Abstract
The spa gene of Staphylococcus aureus encodes protein A and is used for typing of methicillin-resistant Staphylococcus aureus (MRSA). We used sequence typing of the spa gene repeat region to study the epidemiology of MRSA at a German university hospital. One hundred seven and 84 strains were studied during two periods of 10 and 4 months, respectively. Repeats and spa types were determined by Ridom StaphType, a novel software tool allowing rapid repeat determination, data management and retrieval, and Internet-based assignment of new spa types following automatic quality control of DNA sequence chromatograms. Isolates representative of the most abundant spa types were subjected to multilocus sequence typing and pulsed-field gel electrophoresis. One of two predominant spa types was replaced by a clonally related variant in the second study period. Ten unique spa types, which were equally distributed in both study periods, were recovered. The data show a rapid dynamics of clone circulation in a university hospital setting. spa typing was valuable for tracking of epidemic isolates. The data show that disproval of epidemiologically suggested transmissions of MRSA is one of the main objectives of spa typing in departments with a high incidence of MRSA.
Staphylococcus aureus is a major human pathogen causing skin and tissue infections, pneumonia, septicemia, and device-associated infections. The emergence of strains resistant to methicillin and other antibacterial agents has become a major concern especially in the hospital environment, because of the higher mortality due to systemic methicillin-resistant Staphylococcus aureus (MRSA) infections (2). Typing of MRSA is used to support infection control measures. While pulsed-field gel electrophoresis (PFGE) is a “gold standard” for strain typing of MRSA (20), DNA sequence-based approaches are becoming more frequently used because sequence data can easily be transferred between laboratories via the Internet. Multilocus sequence typing (MLST), which was developed by using Neisseria meningitidis as the model species (9, 18), has been successfully adapted to S. aureus (7, 8). However, MLST is not suitable for routine surveillance of MRSA because of the high cost and the necessity of access to a high-throughput DNA sequencing facility.
Although there is evidence for recombination in S. aureus (10), it has been shown that point mutations by far exceed recombination events, in contrast to N. meningitidis or Streptococcus pneumoniae (11). Furthermore, there is only a small number of clonal groupings of MRSA circulating worldwide (7). Therefore, single-locus DNA sequencing of repeat regions of the coa (coagulase) gene and the spa gene (protein A), respectively, could be used for reliable and accurate typing of MRSA (12, 13, 26-29). spa typing is especially interesting for rapid typing of MRSA in a hospital setting since it offers higher resolution than coa typing (27). The repeat region of the spa gene is subject to spontaneous mutations, as well as loss and gain of repeats. Repeats are assigned an alpha-numerical code, and the spa type is deduced from the order of specific repeats. There is a good correlation between clonal groupings determined by MLST and the respective spa types (3, 4, 23, 24). Examples have been reported of isolates with the same spa type belonging to related MLST sequence types that arose by single-locus variation (5). On the other hand, there seems to be a considerable degree of spa gene repeat number variation within a given sequence type, suggesting that spa typing in some instances provides greater resolution than MLST (3, 23). Nevertheless, there is a consensus that pulsed-field gel electrophoresis (PFGE) is superior to spa typing and probably also MLST in its discriminatory power (14, 15, 19, 26, 29). Therefore, PFGE is still considered a valuable tool for MRSA typing, although it is time-consuming and the interlaboratory comparability of results requires extensive effort for protocol harmonization (20).
For a variety of bacterial species, MLST protocols have been developed during the past 5 years because the method allows the creation of Internet-based curated databases that represent virtual strain collections accessible to the scientific community both for entry of data and for their retrieval (www.mlst.net). Despite being a DNA sequence-based method, spa typing, to the best of our knowledge, is hampered by a lack of generally available software tools for repeat identification and by the lack of a consensus on assignments of new repeats and spa types. Therefore, spa typing cannot be considered a portable tool.
In the present study, the dynamics of MRSA spa types at a single hospital was followed in two study periods. A novel software tool was used for spa type determination. This specialized software meets the requirements for modern, Internet-based management of genotyping data.
MATERIALS AND METHODS
Strains.
MRSA strains were isolated and identified from various clinical specimens sent to the Institute for Hygiene and Microbiology at the University of Würzburg. Only patients and staff members of the Würzburg University Clinic were included. This hospital is the largest referral center in the southern German region of Lower Franconia, with a population size of about 1,300,000. Copy strains were excluded. Final identification and antimicrobial resistance testing were performed with Vitek 2, an automated bacteriology system that performs bacterial identification and susceptibility testing analyses (BioMérieux, Marci l'Etoile, France). MICs of mupirocin were determined by E test on Mueller-Hinton agar plates after 24 h of incubation as described by the manufacturer (AB Biodisk, Solna, Sweden). Two study periods were included; period 1 (107 isolates) was June 2001 to May 2002, and period 2 (84 isolates) was January 2003 to April 2003. The staff in charge of the diagnostic laboratories was instructed to submit every MRSA isolate first isolated from a patient to spa typing. Retrospective examination of the database at the Institute for Hygiene and Microbiology revealed that in period 1, 107 (46.5%) of 230 isolates were subjected to spa typing, whereas in period 2, this was the case for 84 (56%) of 150 isolates. SmaI macrorestriction patterns were obtained by use of the harmonized European protocol for typing of S. aureus by PFGE (20). For cluster analysis, the algorithm described by Claus et al. was used (1).
PCR and DNA sequence analysis.
The x region of the spa gene was amplified by PCR with primers 1095F (5′-AGACGATCCTTCGGTGAGC-3′) and 1517R (5′-GCTTTTGCAATGTCATTTACTG-3′) (26). DNA sequences were obtained with an ABI 377 sequencer (Applied Biosystems, Foster City, Calif.). spa types were determined with the Ridom StaphType software described below (Ridom GmbH, Würzburg, Germany). MLST was performed as described recently (7). Sequence types were determined with the database accessible via http://www.mlst.net/dbqry/saureus.htm.
Ridom StaphType.
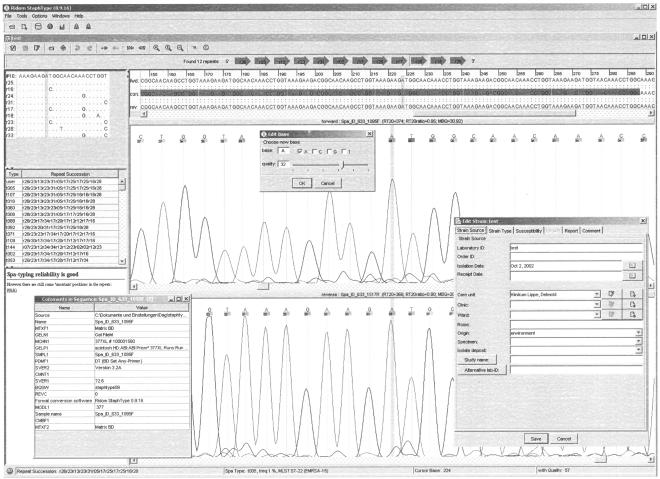
spa types were determined with the novel software Ridom StaphType (Ridom GmbH). Basically, the software consists of three modules: a sequence editor, a database, and a report generator module (Fig. 1). After providing the input sequences (FASTA format or preferably ABI and SCF chromatograms), Ridom StaphType attaches to each called base a quality value that corresponds to a sequence error probability. Taking the quality values into consideration, the software constructs a consensus sequence, automatically detects the spa repeats, and assigns a spa type. In at least 90% of all cases, no further manual editing is necessary. For the remaining sequences, a versatile graphic user interface allows the user to manually edit sequences with the help of an integrated expert system. No sequence information up- or downstream of the repeat region is taken into consideration for spa type coding. However, the software searches for 5′ and 3′ signature sequences at the correct distance to ensure that no leading or ending repeat is missed. Once sequence editing is finished, the spa typing results and additional epidemiological relevant data can be stored in a relational database system. Information from the database can be easily retrieved by Boolean searches and exported in tab-delimited spreadsheet format. Furthermore, the database's integrity is checked on a regular basis and the database content can be backed up to protect from data leakage. For privacy, the content is cryptographically secured. Finally, different configurable reports can be created. These reports are stored internally as read-only encrypted tamperproof pdf files. To view and print these files, the freely available Adobe Acrobat Reader software (version 5.0 or higher) must be installed.
FIG. 1.
Screen shot of the novel Ridom StaphType software featuring a base quality-based sequence editor, a database, and a report generator (not shown) module. This client software synchronizes with an accompanying website to ensure uniform spa code terminology usage.
Numeric spa repeat and type codes are used by Ridom StaphType. To ensure uniform code terminology usage, the software synchronizes either directly via the http protocol or file based (e.g., via e-mail) with an accompanying website that functions as the operative source for all new spa repeat and type codes. If wished, all new spa repeats and types that meet quality criteria (i.e., spa types are deduced from chromatograms and 5′ and 3′ signatures are unambiguously detected) can be transferred during synchronization to the server to obtain a final designation. In exchange, repeats and types detected by others since the last synchronization are transferred to the Ridom StaphType client software. Furthermore, if allowed by the user, the local spa type frequencies are also transmitted to the server, which always returns global frequency data. The website (http://www.ridom.de/spaserver/) is accessible to everyone, and spa repeat sequences (FASTA format) and spa types can be downloaded. Submission of chromatograms of new spa repeats and types for inclusion in the reference database is possible. Therefore, users not working with Ridom StaphType have access to the same uniform terminology.
RESULTS
Design of the study.
MRSA strains were collected at the University of Würzburg during two study periods (Table 1). In both periods, the number of isolates and the number of departments and wards contributing strains were comparable. There was a twofold difference in the MRSA isolation rate per month between the two study periods, which could not be explained by changes in culture submission criteria or MRSA screening procedures (Table 1). For spa type designation, we used newly developed software (Ridom StaphType; for a detailed description, see Materials and Methods).
TABLE 1.
Comparison of the two study periods
| Study period | Duration (mo) | No. of MRSA isolates | No. of patients | No. of staff members | No. of spa types | No. of unique spa types | Predomi- nant spa types | No. of departments involved | No. of wards involved | No. of outpatient clinics | No. of clustersa | No. of wards involved in clusters | No. of patients involved in clusters | spa types involved in clusters |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 10 | 107 | 106 | 1 | 17 | 7 | 1, 3 | 13 | 29 | 7 | 11 | 7 | 33 | 1, 3, 5, 8, 10, 13 |
| 2 | 4 | 84 | 82 | 2 | 14 | 9 | 3, 23 | 14 | 29 | 6 | 8 | 6 | 19 | 1, 3, 23 |
A cluster was defined as the identification, within 9 days, of two or more patients on the same ward who harbored MRSA strains with the same spa type.
Typing of MRSA.
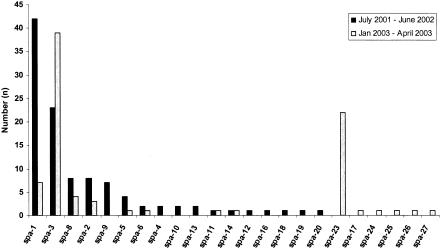
Seventeen and 14 spa types were observed in study periods 1 and 2, respectively (Tables 1 and 2). There were 10 spa types that were found only once in all of the 191 strains analyzed (Fig. 2). Five of those occurred in the first period, and five occurred in the second. This finding indicates either permanent import of novel spa types or in-house microevolution of spa repeats. Microevolution of the spa gene was evident for several spa types, e.g., deletion-insertion events in spa-8 and -9, and repeat exchange in spa-1 and -23 (Table 2). The rank abundance curve in Fig. 2 shows that spa-1 and -3 dominated in period 1, whereas spa-1 was replaced by spa-23 in period 2. This observation indicates that the circulation of epidemic clones within a hospital is not static but a matter of short-term changes.
TABLE 2.
spa types
| spa type | Repeatsa |
|---|---|
| 01 | r26r30r17r34r17r20r17r12r17r16 |
| 02 | r26r23r17r34r17r20r17r12r17r16 |
| 03 | r26r17r20r17r12r17r17r16 |
| 04 | r09r02r16r13r13r17r34r16r34 |
| 05 | r26r23r13r23r31r05r17r25r17r25r16r28 |
| 06 | r26r23r13r23r31r05r17r17r25r16r28 |
| 08 | r11r19r12r21r17r34r24r34r22r25 |
| 09 | r11r12r21r17r34r24r34r22r24r34r22r33r25 |
| 10 | r26r17r34r17r20r17r12r17r16 |
| 11 | r08r16r02r25r34r24r25 |
| 12 | r15r12r16r02r16r02r25r17r24r24 |
| 13 | r26r30r17r34r17r20r17r12r17 |
| 14 | r26r17r20r17r12r17r17r17r16 |
| 16 | r26r23r13r23r31r05r17r25r16r16r28 |
| 17 | r15r12r16r16r02r16r02r25r17r24r24 |
| 18 | r15r12r16r02r16r02r25r17r24r24r24 |
| 19 | r08r16r02r16r02r25r17r24 |
| 20 | r26r23r31r29r17r31r29r17r25r17r25r16r28 |
| 23 | r26r37r17r12r17r16 |
| 24 | r11r12r21r17r34r24r34r22r25 |
| 25 | r26r23r23r13r23r29r17r31r29r17r25r17r25r16r28 |
| 26 | r08r16r34 |
| 27 | r26r17r13 |
A numerical code was chosen for Ridom StaphType. A comprehensive collection of spa types and repeat sequences is available at http://www.ridom.de/spaserver/.
FIG. 2.
Rank abundance graph demonstrating the frequencies of spa types collected during two study periods.
The epidemic strains were analyzed further by MLST, antimicrobial resistance patterns, hemolysis, and PGFE. Three strains each of spa-1, -3, and -23 were subjected to MLST. All were derivatives of the ST-5 complex. spa-1 and -23 were associated with ST-228 (double-locus variant of ST-5; ST-228 strains belong to the southern German epidemic clone) (8). Allele spa-3, which was present in both periods, was associated with ST-225. ST-225 is a single-locus variant of ST-5 (EMRSA-3, New York clone; German designation, Rhine-Hesse clone) (8). We also performed MLST on two of five strains with spa-5 because this spa type was exclusively isolated at a surgical department located outside the campus. MRSA with spa-5 were shown to be ST-22, which corresponds to EMRSA-15 or the Barnim clone (8, 17, 33).
All 60 spa-3 isolates tested for gentamicin susceptibility were gentamicin susceptible, whereas a total of 70 spa-1 and spa-23 isolates tested were gentamicin resistant. Elevated MICs of mupirocin were only observed for spa-1 and -23 strains. For spa-23 strains, the median mupirocin MIC was 24 μg/ml (range, 16 to 48 μg/ml). With regard to mupirocin resistance, two populations of spa-1 isolates were observed, one for which the MICs were high (median, 28 μg/ml; range, 16 to 48 μg/ml; n = 14) and one for which the MICs were low (median, 0.25 μg/ml; range, 0.064 to 0.38 μg/ml; n = 35). Forty-nine (79%) of 62 spa-3 isolates exhibited hemolysis on blood agar plates, whereas hemolysis was observed in only 9 (13%) of a total of 71 spa-1 and -23 isolates.
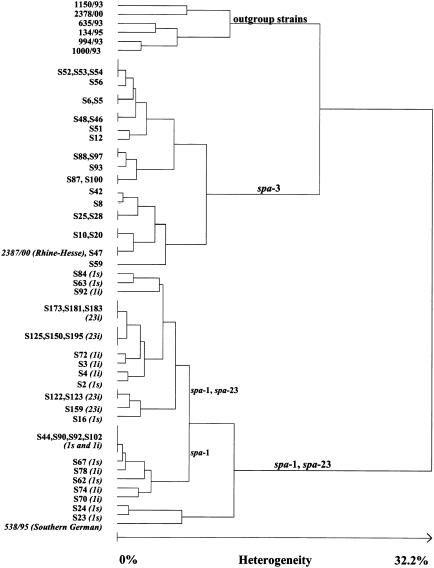
We further analyzed a selection of spa-1, -3, and -23 strains by PFGE. A dendrogram deduced from the cluster analysis of PFGE macrorestriction patterns is shown in Fig. 3. spa-3 strains were clearly distinguished from spa-1 and -23 strains by PFGE. This finding was in accordance with those based on MLST, gentamicin resistance, and hemolysis. As expected, spa-3 isolates were grouped together with Rhine-Hesse epidemic strain 2387/00 and spa-1 and -23 strains were grouped together with southern German epidemic strain 538/95. Fifteen PFGE patterns were detected for the spa-3 group of strains, and 20 patterns were detected for the spa-1-spa-23 group of strains. This finding confirmed the higher resolution of epidemic strains by PFGE compared to spa typing. There was no clear distinction between spa-1 and -23 strains by PFGE, underlining their relatedness. Furthermore, strains for which the mupirocin MICs were low could not be distinguished from strains for which the mupirocin MICs were high.
FIG. 3.
Dendrogram deduced from the cluster analysis of PFGE macrorestriction patterns of spa-1, -3, and -23 strains as determined by the algorithm described by Claus et al. (1). Strains from this study are designated by the letter S followed by a strain number. Additional information was added to spa-1 and -23 strains as follows: 1s, spa-1 and low mupirocin MIC; 1i, spa-1 and high mupirocin MIC; 23i, spa-23 and high mupirocin MIC. As a control, patterns determined previously from diverse other clones at the German reference laboratory for staphylococci were included for cluster analysis (outgroup strains, i.e., 1150/93, Berlin epidemic MRSA; 2378/00, Barnim epidemic MRSA; 635/93, Vienna epidemic MRSA; 134/93, northern German epidemic MRSA; 1000/93, Hannover epidemic MRSA).
Local epidemiology of MRSA.
Table 3 demonstrates the frequent detection of epidemic spa types in different departments during the second study period. It is noteworthy that there were six incidences of detection of different MRSA types within 1 week from patients taken care of at the same ward. This finding indicates that simultaneous but independent transmission events are not rarely encountered, which supports the use of spa typing in a university hospital setting even in periods of a high frequency of isolation of epidemic strains.
TABLE 3.
spa types isolated at the university hospital in weeks 1 to 18 of 2003 (study period 2)
| Departmenta |
spa type(s) isolated during wk:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
| A | 3 | 23 | 3 | 3, 3 | 3 | 3, 3 | 3, 3 | |||||||||||
| B1 | 27 | |||||||||||||||||
| B2 | 3 | |||||||||||||||||
| B3 | 2, 3 | |||||||||||||||||
| C1 | 3, 3 | 3 | 3, 23 | |||||||||||||||
| C2 | 26 | |||||||||||||||||
| C3 | 17 | 3 | 23 | 23 | 23 | 23 | ||||||||||||
| C4 | 3 | 23 | ||||||||||||||||
| C5 | 23 | |||||||||||||||||
| C6 | 23 | 3 | 23 | 3 | ||||||||||||||
| C7 | 3 | 3 | 23, 3 | 1, 1 | 23 | 23 | ||||||||||||
| C8 | 3 | 24 | ||||||||||||||||
| C9 | 3 | |||||||||||||||||
| D | 11 | |||||||||||||||||
| E | 8 | 2 | 6 | 3 | 8 | |||||||||||||
| F | 14 | |||||||||||||||||
| G | 8 | |||||||||||||||||
| H1 | 3 | |||||||||||||||||
| H2 | 23 | |||||||||||||||||
| H3 | 3 | |||||||||||||||||
| H4 | 1 | 3 | ||||||||||||||||
| H5 | 3, 1 | 1 | 23, 3 | 3 | 25 | |||||||||||||
| H6 | 3 | |||||||||||||||||
| H7 | 2 | |||||||||||||||||
| I1 | 23 | |||||||||||||||||
| I2 | 1 | |||||||||||||||||
| J1 | 8 | |||||||||||||||||
| K1 | 23 | |||||||||||||||||
| K2 | 23 | 23 | ||||||||||||||||
| K3 | 23 | |||||||||||||||||
| L1 | 3 | |||||||||||||||||
| L2 | 3 | 3 | 3 | 3 | ||||||||||||||
| M1 | 5 | |||||||||||||||||
| M2 | 3, 23 | 23 | 3 | |||||||||||||||
Capital letters represent departments of the hospital, and numbers represent different wards.
DISCUSSION
We present a novel tool for rapid determination of spa repeats in S. aureus. An important feature of this software is automated data submission via the Internet. Thus, the server can be used to collate and harmonize data from various geographic regions. Furthermore, DNA sequences are automatically subjected to quality control. This feature greatly facilitates the implementation of centralized servers since data need not to be checked by a curator. The software is designed in a way that it can be adapted to single-locus typing schemes relevant to other genetically variable pathogens responsible for nosocomial infections, e.g., vancomycin-resistant enterococci, for which MLST proved to be highly discriminatory (16, 21). In order to simplify spa type nomenclature, a numerical repeat code was established in this study. This approach was chosen despite the current existence of an alpha-numerical repeat nomenclature because numerical codes are now widely used for MLST and because Ridom StaphType is the first Internet-based tool available for assignment of spa types. This tool now provides the opportunity to harmonize spa type designations.
It has long been established that spa typing is less discriminatory than PFGE (26, 29). Tang et al. showed that 20 strains with the same spa type that were collected during an outbreak that lasted 107 weeks exhibited several related but distinguishable PFGE patterns. This finding corresponds to the PFGE analysis performed in this study. The significance of subtle changes of one band in the PFGE patterns of related strains, e.g., with allele spa-3, may be a matter of debate. In N. meningitidis, subtle changes in PFGE patterns could be identified if the nasopharyngeal and clonally identical blood isolates of a patient were compared or if the isolate of a patient and that of the clonally identical one of the closest contact were compared (30; U. Vogel, H. Claus, and M. Frosch, Letter, N. Engl. J. Med. 342:219-220, 2000). In a study on S. aureus carriage, several nasal isolates did not differ from clonally identical isolates consecutively obtained from the blood, which might indicate a higher stability of PFGE patterns in S. aureus than in N. meningitidis (31). Peacock et al., on the other hand, in their study comparing PFGE and MLST, found four patients whose S. aureus isolates, which were recovered from the same patient, differed by one or two bands. This finding suggests that differences in a single PFGE band may be considered hyperdiscriminatory (25). Despite being less discriminatory than PFGE, spa typing will certainly help to disprove epidemiological linkage between MRSA-colonized persons in periods with a high incidence of epidemic strains, as shown for several incidences in Table 3.
We are intuitively aware that the isolation of two MRSA strains with the same spa type is highly suggestive of person-to-person transmission if there is a low incidence of MRSA isolation. However, wards with a high rate of epidemic spa type isolation will suffer from uncertainties about the possibility of direct transmission. It will therefore be desirable to develop algorithms on the basis of spa types and their local isolation frequencies to assess the probability of person-to-person transmission each time two or more MRSA strains with the same spa type are recovered. Furthermore, novel DNA-based typing schemes might increase resolution within epidemic spa types. A possible candidate fulfilling this requirement might be the clumping factor B (clfB) gene, which was recently reported to be a highly stable marker detecting differences between strains with the same spa type (L. Koreen, S. Ramaswamy, S. Naidich, E. A. Graviss, and B. Kreiswirth, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. C-415, 2003).
We determined by MLST the sequence types of dominant spa types isolated at the hospital during two study periods. spa-1 and -23 turned out to be linked with ST-228, which resembles the southern German clone, a multiresistant clone that spread all over Germany (32). spa-1 was found predominantly in period 1, whereas spa-23 expanded at the hospital in period 2. This finding indicates that the allele spa-23 either derived from pre-existing spa-1 strains by spa gene repeat replacement or entered the hospital as a novel clone after the first study period. The first hypothesis is supported by the finding of highly related PFGE patterns of spa-1 and -23 strains (Fig. 3). A similar clonal dynamics has been recently shown by de Sousa et al., who reported a steady decline at a single hospital of ST-30 strains, which were replaced by ST-239 strains (4). In our study, type spa-1 and -23 isolates, in contrast to type spa-3 isolates, were mostly nonhemolytic and resistant to gentamicin. Furthermore, elevated mupirocin MICs were observed only for spa-1 and -23 isolates. These findings provide further support for a close relationship between spa-1 and -23. Interestingly, there were two spa-1 populations with regard to mupirocin sensitivity, a sensitive one and another for which the MICs ranged from 16 to 48 μg/ml. One might speculate that spa-23 strains, which dominated in the second study period, evolved from the mupirocin-resistant subset of the spa-1 population. However, PFGE provided no evidence supporting this suggestion.
Both of the epidemic sequence types identified at the hospital were derivatives of ST-5, i.e., ST-225, a single-locus variant of ST-5, and ST-228, a double-locus variant. ST-5, ST-225, and ST-228 have been assigned to clonal complex 5 (8). The close relationship of ST-5 and ST-225 was also shown by PFGE in our study, which clearly identified the ST-225 strains from the hospital as derivatives of the Rhine-Hesse clone, which is prevalent in Germany and was shown to be ST-5 (W. Witte and M. Enright, unpublished observation). In the MLST database (http://www.mlst.net/dbqry/saureus.htm), there is only a single ST-225 isolate, which was collected in the United States (strain cdc12). A more detailed picture of the microevolution of clonal complex 5 isolates circulating at the hospital might be achieved, e.g., by determination of SCCmec types (8, 22). According to these data, S. aureus with the ST-5 genomic background had acquired different types of SCCmec.
It was of interest that spa-5 strains (ST-22) circulated exclusively at a surgical department located outside of the campus. Although the patients there are in close contact with the university hospital, e.g., for postoperative care, these ST-22 strains did not start to circulate in other departments. Interestingly, ST-22 strains (EMRSA-15) have been shown to be ubiquitous and frequently cause outbreaks (17, 33). It may be that the spa-5 strains in that particular department have been adapted to the special cohort of cancer patients treated there. Reasons for such behavior are unclear, but adaptation processes of MRSA have been demonstrated, e.g., for community-acquired MRSA strains causing skin infections (6).
In conclusion, we reported on two phases of surveillance of MRSA by spa typing. The method was valuable for tracking of epidemic isolates, elucidation of the rate of import of sporadically occurring clones, and disproof of person-to-person transmission in hospitals with high rates of epidemic MRSA infection. The presentation of software for automatic repeat identification, together with an accompanying Internet database, will help to disseminate spa typing to a larger community. Future comparative studies will be greatly facilitated by this, and national and international surveillance of MRSA will be supported.
Acknowledgments
We acknowledge expert technical assistance by Stefanie Gerngras, Marion Patzke-Öchsner, Angelika Hansen, Ines Aulkemeier, and Susanne Ebner. We thank our colleagues at the Institute for Hygiene and Microbiology, especially Oliver Kurzai, for help with the study. Matthias Frosch is thanked for helpful discussions and support.
This investigation made use of the MLST website (http://mlst.net) developed by Man-Suen Chan and David Aanensen. The development of this site is funded by the Wellcome Trust.
REFERENCES
- 1.Claus, H., C. Cuny, B. Pasemann, and W. Witte. 1998. A database system for fragment patterns of genomic DNA of Staphylococcus aureus. Zentbl. Bakteriol. 287:105-116. [DOI] [PubMed] [Google Scholar]
- 2.Cosgrove, S. E., G. Sakoulas, E. N. Perencevich, M. J. Schwaber, A. W. Karchmer, and Y. Carmeli. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 36:53-59. [DOI] [PubMed] [Google Scholar]
- 3.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Sousa, M. A., C. Bartzavali, I. Spiliopoulou, I. S. Sanches, M. I. Crisostomo, and H. de Lencastre. 2003. Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in Patras, Greece. J. Clin. Microbiol. 41:2027-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Sousa, M. A., M. I. Crisostomo, I. S. Sanches, J. S. Wu, J. Fuzhong, A. Tomasz, and H. de Lencastre. 2003. Frequent recovery of a single clonal type of multidrug-resistant Staphylococcus aureus from patients in two hospitals in Taiwan and China. J. Clin. Microbiol. 41:159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dufour, P., Y. Gillet, M. Bes, G. Lina, F. Vandenesch, D. Floret, J. Etienne, and H. Richet. 2002. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin. Infect. Dis. 35:819-824. [DOI] [PubMed] [Google Scholar]
- 7.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 10.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. J. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frenay, H. M., A. E. Bunschoten, L. M. Schouls, W. J. van Leeuwen, C. M. Vandenbroucke-Grauls, J. Verhoef, and F. R. Mooi. 1996. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis. 15:60-64. [DOI] [PubMed] [Google Scholar]
- 13.Frenay, H. M., J. P. Theelen, L. M. Schouls, C. M. Vandenbroucke-Grauls, J. Verhoef, W. J. van Leeuwen, and F. R. Mooi. 1994. Discrimination of epidemic and nonepidemic methicillin-resistant Staphylococcus aureus strains on the basis of protein A gene polymorphism. J. Clin. Microbiol. 32:846-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundmann, H., S. Hori, M. C. Enright, C. Webster, A. Tami, E. J. Feil, and T. Pitt. 2002. Determining the genetic structure of the natural population of Staphylococcus aureus: a comparison of multilocus sequence typing with pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, and phage typing. J. Clin. Microbiol. 40:4544-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heym, B., M. Le Moal, L. Armand-Lefevre, and M. H. Nicolas-Chanoine. 2002. Multilocus sequence typing (MLST) shows that the ‘Iberian’ clone of methicillin-resistant Staphylococcus aureus has spread to France and acquired reduced susceptibility to teicoplanin. J. Antimicrob. Chemother. 50:323-329. [DOI] [PubMed] [Google Scholar]
- 16.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. van Embden, and R. J. Willems. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, A. P., H. M. Aucken, S. Cavendish, M. Ganner, M. C. Wale, M. Warner, D. M. Livermore, and B. D. Cookson. 2001. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS). J. Antimicrob. Chemother. 48:143-144. [DOI] [PubMed] [Google Scholar]
- 18.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montesinos, I., E. Salido, T. Delgado, M. Cuervo, and A. Sierra. 2002. Epidemiologic genotyping of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis at a university hospital and comparison with antibiotyping and protein A and coagulase gene polymorphisms. J. Clin. Microbiol. 40:2119-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nallapareddy, S. R., R. W. Duh, K. V. Singh, and B. E. Murray. 2002. Molecular typing of selected Enterococcus faecalis isolates: pilot study using multilocus sequence typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 40:868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 25.Peacock, S. J., G. D. de Silva, A. Justice, A. Cowland, C. E. Moore, C. G. Winearls, and N. P. Day. 2002. Comparison of multilocus sequence typing and pulsed-field gel electrophoresis as tools for typing Staphylococcus aureus isolates in a microepidemiological setting. J. Clin. Microbiol. 40:3764-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shopsin, B., M. Gomez, M. Waddington, M. Riehman, and B. N. Kreiswirth. 2000. Use of coagulase gene (coa) repeat region nucleotide sequences for typing of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 38:3453-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shopsin, B., and B. N. Kreiswirth. 2001. Molecular epidemiology of methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 7:323-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang, Y. W., M. G. Waddington, D. H. Smith, J. M. Manahan, P. C. Kohner, L. M. Highsmith, H. Li, F. R. Cockerill III, R. L. Thompson, S. O. Montgomery, and D. H. Persing. 2000. Comparison of protein A gene sequencing with pulsed-field gel electrophoresis and epidemiologic data for molecular typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 38:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel, U., G. Morelli, K. Zurth, H. Claus, E. Kriener, M. Achtman, and M. Frosch. 1998. Necessity of molecular techniques to distinguish between Neisseria meningitidis strains isolated from patients with meningococcal disease and from their healthy contacts. J. Clin. Microbiol. 36:2465-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Eiff, C., K. Becker, K. Machka, H. Stammer, and G. Peters. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344:11-16. [DOI] [PubMed] [Google Scholar]
- 32.Witte, W., C. Cuny, C. Braulke, D. Heuck, and I. Klare. 1997. Widespread dissemination of epidemic MRSA in German hospitals. Eur. Surveill. 2:25-28. [DOI] [PubMed] [Google Scholar]
- 33.Witte, W., M. Enright, F. J. Schmitz, C. Cuny, C. Braulke, and D. Heuck. 2001. Characteristics of a new epidemic MRSA in Germany ancestral to United Kingdom EMRSA 15. Int. J. Med. Microbiol. 290:677-682. [DOI] [PubMed] [Google Scholar]