Abstract
Forty-nine influenza B virus isolates collected in Belgium, Finland, Spain, and Israel during the 2001-2002 winter season were categorized into either of two lineages, B/Yamagata/16/88 or B/Victoria/2/87, based on the phylogenetic studies of HA1 sequences. The data trace the geographic spread of B/Victoria/2/87-like viruses and support the emergence of B/Hong Kong/1351/02-like viruses, possibly due to selective advantages of reassortment.
Influenza epidemics are caused by rapid evolution of the viral genome and continue to play a significant role in the annual frequency of mortality and morbidity as a result of respiratory tract infection. Both influenza A virus and influenza B virus undergo antigenic drift, and influenza A virus is capable of antigenic shift resulting from genetic reassortment between different subtypes (2). On the other hand, reassortment of influenza B viruses has been observed between cocirculating influenza B virus strains of different lineages (5, 6, 12, 17). Monitoring antigenic and genetic variations of circulating influenza viruses is critical for the selection of annual vaccine strains. Recent isolates of influenza B viruses, for example, fall into two major phylogenetic lineages: the B/Victoria/2/87 lineage or the B/Yamagata/16/88 lineage (12). The former group of viruses was found primarily in East Asia during the last decade. In contrast, the B/Yamagata/16/88-like viruses have been found worldwide during the same period (11). Increased incidence of B/Victoria/2/87 viruses in the regions beyond Asia in the 2001-2002 season resulted in the World Health Organization recommendation of B/Hong Kong/330/01 (a member of the B/Victoria/2/87 lineage) for the vaccine strain for both the Northern and Southern Hemispheres in the 2002-2003 season (14, 15).
The hemagglutinin (HA) of an influenza virus surface antigen is synthesized as a single polypeptide, which is subsequently cleaved into two chains, HA1 and HA2. The antigenic variation of the HA protein of influenza viruses occurs predominantly in the HA1 domain (13). In this report, we determined the nucleotide sequence of the HA1 gene segment of 49 influenza B viruses circulating in Europe and Israel during the 2001-2002 winter season. The isolates were derived from respiratory specimens in the placebo group, collected during an experimental influenza vaccine efficacy trial conducted concurrently in three European countries, Spain, Belgium, and Finland, as well as in Israel. Briefly, viral RNA was extracted from influenza viruses propagated once in MDCK cells with the aid of a 6700 automatic nucleic acid workstation (Applied Biosystems Inc., Foster City, Calif.). The type and quantities of the viruses were determined for all the specimens using a real-time PCR method (data not shown). Coinfection of influenza A and B was found only in a very few cases (S.-M. Cheng, unpublished data), and the dominant type was chosen to be characterized further. HA1 fragments were amplified by reverse transcription (RT)-PCR using the Titan One tube RT-PCR system (Roche Diagnostics Corp., Indianapolis, Ind.) with oligonucleotides conserved in several influenza B virus strains: 5′-ATAACATCGTCAAACTCACC-3′ and 5′-GCACCATGTAATCAACAACA-3′. DNA sequencing was performed using a 3100 genetic analyzer and BigDye Terminator Mix v3.0 (Applied Biosystems Inc.) with both PCR primers and two additional primers, 5′-GTTCTGTCGTGCATTATAGG-3′ and 5′-GCAACAATGGCTTGGGCTGT-3′. This fragment (from nucleotide 98 to 836) corresponds to the coding region of amino acids 7 to 253 of the HA1 protein. We also selectively amplified and sequenced HA1 fragments directly from five nasal swab specimens, and the result shows a 100% match of the nucleotide sequence with that from cultures (data not shown).
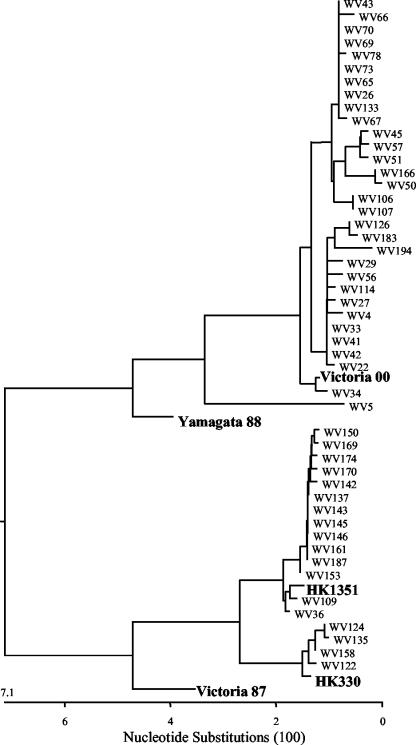
Alignment of multiple sequences was performed by the ClustalV method, which groups sequences into clusters by examining sequence distances between all pairs (4). The phylogenetic tree was then constructed by a neighbor-joining method using MegAlign5.03 software (DNASTAR, Inc., Madison, Wis.). The result indicated that there were two genetically distinct lineages of influenza B viruses among the specimens (Fig. 1). The majority of the B viruses (63%) belonged to the B/Yamagata/16/88 lineage (represented by our reference strain, B/Victoria/504/00, to which the nucleotide sequence identity of the specimens ranged from 94.9 to 99.7%). The remaining viruses (37%) belonged to the B/Victoria/2/87 lineage (represented by our two reference strains B/Hong Kong/330/01 and B/Hong Kong/1351/02, to which the sequence identity of the specimens ranged from 98.9 to 99.7%).
FIG. 1.
Phylogenetic tree based on the nucleotide sequences of HA fragments (nucleotides 98 to 836) of influenza B viruses. Reference strains are indicated by bold letters, and the rest represent all 49 clinical isolates studied. Abbreviations: Victoria 00, B/Victoria/504/00; Yamagata 88, B/Yamagata/16/88; HK1351, B/Hong Kong 1351/02; HK330, B/Hong Kong/330/01; Victoria 87, B/Victoria/2/87.
The appearance of B/Victoria/2/87 lineage in these European countries and Israel documented the spread of this lineage beyond Asia during the 2001-2002 influenza season. This was also observed in Northern Italy (1) and several other countries in Europe (10) as well as in North America (12). It is noteworthy that Israel, the country with the earliest and greatest incidence of B/Hong Kong-like viruses, is located closest to East Asia. On the other hand, the countries with lower incidences (Spain and Belgium) or even an absence (Finland) of B/Hong Kong-like viruses are progressively farther away from East Asia, suggesting the geographic trail of viral spreading. These data suggest that Israel may have been one possible route accounting for the spread of the B/Hong Kong viruses through Europe. Among the B/Victoria/2/87-like group of specimens (n = 18), B/Hong Kong/1351/02-like viruses (n = 14) were found to be at least three times more prevalent than B/Hong Kong/330/01-like viruses (n = 4). We also observed that as the flu season progressed, B/Hong Kong/1351/02-like viruses became more dominant (data not shown). Although the number of specimens in this study is relatively small, we have data from other regions of the world that suggested a similar trend (X. S. Chi, unpublished data).
We also selectively analyzed neuraminidase gene segments from seven clinical specimens, amplified by RT-PCR with oligonucleotides 5′-GCTACCTTCAACTATACAAACG-3′ and 5′-AACGAGGGTATGTCCACTCC-3′ and found that B/Hong Kong/1351/02 is a reassortant of two influenza B lineages, B/Victoria/2/87 and B/Yamagata/16/88 (Table 1). The HA gene of B/Hong Kong/1351/02 derives from the B/Victoria/2/87 lineage, similar to its predecessor, B/Hong Kong/330/01, whereas the neuraminidase gene derives from the B/Yamagata/16/88 lineage, similar to its predecessor, B/Victoria/504/00. These results are in agreement with those reported by Shaw et al. (12). The reassortment between two lineages of influenza B viruses has been reported previously (6, 16), and it is thought that such influenza reassortants have selective advantages in viral growth, replication, or stability. The pattern of spreading of B/Hong Kong/1351/02-like viruses in our study and others (10) suggests that this reassortant may become more dominant worldwide in the 2002-2003 season.
TABLE 1.
Analysis of HA and NA genes of influenza B viruses from clinical specimens
| Group | Subject IDa | Country | Collection dateb | HA type | NA type |
|---|---|---|---|---|---|
| 1 | WV56 | Spain | 1/14/02 | B/Victoria/504/00-like | B/Yamagata/16/88-like |
| 1 | WV183 | Israel | 4/16/02 | B/Victoria/504/00-like | B/Yamagata/16/88-like |
| 2 | WV122 | Belgium | 3/25/02 | B/Hong Kong/330/01-like | B/Victoria/2/87-like |
| 2 | WV135 | Israel | 3/18/02 | B/Hong Kong/330/01-like | B/Victoria/2/87-like |
| 2 | WV124 | Israel | 3/24/02 | B/Hong Kong/330/01-like | B/Victoria/2/87-like |
| 3 | WV145 | Israel | 2/18/02 | B/Hong Kong/1351/02-like | B/Yamagata/16/88-like |
| 3 | WV36 | Spain | 5/14/02 | B/Hong Kong/1351/02-like | B/Yamagata/16/88-like |
Identification for clinical specimen.
Month/day/year.
In recent years, molecular methods have been developed and successfully applied in the diagnosis and surveillance of influenza viruses (3). Here, we employed a high-throughput molecular method combining RT-PCR and DNA sequencing, which provides an alternative to the classical hemagglutinin inhibition assay with specific antisera for determining the phylogenetic relationship of influenza virus isolates. Furthermore, our results reveal detailed genetic information of viral surface protein. We found, for example, some variations in the deduced amino acid sequence of the HA1 protein in our specimens compared to that of reference strains obtained from the Influenza Sequence Database of the Los Alamos National Laboratory. Table 2 shows a summary of all the amino acid variations and the estimated frequencies of the dominant changes in the HA1 sequence of our clinical specimens. Among the B/Victoria/504/00-like group of specimens, there are 28 variations in the HA1 sequence compared with that of the reference strain, with 6 of them being dominant (>35%). One particular specimen, WV5, has the most variations compared to the reference strain. A BLAST search in the database revealed that WV5 has a close relationship with a subgroup of the Yamagata/16/88 lineage, which includes recent circulating strains, such as B/New York/47/01, B/Hong Kong/557/00, and B/Argentina/69/01. It is apparent that there are more variations in the HA1 sequence of the B/Victoria/504/00-like group than in the two B/Hong Kong-like groups, indicating more extensive antigenic drift in the former group of viruses. This observation is in agreement with the hypothesis that new antigenic variants are generated in order to escape existing human immunity (5, 8). A notable variation among the sequences of the B/Yamagata lineage (group 1) is Asp126, which was found in 58% of the isolates studied. The amino acid residue Asn at position 126 had been conserved in B/Yamagata strains for 10 years (5), and a previous study by Nakagawa et al. suggested that a point mutation at position 126 resulted in altered viral antigenicity (9). The variations at position 58, 126, and 175 were also reported recently by Ansaldi et al. (1). However, the variation at position 88 was not found in their specimens. In the group of B/Victoria lineage, nearly all sequences from clinical specimens contain the amino acid Asn at position 197. Interestingly, as a result of this substitution, the stretch of three amino acid residues changes from XXT to NXT (where X is any amino acid), and the latter becomes a possible N-glycosylation site. Such an amino acid substitution at position 197 has also been observed before and was suggested to play an important role in the determination of antigenicity (5, 7). In addition, a predominant change at position 159 in the B/Hong Kong/1351/02 group was also detected, which has not been reported previously. Three signature amino acids, Arg116, Asn121, and Glu164 of B/Hong Kong/330/01 (12), are all conserved in our specimens.
TABLE 2.
Amino acid variations in HA1 sequences of influenza B viruses
| Group or strain | Amino acid residue at positione:
|
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 29 | 34 | 40 | 48 | 56 | 58 | 75 | 76 | 78 | 87 | 88 | 116 | 126 | 127 | 148 | 159 | 162 | 164 | 168 | 175 | 176 | 179 | 180 | 182 | 183 | 184 | 197 | 199 | 220 | 225 | 233 | |
| Group 1a | |||||||||||||||||||||||||||||||
| B/Victoria/504/00b | A | T | H | K | T | L | I | T | S | V | K | K | N | A | S | A | R | D | T | V | E | H | I | T | K | E | D | T | I | V | E |
| WV5 | V | — | Y | R | D | — | T | — | — | — | R | N | D | — | — | — | K | — | N | — | — | Y | V | K | E | G | N | — | V | — | D |
| WV27 | — | I | — | — | — | — | — | — | — | — | R | — | — | — | — | — | — | — | — | I | — | — | — | — | — | — | N | — | — | — | D |
| WV29 | — | — | — | — | — | — | — | — | — | — | R | — | — | — | — | — | — | G | — | I | — | — | — | — | — | — | N | — | — | — | D |
| WV34 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | M | — | — | — | — | — | — | — | — | — | Y | — | — | — | — |
| WV50 | — | — | — | — | — | — | — | — | P | — | R | — | D | — | N | — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | D |
| WV56 | — | — | — | — | — | — | — | — | — | A | R | — | — | — | — | — | — | — | — | I | — | — | — | — | — | — | N | — | — | — | D |
| WV57 | — | — | — | — | — | F | — | — | — | — | R | — | D | — | N | — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | D |
| WV78 | — | — | — | — | — | — | — | — | — | — | R | — | D | — | — | — | — | — | — | — | K | — | — | — | — | — | N | — | — | — | D |
| WV114 | — | — | — | — | — | F | — | — | P | — | R | — | D | — | N | — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | D |
| WV166 | — | — | — | — | — | — | — | — | — | — | R | — | — | T | — | — | — | — | — | I | — | — | — | — | — | — | N | — | — | — | D |
| WV194 | — | — | — | — | — | F | — | — | — | — | R | — | D | — | N | — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | D |
| WV45,51 | — | — | — | — | — | F | — | — | — | — | R | — | D | — | N | — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | D |
| WV22,33,126, 183,4,41,42 | — | — | — | — | — | — | — | — | — | — | R | — | — | — | — | — | — | — | — | I | — | — | — | — | — | — | N | — | — | — | D |
| WV106,107,133, 26,43,65,66,67, 69,70,73 | — | — | — | — | — | F | — | — | — | — | R | — | D | — | — | — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | D |
| Group 2c | |||||||||||||||||||||||||||||||
| B/Hong Kong/330/01b | V | T | H | K | K | L | N | I | S | V | R | R | N | A | V | A | K | D | T | I | E | Y | I | T | E | G | S | T | V | V | N |
| WV122 | — | — | — | — | — | — | T | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | N | A | — | — | — | |
| WV124,135,158 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — |
| Group 3d | |||||||||||||||||||||||||||||||
| B/Hong Kong1351/02b | V | T | H | K | K | L | N | I | S | V | R | H | N | A | V | A | K | D | T | I | E | Y | I | T | E | G | K | T | V | V | N |
| WV36,109 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — |
| WV137,142,143,145, 146,150,153,161, 169,170,174,187 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | V | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — |
n = 31 specimens. Amino acid substitution frequencies are 55% (position 58), 97% (position 88), 58% (position 126), 39% (position 175), 97% (position 197), and 97% (position 233).
Reference strain.
n = 4 specimens. Amino acid substitution frequency was 100% at position 197.
n = 14 specimens. Amino acid substitution frequencies are 86% (position 159) and 100% (position 197).
—, amino acid residue identical to that in the reference strain. Amin with high frequency variations (>35%) are shown in bold characters.
Nucleotide sequence accession numbers.
Nucleotide sequences of the HA gene can be found in GenBank with the following accession numbers: B/Victoria/2/87, M22943; B/Yamagata/16/88, M36105; B/Victoria/504/00, ISDN20057; B/Hong Kong/330/01, AF532549; B/Hong Kong/1351/02, AF532545; B/Hong Kong/557/00, AF532553; B/Argentina/69/01, AF532525; B/New York/47/01, AY139048. Clinical isolates: WV106, AY375988; WV107, AY375989; WV109, AY375990; WV114, AY375991; WV122, AY375992; WV124, AY375993; WV126, AY375994; WV133, AY375995; WV135, AY375996; WV137, AY375997; WV142, AY375998; WV143, AY375999; WV145, AY376000; WV146, AY376001; WV150, AY376002; WV153, AY376003; WV158, AY376004; WV161, AY376005; WV166, AY376006; WV169, AY376007; WV170, AY376008; WV174, AY376009; WV183, AY376010; WV187, AY376011; WV194, AY376012; WV22, AY376013; WV26, AY376014; WV27, AY376015; WV29, AY376016; WV33, AY376017; WV34, AY376018; WV36, AY376019; WV4, AY376020; WV41, AY376021; WV42, AY376022; WV43, AY376023; WV45, AY376024; WV5, AY376025; WV50, AY376026; WV51, AY376027; WV56, AY376028; WV57, AY376029; WV65, AY376030; WV66, AY376031; WV67, AY376032; WV69, AY376033; WV70, AY376034; WV73, AY376035; WV78, AY376036.
Acknowledgments
We thank Giuseppe Palladino for providing wild-type influenza reference strains and Fenglan Li for technical assistance.
REFERENCES
- 1.Ansaldi, F., P. D'Agaro, D. De Florentiis, S. Puzelli, Y. P. Lin, V. Gregory, M. Bennett, I. Donatelli, R. Gasparini, P. Crovari, A. Hay, and C. Campello. 2003. Molecular characterization of influenza B viruses circulating in northern Italy during the 2001-2002 epidemic season. J. Med. Virol. 70:463-469. [DOI] [PubMed] [Google Scholar]
- 2.Cox, N., and C. Bender. 1995. The molecular epidemiology of influenza viruses. Semin. Virol. 6:359-370. [Google Scholar]
- 3.Ellis, J. S., and M. C. Zambon. 2002. Molecular diagnosis of influenza. Rev. Med. Virol. 12:375-389. [DOI] [PubMed] [Google Scholar]
- 4.Higgins, D. G., and P. M. Sharp. 1989. Fast and sensitive multiple sequence alignments on a microcomputer. Comput. Appl. Biosci. 5:151-153. [DOI] [PubMed] [Google Scholar]
- 5.Lindstrom, S. E., Y. Hiromoto, H. Nishimura, T. Saito, R. Nerome, and K. Nerome. 1999. Comparative analysis of evolutionary mechanisms of the hemagglutinin and three internal protein genes of influenza B virus: multiple cocirculating lineages and frequent reassortment of the NP, M, and NS genes. J. Virol. 73:4413-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCullers, J. A., G. C. Wang, S. He, and R. G. Webster. 1999. Reassortment and insertion-deletion are strategies for the evolution of influenza B viruses in nature. J. Virol. 73:7343-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakagawa, N., R. Kubota, A. Maeda, T. Nakagawa, and Y. Okuno. 2000. Heterogeneity of influenza B virus strains in one epidemic season differentiated by monoclonal antibodies and nucleotide sequences. J. Clin. Microbiol. 38:3467-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakagawa, N., R. Kubota, T. Nakagawa, and Y. Okuno. 2001. Antigenic variants with amino acid deletions clarify a neutralizing epitope specific for influenza B virus Victoria group strains. J. Gen. Virol. 82:2169-2172. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa, N., S. Nukuzuma, S. Haratome, S. Go, T. Nakagawa, and K. Hayashi. 2002. Emergence of an influenza B virus with antigenic change. J. Clin. Microbiol. 40:3068-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paget, W. J., T. J. Meerhoff, and N. L. Goddard. 2002. Mild to moderate influenza activity in Europe and the detection of novel A(H1N2) and B viruses during the winter of 2001-02. Eurosurveillance 7:147-157. [DOI] [PubMed] [Google Scholar]
- 11.Rota, P. A., T. R. Wallis, M. W. Harmon, J. S. Rota, A. P. Kendal, and K. Nerome. 1990. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 175:59-68. [DOI] [PubMed] [Google Scholar]
- 12.Shaw, M. W., X. Xu, Y. Li, S. Normand, R. T. Ueki, G. Y. Kunimoto, H. Hall, A. Klimov, N. J. Cox, and K. Subbarao. 2002. Reappearance and global spread of variants of influenza B/victoria/2/87 lineage viruses in the 2000-2001 and 2001-2002 seasons. Virology 303:1-8. [DOI] [PubMed] [Google Scholar]
- 13.Wiley, D. C., and J. J. Skehel. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56:365-394. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization. 2002. Recommended composition of influenza virus vaccines for use in the 2002-2003 influenza season. Wkly. Epidemiol. Rec. 77:57-68. [Google Scholar]
- 15.World Health Organization. 2002. Recommended composition of influenza virus vaccines for use in the 2003 influenza season. Wkly. Epidemiol. Rec. 77:341-348.12407831 [Google Scholar]
- 16.Xu, X., J. Shaw, C. B. Smith, N. J. Cox, and A. I. Klimov. 2001. Multiple lineages co-circulation and genetic reassortment of the neuraminidase and hemagglutinin genes within influenza viruses of the same type/subtype, p. 383-387. In A. Osterhaus, N. Cox, and H. A. (ed.), Options for the control of influenza IV. Elsevier Science, New York, N.Y.
- 17.Yamashita, M., M. Krystal, W. M. Fitch, and P. Palese. 1988. Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology 163:112-122. [DOI] [PubMed] [Google Scholar]