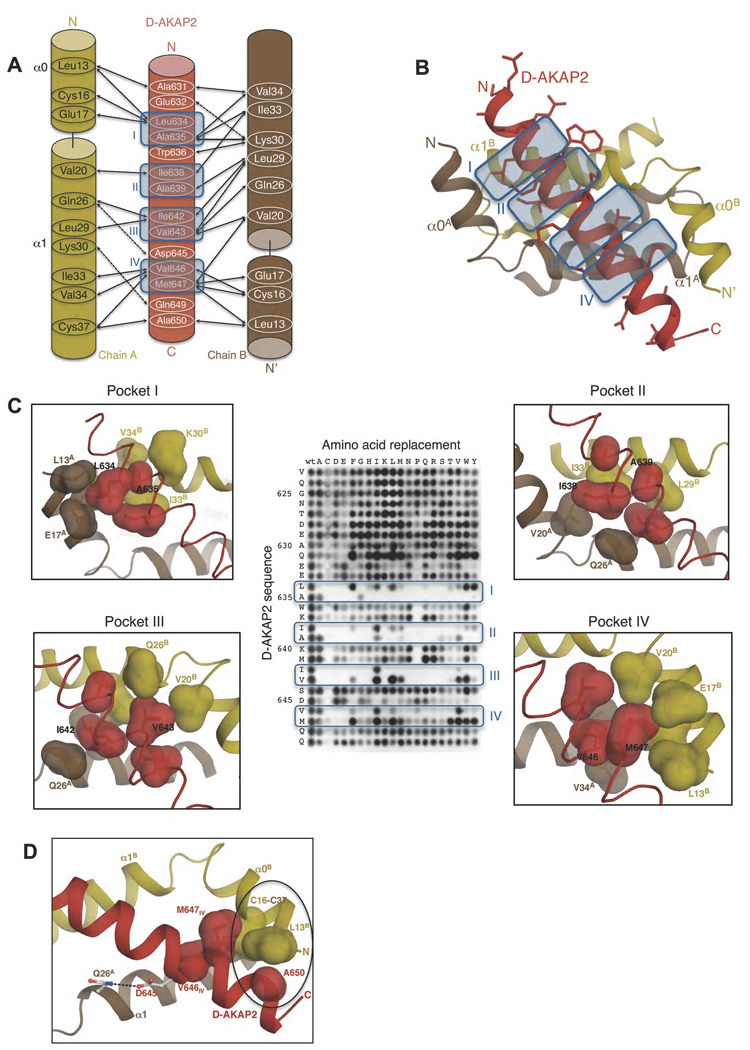
Figure 3. Interactions between RIα D/D and D-AKAP2.
(A) Schematic figure detailing the interactions between the side chains of D-AKAP2 and RIα D/D monomers. Solid and dashed arrows indicate hydrophobic and polar interactions respectively. Pockets I-IV are indicated by blue-colored boxes and numbered. (B) Overall structure showing the interaction. The structures are colored as panel (A). The side chains of D-AKAP2 are also shown to indicate the interface of binding. The pockets are indicated by blue-colored boxes and numbered. (C) Close-up views of pockets I through IV correlated with peptide array data. The coloring scheme is similar to Figure 3A. Previous peptide array data (Burns-Hamuro et al., 2003) are shown to highlight the stringent requirements for D-AKAP2 binding to RIα D/D (Copyright 2003, National Academy of Sciences, U.S.A). The surfaces of residues involved in each pocket are shown to highlight the tight packing interactions at the interface. (D) View of additional hydrophobic and polar interactions that stabilize RIα D/D:D-AKAP2 interaction. The coloring scheme is similar to Figure 3A. The residues involved in the hydrophobic interaction are represented as surfaces and circled. Hydrogen bonds between residues are indicated by dashed lines. For orientation, pocket IV residues, V646 and M647, are also shown.
There are four main regions or pockets at the interface, pockets I through IV (Figure 3). The residues of the D/D domain lining the pockets define the amino acids that can bind to the pockets and which residues are precluded. Correspondingly, binding specificity is achieved by the specific AKAP sequence that is bound to the pocket. This complementarity defines the specificity of the D/D domain: AKAP interaction.
Pocket I, comprised of Leu13A, Cys16A, Glu17A, Val34B and Ile33B (Chains A and B of the D/D domain are denoted as a superscript), is occupied by two hydrophobic residues, Leu634 and Ala635, from the second turn of the AKAP helix (Figure 3C). Two residues of pocket I, Val34B and Ile33B, are tightly packed such that the space can accommodate only an Ala residue; any other residue would be sterically occluded.
Pockets II and III lie along the non-crystallographic 2-fold axis, and therefore identical residues from both the chains of the D/D domains are involved in the interaction. The local conformational differences, however, provide variability in binding. Pocket II is occupied by Ile638 and Ala639 from the third turn of the AKAP helix, which pack against Val20A, Ile33B and Leu29B (Figure 3C). Pocket III is occupied by Ile642 and Val643 from the fourth turn of the AKAP helix which interact with Leu29A, Ile33A and Val20B (Figure 3C). Ile642, in particular, fits very snugly into the hydrophobic pocket and is therefore likely to be a unique fit to the pocket.
Pocket IV, occupied by Val646 and Met647 from the fifth turn of the AKAP helix, has similar residues lining it as pocket I. Unique to his pocket is the involvement of Cys37A, the Cβ atom of which packs against Val646. In contrast to pocket I, the residues in this pocket are not as tightly packed as compared to the other pockets and therefore would offer more flexibility in accommodating other residues (Figure 3C). For example, in order to accommodate Val646, Ile33A undergoes a conformational change compared to the apo structure and to Ile33B of the other monomer.
Additional hydrophobic interactions and salt bridges increase the stability of the interaction. Toward the N-terminus, Trp636 makes hydrophobic interactions with the aliphatic chain of Lys30B. Similarly, at the C-terminus, Ala650 packs against Cys37A and Leu13B (Figure 4). Exposed AKAP side chains interact with either solvent molecules or D/D domain side chains. For example, Glu632, Asp645 and Gln649 form hydrogen bonds with Lys30B, Gln26A and Lys30A respectively (Figure 3D).
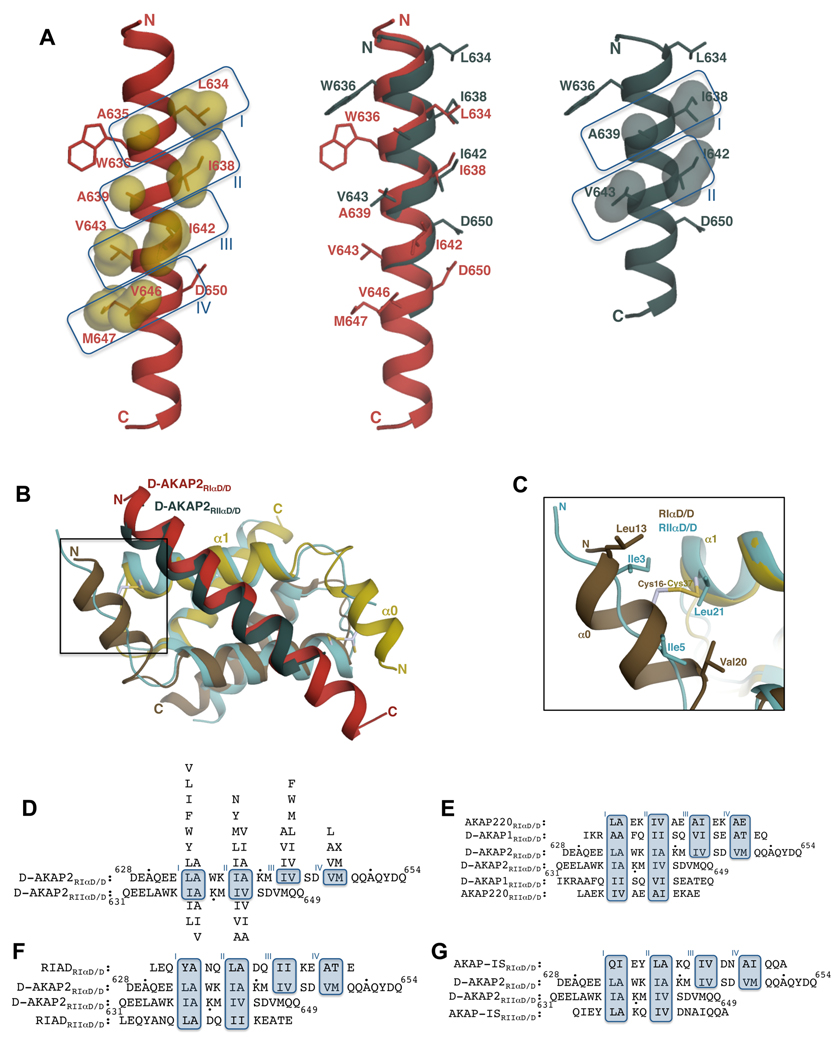
Figure 4. Comparison of RIα D/D:D-AKAP2 and RIIα D/D:D-AKAP2 structures revealing D-AKAP2 specificity.
(A) Structural overlay of D-AKAP2 bound to RIα D/D (in red) and to RIIα D/D (in teal; PDB code: 2HWN) (Kinderman et al., 2006) highlighting the shift in the helical register. Pocket residues are labeled. Residues Trp636 and Asp650 at the termini are also shown to facilitate visualization of the register shift. D-AKAP2 bound to RIα (left) and RIIα (right) are also shown to indicate the pocket positions. Pocket residues are represented as surfaces. (B) Structural overlay of RIα D/D:D-AKAP2 complex with the RIIα D/D:D-AKAP2 complex. The RIα D/D complex is colored as Figure 3A. The D/D domain of RIIα is colored light blue and the D-AKAP2 peptide is colored teal. The helices and the termini are labeled with the respective colors. The N-terminal region that houses the disulfide bond in RIα is boxed. (C) A close-up view of the N-terminal region showing the disulfide bond region of RIα D/D and the equivalent residues from RIIα D/D. For clarity, the D-AKAP2 peptides bound to the two structures are not shown. Cys37 is structurally equivalent to Leu21 of RIIα whereas Cys16 does not have any structurally equivalent counterparts. (D) Structure-based alignment of D-AKAP2 sequence bound to RIα and RIIα. The residue numbers of the N- and C-termini of D-AKAP2 are shown and every tenth residue indicated by a black dot. Due to the shift in the helical register, different residues occupy structurally equivalent pockets of RIα and RIIα. Binding pockets are boxed and labeled. Residues that can replace the pocket residues without disrupting the interaction are shown above (for RIα) and below (for RIIα) the alignment in order of preference. (E–G) Alignment of dual-specific (E), RI-specific (F) and RII-specific (G) AKAPs to the aligned D-AKAP2 sequences according to their expected mode of binding to both RIα and RIIα. Dual-specific AKAPs satisfy the requirements for binding to both R subunits whereas RI- or RII-specific AKAPs satisfy criteria to binding to RI or RII subunits.
Significance of the Disulfide Bond in AKAP Binding
Recently, Brennan et al. reported that treatment with H2O2 resulted in increased disulfide formation and nuclear localization of PKA (Brennan et al., 2006). The disulfide bond was suggested to play a role in the anchoring of AKAPs, which in turn promoted the increased nuclear localization. As mentioned earlier, the D/D dimer exists in the oxidized state in the complex and is therefore, a physiologically relevant AKAP-bound form. In order to understand the effect of the disulfide bond and other structurally close residues on AKAP binding, we have generated several mutants.
Tyr19 and His23, which lie in close proximity to the disulfide bond, when mutated to Ala interfered with disulfide bond formation without disrupting the dimer (Figure 2B) (Figure S2). This is similar to the effects of mutating the two Cys residues to Ala (Banky et al., 1998). The Y19A mutant protein is predominantly reduced and the H23A mutant exists as a mixture of reduced and oxidized proteins suggesting that its role in maintaining the disulfide bond integrity is not as important as Tyr19 (Figure S2). Tyrosines are often found to stabilize disulfide bonds, and our structure shows tight packing of the disulfide bond on the aromatic ring of Tyr19 (Petersen et al., 1999; Zauhar et al., 2000).
Using fluorescence anisotropy experiments, the Cys16 and Cys37 mutants of RIα showed a 3-fold and a 16-fold reduction in binding to D-AKAP2 compared to the wild-type protein (Table 2). On the other hand, the Y19A mutant showed a 27-fold decrease and the H23A mutant a 4-fold decrease in affinity. The fact that the Y19A mutant has the greatest effect on D-AKAP2 binding clearly suggests that its role in AKAP binding is greater than just disulfide stabilization.
Table 2.
Effect of RIα mutations on D-AKAP2 bindinga
| RIα construct | KD (nM) |
|---|---|
| Wild type | 48 |
| C16A | 126 (3-fold)b |
| C37A | 745 (16-fold) |
| Y19A | 1329 (27-fold) |
| H23A | 198 (4-fold) |
KD=EC50 calculated from curves fit to 1:1 binding model.
Values in parentheses denote the relative decrease in binding.
Disulfide bond formation upon H2O2 treatment would decrease the flexibility of the α0 helix relative to the core, and more importantly, it would result in the assembly of pockets I and IV (Figure 3C). This assembly would partly be stabilized by Tyr19 and His23. This ordering of the pockets would position critical residues such as Leu13 and Cys16 in close proximity to the AKAP, resulting in improved binding of AKAPs to RIα.
Structure Confirms Biophysical and Biochemical Studies
Mutational Data
Mutation of Val20 and Ile25 to Ala disrupts binding to AKAPs (Banky et al., 1998). Val20 is part of pockets II and III and is therefore important for D-AKAP2 binding (Figures 3A and 3B). Interestingly, Ile25 does not directly interact with D-AKAP2. A comparison with the apo structure reveals that in order to accommodate D-AKAP2, Ile33 (pockets I and IV) undergoes a conformational change that is facilitated by a shift in the position of Ile25 (Figure 3C). Thus, an I25A mutation would disrupt the packing of Ile33, which in turn would disrupt the integrity of pockets I and IV.
Peptide Array Information
Peptide array analyses first highlighted the importance of the four turns of the helix for D-AKAP2 binding to the D/D domain of RIα (Burns-Hamuro et al., 2003) and our structure fully supports this conclusion. In general, there is decreased tolerance for mutations in pockets I, II and III. Ala635, as noted earlier, is tightly nestled in pocket I so that any mutation would result in disruption of binding. Indeed, this is confirmed by the peptide array, which shows that any mutation to the Ala635 position results in the loss of binding with RIα (Figure 3C). In pocket II, Ala639 can be changed only to a Val or an Ile (Figure 3C). The side chain of Trp636 packs against the AKAP helix and the structure suggests that in its absence more changes could be tolerated at position 639 due to increased flexibility. The availability of structures allows us to deduce such correlated changes and also provides us with the ability to predict specificity for unknown AKAP peptides. Finally, the physiologically relevant Val646 to Ile SNP would result in a local shift of residues in order to accommodate the Cδ1 atom, which would then pack against the disulfide bond. This change to Ile would be tolerated for binding but would likely result in a local disruption of structure leading to reduced affinity (Figure 3C).
In another study, Carlson and co-workers (Carlson et al., 2006) systematically mutated residues of a consensus sequence and tested it for RIα D/D binding. Three residues found to significantly increase binding to RIα compared to RIIα were Glu, Asp and Ile corresponding to positions occupied by Trp636, Ser644 and Val646, respectively, in D-AKAP2. While Ile would make a hydrophobic interaction, the other two residues would be involved in stabilizing the helix through salt bridges or hydrogen-bonding interactions with D/D domain residues. These data highlight the observation that the helix-stabilizing interactions, as well as the core hydrophobic interactions in pockets, are important for high affinity binding of AKAPs to the RIα D/D domains.
Shift in the Helical Register Presents a Novel Interface for Binding
To deduce the isoform-specific differences of AKAP anchoring, in this study we compared the RIα D/D complex structure with the RIIα D/D structure in complex with a similar D-AKAP2 peptide.
An unexpected and significant difference between the two complex structures is the shift in the α-helical register by a complete turn! With RIα D/D:D-AKAP2 structure as a reference, a structural alignment of RIα D/D and RIIα D/D shows that the D-AKAP2 peptide bound to the RIIα structure is shifted toward the N-terminus by a single helical turn (Figure 4A). Thus, pocket I of the RIα D/D is occupied by D-AKAP2 residues Leu634 and Ala635, whereas the structurally equivalent pocket of the RIIα D/D is filled by Ile637 and Ala639 (from the next α-helical turn). Similarly, pocket II is occupied by Ile638 and Ala639 in RIα D/D and by Ile642 and Val643 in RIIα D/D. Based on a structure-based sequence alignment pockets I and II are occupied in both RI and RII but pockets III and IV are unique in RIα D/D (Figures 4A). Both the RIα and RIIα D/D domains present a hydrophobic interface for AKAP interaction, so the determination of only one of the two structures would not have been sufficient to predict this shift of the helix. The implications of the helical shift for AKAP specificity are discussed in the following section.
Overall, the cores of the D/D domains align very well with each other and their similarity extends to the angle of binding to the AKAP (Figure 4B). The AKAP interaction interface is less extensive with RIIα D/D than with RIα D/D. In RIIα D/D, the binding of the asymmetric D-AKAP2 sequence induces a pronounced asymmetry in the monomers; the N-terminus of only one monomer becomes ordered and makes hydrophobic interactions with the AKAP (Figure 4B). This flexibility gives it the ability to bind tightly to a wider variety of AKAP sequences. On the other hand, both of the N-termini of RIα D/D are ordered by virtue of the interchain disulfide bonds. Thus, the asymmetric D-AKAP2 sequence induces subtle changes in the monomers. This inflexibility decreases the potential of RIα to bind to as many AKAPs as RIIα. The ordering of the N-termini results in additional contacts with D-AKAP2, which in turn results in a longer, ordered helix (two additional turns of the helix compared to the RIIα: D-AKAP2 structure).
In the case of RIIα D/D, Ile3 and Ile5 are part of the ordered N-terminus making critical hydrophobic interactions with Leu634 and Ala635 of D-AKAP2 in pocket I of RIIα D/D (nomenclature as defined in this study). The other monomer contributes Leu21 for the stabilization of the AKAP peptide. It was suggested that Ile3 and Leu21 play structurally equivalent roles to Cys16 and Cys37 in the RIα D/D structure (Gold et al., 2006; Kinderman et al., 2006). From the present structure, it is evident that Cys37 is structurally similar to Leu21, as predicted. However, Cys16 is not spatially aligned with Ile3. The only residue that is spatially close to Ile3 is Leu13 from RIα D/D though it does not make equivalent interactions with the AKAP (Figure 4C). Thus, the present structure allows us to validate previous predictions made about the roles of RIα D/D residues in D-AKAP2 binding.
Basis for RI and RII Specificity
The specific and tight binding of AKAPs to the D/D domains is determined by the AKAP sequence and the spatial restrictions placed by the residues lining the pockets. Below, we describe the specific requirements for binding to R subunits and then propose a general model for AKAP binding to R subunits.
Requirements for RI and RII binding
In pocket I of RIα D/D, any hydrophobic residue can replace position Leu634 of D-AKAP2 whereas Ala635 is a unique fit. In pocket II, residues Ile638 and Ala639 fit snugly in the space. Based on the structure, similar residues such as Leu or Val could replace Ile638. Peptide array analyses suggest that larger hydrophobic residues can also occupy this position. Either Val or Ile can replace Ala639 but the longer Leu side chain disrupts the pocket. The restricted space at position Ile642 in pocket III allows it to be replaced only by Val or a smaller amino acid such as an Ala. On the other hand, any hydrophobic residue can replace Val643. In pocket IV, only shorter chain hydrophobic residues (Ile, Ala, and Leu) can occupy the Val646 position and nearly any residue can occupy the Met647 position (Figure 4D).
Based on the structure of the RIIα D/D:D-AKAP2 complex and peptide array analyses, spatial requirements of pockets I and II in RIIα are very stringent. In pocket I, the Ile638 position weakly tolerates Leu, Phe or Val and the Ala639 position can tolerate a change to only Ile or Val. In pocket II, Ile642 and Val643 can tolerate a change to Val and Ile, respectively or to Ala (Figure 4D).
Consistency of dual-specific, RI- and RII-specific AKAP binding to R subunits
With the structure-based alignment of the D-AKAP2 sequence bound to RI and RII subunits and the AKAP sequence requirements based on the properties of the pockets now established, the likelihood of a putative AKAP being RI-, RII- or dual-specific can now be predicted. Indeed, this model correctly identifies the specificity of previously established AKAPs. Further, it confirms that the differential binding of D-AKAP2 to RI and RII subunits is universal to all AKAPs.
An alignment of the AKB regions of two dual-specific AKAPs, D-AKAP1 (Huang et al., 1997b) and AKAP220 (Reinton et al., 2000) with the D-AKAP2 sequence confirms that it matches the requirements of binding to both R subunits. For example, in RI binding, both AKAPs would have an Ala in position Ala635 of pocket I. In pocket II, position Ala639 would have an Ile and Val in D-AKAP1 and AKAP220 respectively. Finally, in pocket III, position Ile642 would be occupied by a Val and Ala in D-AKAP1 and AKAP220 respectively, both of which would be tolerated (Figures 4D and 4E). For RII binding, D-AKAP1 would present two Ile residues in pocket I and Val and Ile in pocket II. These residues would be very well tolerated in these pockets based on the structural and peptide array analyses. Similarly, in the case of AKAP220, Ile and Val (pocket I) and Ala and Ile (pocket II) would fulfill the requirements of binding to RII subunit (Figures 4D and 4E).
RIAD, a peptide that has been shown to preferentially bind to RI, satisfies all the requirements for binding to the four RIα pockets (Carlson et al., 2006). While high specificity binding to RI was achieved by a variety of peptides, three critical mutations made the peptide preferential for RI. Two residues mutated to acidic residues would contribute to the binding with RI through helix-stabilizing interactions. These mutations would disrupt the interaction with the RII subunit. For example, a mutation that results in an Asp residue in position K640 would be expected to make a H-bond interaction with Gln26 of RIα but would clash with a Thr residue in RIIα. Interestingly, within the core region of binding, RIAD satisfies the requirements of RIIα D/D binding (Figures 4D and 4F).
AKAP-IS binds specifically to RIIα and as expected, fulfills the requirements of RII binding (Alto et al., 2003) (Figures 4D and 4G). However, for RI binding this peptide would present an Ile in position 635, a position that can tolerate only an Ala residue. Thus, it is not the specificity for RII subunits but rather the preclusion from RI binding that makes this peptide specific for RII subunits.
Strategy for predicting the specificity of an AKAP
The structures of RIα D/D:D-AKAP2 and RIIα D/D:D-AKAP2 now allow us to propose a strategy for deducing whether an AKAP is likely to be an RI-, RII- or dual-specific AKAP. Since D-AKAP2 is the only AKAP whose structure is known with both R subunits, it can be used as a foundation for comparisons with putative AKAPs. The approach would be to first do a structure-based sequence alignment of the putative AKAP with the D-AKAP2 sequence in a fashion similar to that used in this study (Figure 6). Based on the sequence alignment, it can be deduced whether the putative AKAP satisfies the requirements of RI and RII binding.
Since determinants in the core region for RII binding are fewer than RI binding, consistency with the requirements do not necessarily imply binding. Thus, the second step would be to overlay the sequence onto the structures of RI and RII bound to D-AKAP2. This would be crucial for deciding whether residues outside the core interacting regions would disrupt or enhance binding. As seen for the RIAD peptide, while the core requirements are fulfilled, an acidic residue outside the core region would be expected to disrupt RII binding. Similarly, as shown before, minor changes in the AKAP sequence could make it tolerant as seen in the case of Trp636 of D-AKAP2, the absence of which would make position Ala639 more tolerant to changes.
In summary, the structures presented here establish that a single AKAP peptide binds in a different mode to different R subunits. The shift in the α-helical register of D-AKAP2 also raises interesting questions about the evolution of AKAP sequences. Finally, as a general point, the register shift serves as a cautionary tale for predicting the mode of binding to homologous proteins.
Experimental Procedures
Protein Expression and Purification
Bovine full-length RIα was expressed and purified as described previously (Kim et al., 2007) and mutations were generated as described by Kunkel et al. (Kunkel et al., 1987). Bovine RIα D/D (residues 1–61) was expressed as described earlier (Banky et al., 2000). Prior to purification, the RIα D/D cells were suspended and lysed in 20 mM Tris, pH 8.0, 100 mM NaCl buffer containing protease inhibitors. The AKB region of human D-AKAP2 (residues 623–662) was cloned as a GST-tagged protein in pGEX 4T-1 vector with an internal thrombin cleavage site and expressed in a manner similar to RIα D/D.
Initial purification of His-RIα D/D was carried out using the Profinia Protein Purification System (Bio-Rad Laboratories). Briefly, a pre-programmed method was used to bind the protein to a Ni affinity column, followed by imidazole elution and a desalting step. Trypsin was used to cleave the His-tag and first 11 residues of the protein and then separated by batch binding to Benzamidine Sepharose 6B resin (GE Healthcare). As a final step, RIα D/D was purified by S75 gel filtration. The protein was concentrated to ~30 mg/ ml (ε280 nm=4270 M−1 cm−1) for crystallization. GST-D-AKAP2 was first bound to Glutathione Sepharose 4B resin (GE Healthcare) followed by cleavage of the tag. The resulting protein was purified on an S75 column and concentrated to ~20 mg/ml (ε280 nm=6970 M−1 cm−1).
For complex formation, equimolar quantities of RIα D/D dimer and D-AKAP2 were mixed and placed in ice for 30 minutes. The complex with a final concentration of ~25 mg/ ml was used for crystallization.
Crystallization, Structure Determination and Refinement
Crystals of RIα D/D and RIα D/D:D-AKAP2 complex appeared in a week at room temperature using microbatch crystallization. Apo crystals grew from a 1:1 drop containing 30% PEG 3350, 200 mM sodium formate, and 0.1 M Bis-Tris Propane, pH 9.0. Prior to data collection, the crystals were soaked for 2 min in the reservoir solution containing 10% glycerol and flash-frozen in liquid nitrogen. RIα D/D:D-AKAP2 complex crystals were obtained by mixing equal volumes of protein with 10% PEG 6000, 0.01 M ZnCl2, and 0.1 M MES, pH 5.5 and directly frozen in liquid nitrogen.
For structure determination and refinement, data sets were collected using synchrotron and in-house sources and processed using HKL2000 (Table 1) (Otwinowski and Minor, 1997). The apo RIα D/D structure was determined using the weak anomalous signal arising from inherently present sulfur atoms. The program ShelxD (Sheldrick, 2008) was used to locate the positions of the sulfurs. Heavy atom refinement and phasing followed by density modification were performed using Solve/Resolve (Terwilliger and Berendzen, 1999).
Prior to model building and refinement, 10% of the data were set aside for cross-validation. The structure was built manually using the program, Coot (Emsley and Cowtan, 2004) and water molecules were placed manually using criteria described previously (Sarma et al., 2005). Anistropic TLS refinement as incorporated in Phenix (Adams et al., 2002) followed by positional and individual B-factor refinements resulted in a final R/Rfree of 18.7/24.9%. The final model has 50 residues, 29 waters and 2 formate ions, with alternate conformations for Cys16 and Glu35. Alternate conformations were refined using various occupancies and the ones that yielded similar B-factors for the two conformations, were chosen.
The RIα D/D:D-AKAP2 complex structure was solved by molecular replacement using the program Phaser (McCoy et al., 2007) and the apo RIα D/D monomer (residues 23–61) as the search model. Initial Fo –Fc maps clearly showed the presence of the N-terminal residues and D-AKAP2. The residues were carefully built into maps resulting from the 2.3 Å data. Positional and individual B-factor refinements were carried out using Phenix (Adams et al., 2002) with 10% of the reflections set aside for cross-validation. With the two monomers and the AKB molecule, treated as rigid bodies for TLS refinement, the structure was refined to a final R/Rfree of 18.6/25.4%. The final model includes 30 waters and 5 Zn+2 ions. Occupancies for five Zn+2 ions were refined as described earlier (Sarma et al., 2005) and ranged from 0.4 to 1.0. Alternate conformations were modeled for Asn24A and Cys16B of RIα D/D. Lys61B of RIα D/D and D-AKAP2 residues 623–627, 655–662 were not modeled due to weak density (Figure 1B). Additionally, five D-AKAP2 N-terminal residues that were part of the GST tag were also disordered. Lys22A, Lys61A of RIα D/D and Asp628, Gln631, Gln648, Gln651, Tyr652, Gln654 in D-AKAP2 were modeled as Ala due to weak side chain density (Figure 1B).
Since waters are an integral part of structures, the solvent molecules in each structure were renumbered according to the strength of electron density, with water 1 being the highest. All structural figures were prepared using Pymol (www.pymol.org).
Fluorescence Anistotropy Binding Assays
The 27-amino acid region from the D-AKAP2 AKB (residues 623–650) was synthesized and purified to greater than 95% purity by HPLC (Peptron, Inc). The last residue was changed to Cys for fluorescent labeling. 0.5 mg of protein was incubated overnight with a 2-fold molar excess of fluorescin-5-maleimide or tetramethylrhodamine-5-maleimide (Molecular Probes, Inc.). The labeled peptides were purified by HPLC and excess organic solvent was evaporated on a Speed Vac (Savant, Inc.). Concentrations were determined using absorbance at 485 nm (ε485nm=90000 M−1 cm−1) and 541 nm (ε541nm=91000 M−1 cm−1) for the fluorescein- and rhodamine-labeled peptides, respectively. Binding studies with full-length and mutant proteins were carried out as previously described (Burns et al., 2003).
Supplementary Material
Acknowledgements
Valuable discussions with Drs. P.A. Karplus, H.R. Faber, F. A. Hays, Z.A. Wood, L. Ten Eyck, C.T. Eggers, M. Sastri and P. Ghosh are appreciated. This work was supported by NIH grants DK054441 and GM34921 to S.S.T and an American Heart Association postdoctoral fellowship 0825041F to G.N.S. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Use of the APS is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The coordinates and the structure factors have been deposited in the PDB (www.rcsb.org) as entries 3IM3 (RIα D/D) and 3IM4 (RIα D/D:D-AKAP2).
References
- Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure refinement. Acta Cryst D Biol Cryst. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Alto NM, Soderling SH, Hoshi N, Langeberg LK, Fayos R, Jennings PA, Scott JD. Bioinformatic design of A-kinase anchoring protein-in silico: a potent and selective peptide antagonist of type II protein kinase A anchoring. Proc Natl Acad Sci USA. 2003;100:4445–4450. doi: 10.1073/pnas.0330734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amieux PS, Cummings DE, Motamed K, Brandon EP, Wailes LA, Le K, Idzerda RL, McKnight GS. Compensatory regulation of RIalpha protein levels in protein kinase A mutant mice. J Biol Chem. 1997;272:3993–3998. doi: 10.1074/jbc.272.7.3993. [DOI] [PubMed] [Google Scholar]
- Banky P, Huang LJ, Taylor SS. Dimerization/docking domain of the type Ialpha regulatory subunit of cAMP-dependent protein kinase. Requirements for dimerization and docking are distinct but overlapping. J Biol Chem. 1998;273:35048–35055. doi: 10.1074/jbc.273.52.35048. [DOI] [PubMed] [Google Scholar]
- Banky P, Newlon MG, Roy M, Garrod S, Taylor SS, Jennings PA. Isoform- specific differences between the type Ialpha and IIalpha cyclic AMP-dependent protein kinase anchoring domains revealed by solution NMR. J Biol Chem. 2000;275:35146–35152. doi: 10.1074/jbc.M003961200. [DOI] [PubMed] [Google Scholar]
- Banky P, Roy M, Newlon MG, Morikis D, Haste NM, Taylor SS, Jennings PA. Related protein-protein interaction modules present drastically different surface topographies despite a conserved helical platform. J Mol Biol. 2003;330:1117–1129. doi: 10.1016/s0022-2836(03)00552-7. [DOI] [PubMed] [Google Scholar]
- Brennan JP, Bardswell SC, Burgoyne JR, Fuller W, Schroder E, Wait R, Begum S, Kentish JC, Eaton P. Oxidant-induced activation of type I protein kinase A is mediated by RI subunit interprotein disulfide bond formation. J Biol Chem. 2006;281:21827–21836. doi: 10.1074/jbc.M603952200. [DOI] [PubMed] [Google Scholar]
- Burns LL, Canaves JM, Pennypacker JK, Blumenthal DK, Taylor SS. Isoform specific differences in binding of a dual-specificity A-kinase anchoring protein to type I and type II regulatory subunits of PKA. Biochemistry. 2003;42:5754–5763. doi: 10.1021/bi0265729. [DOI] [PubMed] [Google Scholar]
- Burns-Hamuro LL, Hamuro Y, Kim JS, Sigala P, Fayos R, Stranz DD, Jennings PA, Taylor SS, Woods VL., Jr Distinct interaction modes of an AKAP bound to two regulatory subunit isoforms of protein kinase A revealed by amide hydrogen/deuterium exchange. Protein Sci. 2005;14:2982–2992. doi: 10.1110/ps.051687305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns-Hamuro LL, Ma Y, Kammerer S, Reineke U, Self C, Cook C, Olson GL, Cantor CR, Braun A, Taylor SS. Designing isoform-specific peptide disruptors of protein kinase A localization. Proc Natl Acad Sci USA. 2003;100:4072–4077. doi: 10.1073/pnas.2628038100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CR, Lygren B, Berge T, Hoshi N, Wong W, Tasken K, Scott JD. Delineation of type I protein kinase A-selective signaling events using an RI anchoring disruptor. J Biol Chem. 2006;281:21535–21545. doi: 10.1074/jbc.M603223200. [DOI] [PubMed] [Google Scholar]
- Casey M, Vaughan CJ, He J, Hatcher CJ, Winter JM, Weremowicz S, Montgomery K, Kucherlapati R, Morton CC, Basson CT. Mutations in the protein kinase A R1alpha regulatory subunit cause familial cardiac myxomas and Carney complex. J Clin Invest. 2000;106:R31–R38. doi: 10.1172/JCI10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst D Biol Cryst. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gold MG, Lygren B, Dokurno P, Hoshi N, McConnachie G, Tasken K, Carlson CR, Scott JD, Barford D. Molecular Basis of AKAP Specificity for PKA Regulatory Subunits. Mol Cell. 2006;24:383–395. doi: 10.1016/j.molcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Gronholm M, Vossebein L, Carlson CR, Kuja-Panula J, Teesalu T, Alfthan K, Vaheri A, Rauvala H, Herberg FW, Tasken K, Carpen O. Merlin links to the cAMP neuronal signaling pathway by anchoring the RIbeta subunit of protein kinase A. J Biol Chem. 2003;278:41167–41172. doi: 10.1074/jbc.M306149200. [DOI] [PubMed] [Google Scholar]
- Harris NL, Presnell SR, Cohen FE. Four helix bundle diversity in globular proteins. J Mol Biol. 1994;236:1356–1368. doi: 10.1016/0022-2836(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Huang LJ, Durick K, Weiner JA, Chun J, Taylor SS. D-AKAP2, a novel protein kinase A anchoring protein with a putative RGS domain. Proc Natl Acad Sci USA. 1997a;94:11184–11189. doi: 10.1073/pnas.94.21.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LJ, Durick K, Weiner JA, Chun J, Taylor SS. Identification of a novel protein kinase A anchoring protein that binds both type I and type II regulatory subunits. J Biol Chem. 1997b;272:8057–8064. doi: 10.1074/jbc.272.12.8057. [DOI] [PubMed] [Google Scholar]
- Kammerer S, Burns-Hamuro LL, Ma Y, Hamon SC, Canaves JM, Shi MM, Nelson MR, Sing CF, Cantor CR, Taylor SS, Braun A. Amino acid variant in the kinase binding domain of dual-specific A kinase-anchoring protein 2: a disease susceptibility polymorphism. Proc Natl Acad Sci USA. 2003;100:4066–4071. doi: 10.1073/pnas.2628028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Cheng CY, Saldanha SA, Taylor SS. PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation. Cell. 2007;130:1032–1043. doi: 10.1016/j.cell.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Kinderman FS, Kim C, von Daake S, Ma Y, Pham BQ, Spraggon G, Xuong NH, Jennings PA, Taylor SS. A Dynamic Mechanism for AKAP Binding to RII Isoforms of cAMP-Dependent Protein Kinase. Mol Cell. 2006;24:397–408. doi: 10.1016/j.molcel.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner LS, Carney JA, Svetlana DP, Taymans SE, Giatzkis C, Cho YS, Cho- Chung YS, Stratakis CA. Mutations of the gene encoding the protein kinase A type I-a regulatory subunit in patients with Carney complex. Nature Genetics. 2000;26:89. doi: 10.1038/79238. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Levy FO, Rasmussen AM, Tasken K, Skalhegg BS, Huitfeldt HS, Funderud S, Smeland EB, Hansson V. Cyclic AMP-dependent protein kinase (cAK) in human B cells: co-localization of type I cAK (RI alpha 2 C2) with the antigen receptor during anti- immunoglobulin-induced B cell activation. Eur J Immunol. 1996;26:1290–1296. doi: 10.1002/eji.1830260617. [DOI] [PubMed] [Google Scholar]
- Li H, Degenhardt B, Tobin D, Yao ZX, Tasken K, Papadopoulos V. Identification, localization, and function in steroidogenesis of PAP7: a peripheral-type benzodiazepine receptor- and PKA (RIalpha)-associated protein. Mol Endocrinol. 2001;15:2211–2228. doi: 10.1210/mend.15.12.0736. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon MG, Roy M, Morikis D, Carr DW, Westphal R, Scott JD, Jennings PA. A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes. Embo J. 2001;20:1651–1662. doi: 10.1093/emboj/20.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama K, Yamada Y, Matsuo H, Segawa T, Watanabe S, Kato K, Yajima K, Hibino T, Yokoi K, Ichihara S, et al. Association of gene polymorphisms with myocardial infarction in individuals with or without conventional coronary risk factors. Int J Mol Med. 2007;19:129–141. [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CWJ, Sweet RM, editors. Methods in Enzymology. 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Petersen MT, Jonson PH, Petersen SB. Amino acid neighbours and detailed conformational analysis of cysteines in proteins. Protein Eng. 1999;12:535–548. doi: 10.1093/protein/12.7.535. [DOI] [PubMed] [Google Scholar]
- Reinton N, Collas P, Haugen TB, Skalhegg BS, Hansson V, Jahnsen T, Tasken K. Localization of a novel human A-kinase-anchoring protein, hAKAP220, during spermatogenesis. Dev Biol. 2000;223:194–204. doi: 10.1006/dbio.2000.9725. [DOI] [PubMed] [Google Scholar]
- Sarma GN, Manning VA, Ciuffetti LM, Karplus PA. Structure of Ptr ToxA: an RGD-containing host-selective toxin from Pyrenophora tritici-repentis. Plant Cell. 2005;17:3190–3202. doi: 10.1105/tpc.105.034918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Cryst A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Skalhegg BS, Tasken K. Specificity in the cAMP/PKA signaling pathway. differential expression, regulation, and subcellular localization of subunits of PKA. Front Biosci. 1997;2:d331–d342. doi: 10.2741/a195. [DOI] [PubMed] [Google Scholar]
- Skalhegg BS, Tasken K, Hansson V, Huitfeldt HS, Jahnsen T, Lea T. Location of cAMP-dependent protein kinase type I with the TCR-CD3 complex. Science. 1994;263:84–87. doi: 10.1126/science.8272870. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Kim C, Vigil D, Haste NM, Yang J, Wu J, Anand GS. Dynamics of signaling by PKA. Biochim Biophys Acta. 2005;1754:25–37. doi: 10.1016/j.bbapap.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Cryst D Biol Cryst. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingley WG, Pawlikowska L, Zaroff JG, Kim T, Nguyen T, Young SG, Vranizan K, Kwok PY, Whooley MA, Conklin BR. Gene-trapped mouse embryonic stem cell-derived cardiac myocytes and human genetics implicate AKAP10 in heart rhythm regulation. Proc Natl Acad Sci USA. 2007;104:8461–8466. doi: 10.1073/pnas.0610393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtenberger M, Schmutzhard J, Hemminki K, Meindl A, Sutter C, Schmutzler RK, Wappenschmidt B, Kiechle M, Arnold N, Weber BH, et al. The functional genetic variant Ile646Val located in the kinase binding domain of the A-kinase anchoring protein 10 is associated with familial breast cancer. Carcinogenesis. 2007;28:423–426. doi: 10.1093/carcin/bgl164. [DOI] [PubMed] [Google Scholar]
- Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Yajima K, Hibino T, Kato K, Matsuo H, Segawa T, Watanabe S, Oguri M, Yokoi K, Nozawa Y, et al. Association of gene polymorphisms with myocardial infarction in individuals with different lipid profiles. Int J Mol Med. 2007;20:581–590. [PubMed] [Google Scholar]
- Zauhar RJ, Colbert CL, Morgan RS, Welsh WJ. Evidence for a strong sulfur- aromatic interaction derived from crystallographic data. Biopolymers. 2000;53:233–248. doi: 10.1002/(SICI)1097-0282(200003)53:3<233::AID-BIP3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.