Abstract
Introduction
Prediction of progression to cancer in patients with Barrett’s esophagus is difficult using current techniques. We determined whether DNA promoter hypermethylation of genes frequently methylated in esophageal adenocarcinoma (p16 and APC) could be used as predictors of progression in Barrett’s esophagus.
Methods
We first performed a cross-sectional study to evaluate the prevalence of gene hypermethylation in biopsies from patients with normal esophagus (n=17), Barrett’s esophagus (n=102), and adenocarcinoma (n=42). We then performed a nested case-control study comparing gene hypermethylation in Barrett’s esophagus patients who progressed from baseline pathology to high-grade dysplasia or cancer (n=7) versus patients who did not progress (n=50).
Results
None of the patients with normal esophagus had p16 or APC hypermethylation. Hypermethylation was prevalent in Barrett’s esophagus without dysplasia or low-grade dysplasia (p16=31% and APC=50%; p<0.01) and high-grade dysplasia or adenocarcinoma (p16=54% and APC=68%; p<0.001) compared to normal esophagus (not detected). Patients who progressed from baseline pathology to high-grade dysplasia or cancer had higher prevalence of hypermethylation in their initial esophagus biopsies compared to those who did not progress for both p16 (100% vs. 33%; p=0.008) and APC (86% vs. 40%; p=0.02). Hypermethylation of both p16 and APC was a strong predictor of subsequent progression to cancer during a mean follow-up time of 4.1 years (adjusted OR [95% CI]=14.97 [1.73,inf], p=0.01). Among patients who were negative for both p16 and APC hypermethylation, none progressed from baseline pathology to high-grade dysplasia or cancer.
Conclusions
Hypermethylation of both p16 and APC strongly predicts progression to high-grade dysplasia or cancer in patients with Barrett’s esophagus. Absence of p16 and APC hypermethylation is associated with a benign course.
INTRODUCTION
Patients with Barrett’s esophagus (BE) have a significantly increased risk of esophageal adenocarcinoma, 40–125 times higher than the general population (1). The detection of dysplasia in esophageal biopsies is currently the only standard method used in clinical practice as a marker for increased risk of cancer. However, dysplasia has not been a reliable marker for predicting malignant progression because of poor interobserver agreement among pathologists for diagnosis of dysplasia (2, 3) and the inherent problem of incomplete sampling of the Barrett’s mucosa by standard mucosal biopsy techniques. Since only a small fraction of BE patients will actually progress to adenocarcinoma (4–7), there is a need to develop markers that may accurately predict which patients with BE are likely to have aggressive disease and progress from their baseline pathology to high-grade dysplasia (HGD) or cancer versus patients who will remain histologically stable and have a benign course. This would allow for better risk stratification of patients with BE in order to target aggressive surveillance and intervention towards only those patients at highest risk for neoplastic progression. Predictive biomarkers would thus have significant utility in the management of BE patients.
DNA promoter hypermethylation is an epigenetic phenomenon that holds significant promise as a molecular marker for early detection of malignant transformation, since this alteration is found in pre-invasive neoplastic tissue and involves genes which regulate key pathways in cancer (8–10). Methylation of the promoter region of a gene results in suppression of gene transcription (11) and has been associated with the development of esophageal cancer (12–20). However, to date, few studies have evaluated the role of gene hypermethylation in predicting the progression from BE to esophageal adenocarcinoma in a longitudinal cohort of patients (21). Our study focused on two genes, p16/CDKN2a (22–26) and APC (27) which are important tumor suppressor genes that have been found to be methylated in esophageal adenocarcinoma. The p16 gene is a cyclin-dependent kinase inhibitor and tumor suppressor involved in many solid tumors and lymphomas. APC is the adenomatous polyposis coli tumor suppressor gene.
Our aims were: 1) to perform a cross-sectional study to determine the prevalence of DNA promoter hypermethylation in the premalignant and malignant stages of BE; and 2) to perform a nested case-control study to determine whether DNA promoter hypermethylation is a biomarker that will predict which patients with BE are likely to progress from baseline pathology to HGD or esophageal adenocarcinoma.
METHODS
Study Population
This study was approved by the Johns Hopkins Medicine Institutional Review Board (Baltimore, Maryland, USA). We used archived esophageal biopsy specimens obtained from adult patients in our BE and Esophageal Cancer Registry during outpatient upper endoscopy at the Johns Hopkins Hospital between 1998–2007. The BE and Esophageal Cancer Registry tracks all patients with the diagnosis of GERD, BE, or esophageal cancer who had endoscopy at The Johns Hopkins Hospital since 1996.
Specimens were excluded if they were obtained under the following situations which had potential for DNA damage and thus could confound methylation results: (1) after photodynamic therapy or other types of mucosal ablation; (2) undergoing active chemotherapy; and (3) history of radiation therapy to the chest. Clinical and demographic information was obtained for each patient from the BE and Esophageal Cancer Registry including gender, race, and age. All patients underwent endoscopic surveillance using the Seattle biopsy protocol (28, 29). This involved four quadrant biopsies every 2 cm in the Barrett’s segment for patients without dysplasia or with low-grade dysplasia, and four quadrant biopsies every 1 cm for patients with suspected or known high-grade dysplasia. Jumbo biopsy forceps were used until 2004; thereafter large capacity biopsy forceps were used. Interval of surveillance was every 3 months for patients with high-grade dysplasia, every year for patients with low-grade dysplasia, and every 3 years for patients without dysplasia.
For each patient, we selected one archived esophageal biopsy specimen representative of their diagnosis (highest grade of neoplasia) at the time of endoscopy. In cases where the specimen contained several areas of different grades of neoplasia, classification was based on the highest grade of neoplasia present, according to clinical practice. All specimens were reviewed by an expert gastrointestinal pathologist (EAM) to ensure accurate histological diagnosis, using a method similar to the Vienna classification of gastrointestinal epithelial neoplasia (30). Each specimen was labeled with a study code, which did not include patient identifiers.
For the cross-sectional study, we selected specimens with the pathological diagnosis of BE with no dysplasia, BE with low-grade dysplasia, BE with high-grade dysplasia, or esophageal adenocarcinoma. In cases where the specimen contained several areas of different grades of neoplasia, the specimen was classified based on the highest grade of neoplasia present, since this is most clinically relevant. In addition, control normal esophagus specimens taken from patients without any history of esophagitis, BE, or esophageal cancer who underwent upper endoscopy for evaluation of gastrointestinal symptoms were also examined.
For the nested case-control study, we obtained archived esophageal biopsy specimens from patients who had undergone at least two upper endoscopies between 1998–2007, and had an initial pathological diagnosis from the first endoscopy of BE (at any grade) without evidence of adenocarcinoma. For patients who had undergone multiple upper endoscopies for surveillance, we obtained the initial specimen from the earliest endoscopy during this time period and the clinical outcome was based on the last endoscopy within the time period. Follow-up time was calculated by measuring the time interval between the first and last endoscopies within the study period. We chose patients who had a minimum of 12 months between endoscopies to rule out synchronous cancers. The cases included all patients who progressed from their baseline pathology to HGD or adenocarcinoma within the cohort. The controls were randomly selected from all patients within the cohort who did not develop adenocarcinoma during the study period.
DNA preparation
Archived paraffin-embedded esophageal biopsies were used for this study. For each esophageal biopsy used, a total of ten sequential 10-micron sections were cut, with the first and last sections used for H&E staining and the intervening 8 sections left unstained and used for methylation assays. The first and last H&E sections were examined by a pathologist with expertise in BE (EAM) to confirm the diagnosis of BE and degree of dysplasia. Tissue was first deparaffinized with xylene, digested overnight with proteinase K in 1% SDS, and DNA isolated by phenol-chloroform extraction and ethanol precipitation.
Sodium bisulfite modification
Approximately 3 μg of genomic DNA was treated with sodium bisulfite, which modified the DNA by converting all unmethylated cytosines to uracil, and then subsequently to thymidine during PCR. The modified DNA was purified with the Wizard DNA purification system (Promega), desulfonated with NaOH, precipitated with ethanol, and resuspended in Tris-EDTA buffer as described.(31)
Methylation-specific PCR
The methylation status of the promoter regions of the genes p16 and APC was determined using a nested, 2-stage PCR approach as previously described (31, 32). First, a stage-1 multiplex PCR reaction was performed using primers which flanked the CpG-rich promoter regions for both genes simultaneously. One hundred ng of DNA template was used for stage-1 amplification. PCR products of stage-1 were then diluted 1:500 and a stage-2 PCR performed to discriminate between methylated and unmethylated sequences. All primer sequences for this nested-MSP approach have been published previously (33).
PCR used a 25-μL reaction volume as described (34), and 1.0 U of Jump Start Red Taq DNA polymerase (Sigma, St. Louis, MO). DNA isolated from normal peripheral lymphocytes from healthy individuals served as a negative methylation control. Human lymphocyte DNA treated with SssI methyltransferase (NEB, Beverly, MA) served as a positive methylation control. PCR conditionsfor stage-1 were: 95°C hotstart × 5 min, then 35 repetitive cycles of denaturation (95°C × 30 s), annealing (55°C × 30 s), extension (72°C × 36s) followed by a final 5 min extension at 72°C. The PCR conditionsfor stage-2 of the nested MSP were as follows: 95°C hotstart ×5 min, then repetitive cycles of denaturation [30 cycles for p16; 25 cycles for APC] (95°C × 30 s), annealing (60°C × 30 s), extension (72°C × 30s) followed by a final 5 min extension at 72°C. Ten μL of each PCR reaction was loaded onto non-denaturing 6% polyacrylamide gels, stained with ethidium bromide, and visualized under UV illumination. Methylation status was coded as a dichotomous variable (positive or negative). Specimens that failed to amplify with either PCR reaction signified inadequate DNA and and these were not included in the data analysis. In our experience, this occurred in less than 5% of samples.
Data analysis
Methylation-specific PCR was performed by a researcher blinded to the pathological diagnosis to determine promoter hypermethylation in p16 and APC.
For the cross-sectional study, we determined the prevalence of gene promoter hypermethylation in biopsy specimens as well as odds ratios and corresponding 95% confidence intervals for each gene (p16, APC) using multivariable logistic regression. In specimens with data for both p16 and APC promoter hypermethylation, we also examined whether both genes were methylated. For these two-gene analyses, we only included specimens with adequate PCR amplification for both genes; therefore the total number of specimens in this analysis is smaller than with the individual gene analyses. We adjusted for multiple comparisons using the Bonferroni adjustment for significance level.
For the nested case-control study, we defined cases as BE patients who progressed from baseline pathology to HGD or esophageal adenocarcinoma and controls as BE patients who did not progress from baseline pathology. For each patient, we determined the hypermethylation status of p16 and APC in their initial esophagus biopsy specimens to evaluate whether patients who had presence of gene promoter hypermethylation at initial presentation would be more likely to subsequently progress to HGD or adenocarcinoma over their period of follow-up. The primary outcome variable was a dichotomous variable based on the patient’s last endoscopy during the study period, indicating whether or not there was evidence of progression to HGD or adenocarcinoma. We calculated odds ratios and corresponding 95% confidence intervals for each gene (p16, APC) by case-control status using univariate logistic regression. Exact methods were used when a gene methylation result predicted progression perfectly. We then performed time to event analyses using the method of Kaplan-Meier with the corresponding log rank test.
RESULTS
Cross-sectional study
To investigate whether there was an association of neoplasia grade with gene hypermethylation, we analyzed specimens from 102 patients with BE and 42 patients with esophageal adenocarcinoma. Among the 102 patients with BE, 52 patients had no dysplasia, 30 patients had low-grade dysplasia, and 20 patients had high-grade dysplasia. In addition, we included 17 patients with normal esophagus to determine whether methylation of these genes was found in non-disease states. The demographics of the BE patients included in the cross-sectional study are listed in Table 1. These are similar to other previously published cohorts of BE patients.(35–37) Age and gender were significantly different among patients in the different grades of dysplasia, and subsequent analyses were therefore adjusted for these variables.
Table 1.
Demographics of patients
| Cross-sectional study | Longitudinal nested case-control study | |||||||
|---|---|---|---|---|---|---|---|---|
| Total (n=144) | ND/LGD (n=82) | HGD/CA (n=62) | p | Total (n=57) | Controls (n=50) | Cases (n=7) | p | |
| Mean age (years) | 64.1 | 60.2 | 69.2 | <0.001* | 60.5 | 59.5 | 67.7 | 0.16 |
| Gender = male (%) | 81% | 76% | 89% | 0.05 | 79% | 78% | 86% | 0.64 |
| Race = white (%) | 97% | 96% | 98% | 0.46 | 98% | 98% | 100% | 0.73 |
Denotes statistical significance at p<0.05
BE samples with differing grades of neoplasia were first examined to determine whether specimens with more severe dysplasia or adenocarcinoma had differing prevalence of p16 and/or APC promoter hypermethylation. Specimens were categorized into three main pathology categories for purposes of data analysis: normal esophagus, BE with no dysplasia or low-grade dysplasia, and BE with HGD or esophageal adenocarcinoma (Table 2).
Table 2.
Association of dysplasia grade and promoter hypermethylation
| Barrett’s esophagus stage | p16 | p | APC | p | Both Genes | p |
|---|---|---|---|---|---|---|
| Normal esophagus | 0/17 (0%) | 0/17 (0%) | 0/17 (0%) | |||
| No/low-grade dysplasia | 23/74 (31%) | 0.008a* | 38/76 (50%) | < 0.001a* | 17/68 (25%) | 0.021 |
| No dysplasia | 14/47 (30%) | 24/48 (50%) | 9/43 (21%) | |||
| Low-grade dysplasia | 9/27 (33%) | 14/28 (50%) | 8/25 (32%) | |||
| High-grade dysplasia/adenocarcinoma | 32/59 (54%) | <0.001a* 0.007b* |
34/50 (68%) | <0.001a* 0.046b |
19/47 (40%) | 0.002a* 0.080b |
| High-grade dysplasia | 10/18 (56%) | 14/18 (78%) | 7/16 (44%) | |||
| Adenocarcinoma | 22/41 (54%) | 20/32 (63%) | 12/31 (39%) | |||
| Odds Ratios (95% CI) | 2.63 (1.29–5.35) c 2.12 (0.98–4.61) d |
0.008+ 0.057 |
2.13 (1.01–4.48) c 1.70 (0.76–3.83) d |
0.047+ 0.197 |
2.04 (0.91–4.53) c 1.57 (0.66–3.77) d |
0.082 0.310 |
OR = odds ratio
compared to normal esophagus
compared to Barrett’s esophagus with no or low-grade dysplasia
unadjusted odds ratios compared to no or low-grade dysplasia
adjusted odds ratios compared to no or low-grade dysplasia adjusted for age, gender, and race
denotes statistical significance, set to a level of p < 0.017 using the Bonferonni method to adjust for multiple comparisons
denotes statistical significance at p<0.05
None of the patients with normal esophagus had promoter hypermethylation of p16 or APC. In contrast, those with BE and no dysplasia or low-grade dysplasia had a significantly higher prevalence of promoter hypermethylation for both p16 and APC in their mucosal biopsies (31% and 50%, respectively; p<0.01 for both compared to normal esophagus). Those with BE and HGD or adenocarcinoma had even higher rates of promoter hypermethylation for both p16 and APC (54% and 68%, respectively; p<0.001 for both compared to normal esophagus; p<0.05 for both compared to BE with no or low-grade dysplasia).
We then analyzed the strength of the association of dysplasia grade with gene hypermethylation in patients with BE by calculating odds ratios (Table 2). Individually, p16 and APC hypermethylation were significantly associated with having BE with HGD or adenocarcinoma in the univariate analyses. However, when the odds ratios were adjusted for age, gender, and race, these associations became non-significant.
Nested case-control study
Since hypermethylation of p16 and APC was present at a moderate level even in nondysplastic BE, we next examined whether the presence of these changes in lower grade histology would predict progression to cancer. For this study, we identified 7 patients with BE who subsequently progressed from baseline pathology to HGD or adenocarcinoma (cases), and 50 patients who did not progress from baseline pathology (controls) during a mean follow-up time of 4.1 years.
The demographics of the patients included in the nested case-control study are shown in Table 1. There were no significant differences among cases and controls with respect to age, gender, and race. Overall, patients had baseline diagnoses of BE with no dysplasia (29/57 =51%), low-grade dysplasia (22/57=39%), and high-grade dysplasia (6/57=11%). Among the cases, baseline diagnoses included no dysplasia (n=2), low-grade dysplasia (n=4), and high-grade dysplasia (n=1). Among the controls, baseline diagnoses included no dysplasia (n=28), low-grade dysplasia (n=17), and high-grade dysplasia (n=5). Among the cases, the mean length of time to progression was 24 months.
Patients with BE who subsequently progressed from baseline pathology to HGD or adenocarcinoma had a significantly higher prevalence of hypermethylation in their initial esophagus biopsies compared to those who did not progress with respect to both p16 (100% vs. 33%; p=0.008) and APC (86% vs. 40%; p=0.02) (Table 3). Hypermethylation of p16 was a highly significant predictor of subsequent progression from baseline pathology to HGD or adenocarcinoma (OR [95% CI] = 10.02 [1.18,inf], p=0.03) (Table 3, Figure 1). The association of APC hypermethylation and subsequent progression to adenocarcinoma was also significant (OR [95% CI] =9.00 [1.01,80.52], p=0.049, Table 3). However, the combination of both hypermethylated p16 and APC in baseline esophagus biopsies was the best predictor of subsequent progression to adenocarcinoma (OR [95% CI] =14.97 [1.73,inf], p=0.012). Among patients who were negative for both p16 and APC hypermethylation, none progressed to HGD or cancer.
Table 3.
Association of p16 and/or APC promoter hypermethylation with progression to high-grade dysplasia or cancer
| Clinical Outcome of patients | p16 | p | APC | p | Both genes | p |
|---|---|---|---|---|---|---|
| Progressed from baseline pathology to high-grade dysplasia or cancer | 100% | 86% | 100% | |||
| Did not progress from baseline pathology | 33% | 40% | 24% | |||
| Odds Ratios (95% CI) | 10.02 (1.18–inf) | 0.034* | 9.00 (1.01–80.52) | 0.049* | 14.97 (1.73–inf) | 0.012* |
denotes statistical significance at p<0.05
Figure 1.
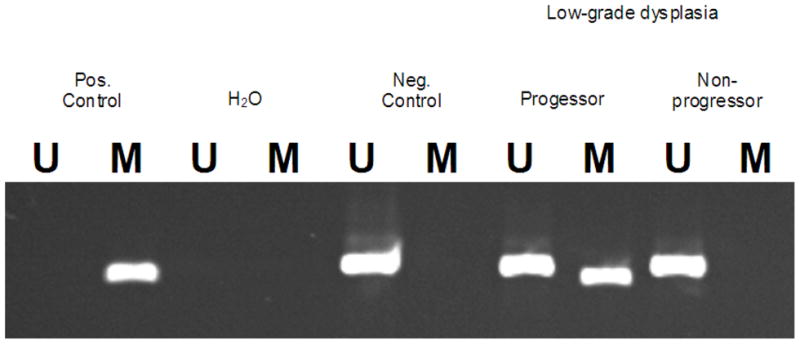
Representative results from the methylation-specific PCR assay can be seen for the p16 gene. Visualization of a band in the U lane indicates the presence of unmethylated DNA in the p16 promoter, whereas a band in the M lane indicates the presence of methylated DNA in the p16 promoter. We confirmed the presence of methylated DNA by sequencing the promoter region amplified by the PCR primers. The representative gel below shows that one BE patient who initially presented with low-grade dysplasia and progressed pathologically to cancer had hypermethylated promoter DNA in the p16 gene in an esophagus biopsy taken at initial presentation. Meanwhile, another BE patient who also initially presented with low-grade dysplasia but did not progress pathologically did not have any promoter hypermethylation detected in the p16 gene on initial esophagus biopsy.
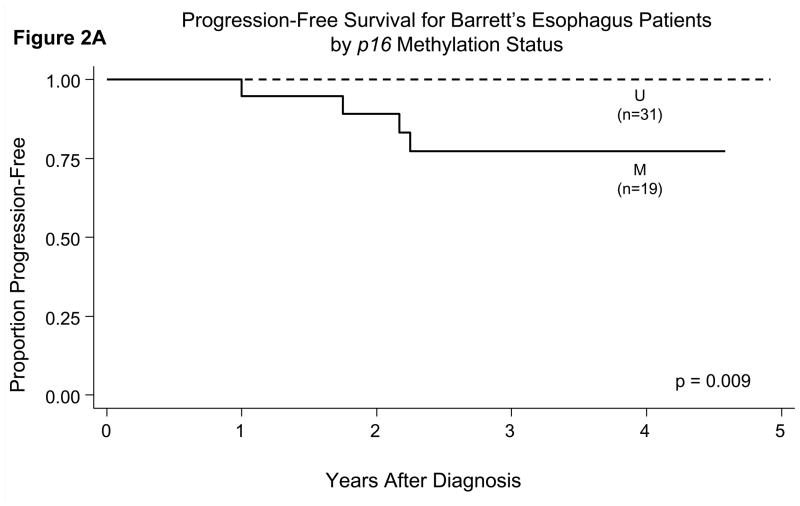
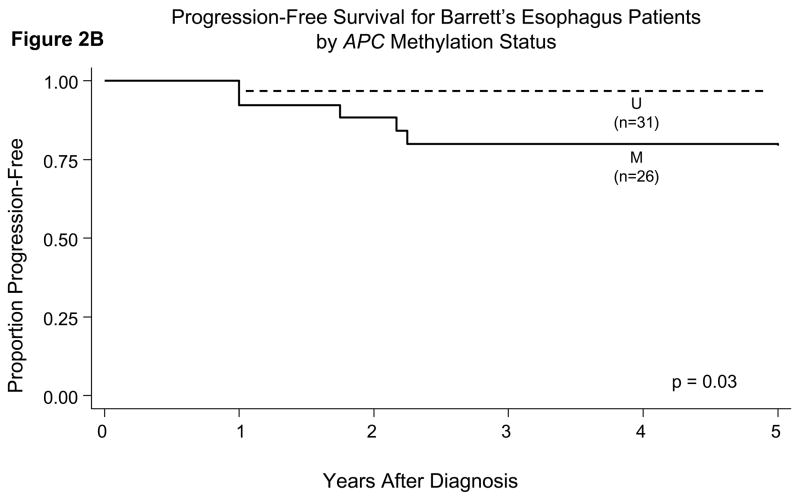
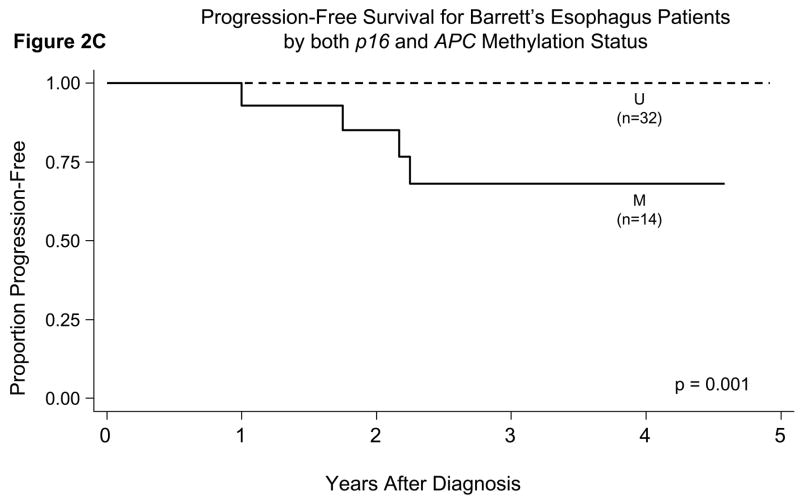
When we incorporated the length of follow-up time in prediction of outcome, we found statistically significant differences in the probability of progression-free survival when comparing patients who had hypermethylation of p16 or APC in their initial esophagus biopsies versus those who did not (Figures 2A, 2B, 2C). Patients who had hypermethylation of p16 or APC individually or both genes all had a significantly lower probability of progression-free survival (p<0.05 for all comparisons of presence versus absence of gene hypermethylation by the log-rank test).
Figure 2.
Probability of progression-free survival in patients with BE in relation to their gene promoter hypermethylation status in initial esophagus biopsies. There were statistically significant differences for p16 (Panel A, p=0.009), APC (Panel B, p=0.03), and both genes (p16 and APC) hypermethylated (Panel C, p=0.001) by the log-rank test.
DISCUSSION
There was a moderate prevalence of p16 and APC hypermethylation in BE patients at all grades of dysplasia. These results suggest that p16 and APC hypermethylation are early events in the neoplastic progression of BE, since even BE patients with no dysplasia or low-grade dysplasia had hypermethylation of p16 and APC ranging from 30–50%. In our longitudinal nested case-control study, we therefore sought to determine whether the presence of these changes in lower grade histology would predict aggressive behavior and subsequent progression from baseline pathology to HGD or adenocarcinoma.
We felt it was important to include patients with a baseline diagnosis of HGD in our study because management of these patients is controversial. The risk of progression to cancer in patients with HGD has been estimated to range from 16–61% based on published studies (2, 38–41). Therefore, a large proportion of patients with HGD will remain stable with HGD long-term, and it is more important to distinguish those with aggressive disease from those with stable disease, even among those with an initial diagnosis of HGD. Thus, we sought to determine whether DNA hypermethylation could differentiate between aggressive and stable Barrett’s esophagus, regardless of the initial baseline diagnosis. We are confident that in patients who have undergone endoscopic surveillance with the Seattle endoscopic biopsy protocol at our institution, we are not missing any prevalent cancers at the time of endoscopy. We base this statement on the results of a separate study performed at our institution (42), where we reviewed patients with Barrett’s esophagus and HGD who subsequently underwent esophagectomy and found that among patients who had undergone the Seattle endoscopic biopsy protocol (as these patients had) there were no occult cancers found at the time of surgery.
We found that promoter hypermethylation of both p16 and APC was the best predictor of subsequent progression from baseline pathology to HGD or adenocarcinoma. Importantly, a negative methylation result may be even more clinically useful since our results showed that no patient without hypermethylation of both p16 and APC progressed from baseline pathology to HGD or cancer.
Our results are consistent with a previous study which found p16, RUNX3, and HPP1 hypermethylation to be predictors of progression to the combined endpoint of high-grade dysplasia or adenocarcinoma in BE patients, with the odds ratios for risk of progression being 1.74, 1.80, and 1.77, respectively (21). However, our study differs from this previous study in that our study results show a much stronger association between promoter hypermethylation of both p16 and APC and subsequent progression to HGD or cancer with much higher odds ratios than previously reported (21). These high odds ratios may be due to differences in the assays used for methylation assessment. The use in our study of the highly sensitive nested methylation-specific PCR assay for detection of gene promoter hypermethylation may explain the high odds ratios and strong associations between promoter hypermethylation of p16 and APC and subsequent progression to cancer which we observed.
Our study results vary somewhat from a previous study which reported that APC, TIMP3, and TERT hypermethylation were predictive markers of progression to adenocarcinoma (43). APC, TIMP3, and TERT promoters were hypermethylated in 100%, 91%, and 92% of cases in BE mucosa from patients who had adenocarcinoma. Meanwhile, in contrast to our study, p16 hypermethylation was not reported to be a predictive marker. There are several possible reasons for the difference in results, which are most likely attributable to differences in the study design and methodology. First, the BE specimens for the cases in this previous study were taken from BE mucosa adjacent to adenocarcinoma in esophagectomy specimens. Meanwhile, in the cases for our study, we used BE specimens from endoscopic biopsies which were obtained temporally at least 12 months prior to the diagnosis of adenocarcinoma. This previous study also used a different assay for assessing methylation, the methylation-sensitive dot-blot assay. Our use of the highly sensitive nested methylation-specific PCR assay for detection of gene promoter hypermethylation may explain why we were able to detect p16 hypermethylation as a predictor of progression.
A limitation of our study is that biopsy selection was dependent on dysplasia classification and therefore is subject to the known problems of inter-observer variability in dysplasia. Although we had our specimens examined by an expert pathologist in Barrett’s esophagus, future cross-sectional study could be more rigorously performed by having multiple pathologists examine each specimen and to only include those specimens for which the diagnosis was universally agreed upon.
In the longitudinal nested case-control study, we do recognize that our confidence intervals are very large due to the small number of cases, and therefore the exact point estimates may not be accurate. Future studies should be done with larger populations and longer follow-up time to validate our study results. This will likely require multi-center collaboration in order to pool BE populations across institutions to increase the number of cases captured.
In conclusion, we report that promoter hypermethylation of p16 and APC is a frequent and early event in the stepwise progression from normal esophagus through BE to esophageal adenocarcinoma. Regardless of baseline dysplasia grade, hypermethylation of both p16 and APC is a strong predictor of subsequent progression from baseline pathology to HGD or esophageal adenocarcinoma in patients with BE even before pathologic changes are evident. Meanwhile, the absence of p16 and APC hypermethylation may be equally important in predicting a benign course. These results could have significant clinical implications for patient management by using methylation profiling to identify BE patients who are at the highest and lowest risk for progression to cancer. If these results are confirmed, follow-up of BE patients could then be adjusted accordingly, with the highest risk patients who have both p16 and APC hypermethylated undergoing early intervention in the form of more frequent endoscopic surveillance, chemoprevention, ablative therapy, or surgery. Meanwhile, the lowest risk patients with BE unmethylated for both p16 and APC could be offered more conservative management.
Our study results suggest that the presence or absence of p16 and APC promoter methylation in esophageal tissue may potentially be used as biomarkers to help in the risk stratification of patients with BE. The triggers for the neoplastic progression of BE are likely multifactorial in nature, involving both epigenetic and genetic events, as well as environmental factors. Over time, all of these events may have cumulative effects that ultimately lead to the development of cancer. A multitude of epigenetic and genetic factors have been studied as potential biomarkers including p53 protein overexpression and allelic loss (44, 45), aneuploidy or tetraploidy (40, 46), and gains of proto-oncogenes (47). Meanwhile, environmental exposures such as obesity (48), diet (49), and nonsteroidal anti-inflammatory drugs (50) have also been postulated to influence the development or progression of BE. It is likely that the best model for predicting clinical outcome in BE patients will ultimately involve a combination of epigenetic and genetic factors, dysplasia grade and other pathological characteristics, and clinical and demographic attributes.
Acknowledgments
Financial support: American College of Gastroenterology Clinical Research Award, American Gastroenterological Association Esophageal Clinical Research Award, National Institutes of Health 1K23DK068149-01A1 and 5K12RR017627-02, Roy L. Jeannotte Memorial Foundation, The Jerry D’Amato Foundation, Early Detection Research Network. None of the study sponsors had any role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
We would like to thank Frank Giardiello, Marie Diener-West, Neil Powe, Frederick Brancati, Christine Smith, Laurie McClelland, and Craig Hooker for their helpful advice and discussions in the development of this research study.
Footnotes
Potential competing interests: JGH isa consultant for OncoMethylome Sciences
Specific author contributions:
All of the authors listed below have approved the final draft submitted.
Jean S. Wang – planning and conducting the study, collecting and interpreting data, drafting the manuscript
Mingzhou Guo – conducting the study, critical revision of manuscript, technical support
Elizabeth A. Montgomery – analysis of data, critical revision of manuscript, technical support
Richard E. Thompson – analysis of data, critical revision of manuscript, statistical analysis
Hilary Cosby – collecting data, critical revision of manuscript, administrative support
Lisa Hicks – collecting data, critical revision of manuscript, administrative support
Shelun Wang – conducting the study, critical revision of manuscript, technical support
James G. Herman – planning the study, interpreting data, critical revision of manuscript, supervision
Marcia I. Canto - planning the study, interpreting data, critical revision of manuscript, supervision
Guarantor of the article: Jean S. Wang
References
- 1.Shaheen N, Ransohoff DF. Gastroesophageal reflux, Barrett esophagus, and esophageal cancer: clinical applications. Jama. 2002;287:1982–6. doi: 10.1001/jama.287.15.1982. [DOI] [PubMed] [Google Scholar]
- 2.Montgomery E, Goldblum JR, Greenson JK, et al. Dysplasia as a predictive marker for invasive carcinoma in Barrett esophagus: a follow-up study based on 138 cases from a diagnostic variability study. Hum Pathol. 2001;32:379–88. doi: 10.1053/hupa.2001.23511. [DOI] [PubMed] [Google Scholar]
- 3.Montgomery E, Bronner MP, Goldblum JR, et al. Reproducibility of the diagnosis of dysplasia in Barrett esophagus: a reaffirmation. Hum Pathol. 2001;32:368–78. doi: 10.1053/hupa.2001.23510. [DOI] [PubMed] [Google Scholar]
- 4.Cameron AJ, Ott BJ, Payne WS. The incidence of adenocarcinoma in columnar-lined (Barrett’s) esophagus. N Engl J Med. 1985;313:857–9. doi: 10.1056/NEJM198510033131404. [DOI] [PubMed] [Google Scholar]
- 5.Williamson WA, Ellis FH, Jr, Gibb SP, et al. Barrett’s esophagus. Prevalence and incidence of adenocarcinoma. Archives of internal medicine. 1991;151:2212–6. doi: 10.1001/archinte.151.11.2212. [DOI] [PubMed] [Google Scholar]
- 6.Spechler SJ, Robbins AH, Rubins HB, et al. Adenocarcinoma and Barrett’s esophagus. An overrated risk? Gastroenterology. 1984;87:927–33. [PubMed] [Google Scholar]
- 7.Reid BJ, Weinstein WM. Barrett’s esophagus and adenocarcinoma. Annual review of medicine. 1987;38:477–92. doi: 10.1146/annurev.me.38.020187.002401. [DOI] [PubMed] [Google Scholar]
- 8.Licchesi JD, Westra WH, Hooker CM, et al. Promoter hypermethylation of hallmark cancer genes in atypical adenomatous hyperplasia of the lung. Clin Cancer Res. 2008;14:2570–8. doi: 10.1158/1078-0432.CCR-07-2033. [DOI] [PubMed] [Google Scholar]
- 9.Licchesi JD, Westra WH, Hooker CM, et al. Epigenetic alteration of Wnt pathway antagonists in progressive glandular neoplasia of the lung. Carcinogenesis. 2008;29:895–904. doi: 10.1093/carcin/bgn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eads CA, Lord RV, Kurumboor SK, et al. Fields of aberrant CpG island hypermethylation in Barrett’s esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021–6. [PubMed] [Google Scholar]
- 11.Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–9. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 12.Bachman KE, Herman JG, Corn PG, et al. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 13.Baylin S. DNA methylation and epigenetic mechanisms of carcinogenesis. Dev Biol (Basel) 2001;106:85–7. discussion 143-60. [PubMed] [Google Scholar]
- 14.Baylin SB, Belinsky SA, Herman JG. Aberrant methylation of gene promoters in cancer---concepts, misconcepts, and promise. J Natl Cancer Inst. 2000;92:1460–1. doi: 10.1093/jnci/92.18.1460. [DOI] [PubMed] [Google Scholar]
- 15.Corn PG, Kuerbitz SJ, van Noesel MM, et al. Transcriptional silencing of the p73 gene in acute lymphoblastic leukemia and Burkitt’s lymphoma is associated with 5′ CpG island methylation. Cancer Res. 1999;59:3352–6. [PubMed] [Google Scholar]
- 16.Esteller M, Catasus L, Matias-Guiu X, et al. hMLH1 promoter hypermethylation is an early event in human endometrial tumorigenesis. Am J Pathol. 1999;155:1767–72. doi: 10.1016/S0002-9440(10)65492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esteller M, Corn PG, Baylin SB, et al. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–9. [PubMed] [Google Scholar]
- 18.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 19.Esteller M, Risques RA, Toyota M, et al. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61:4689–92. [PubMed] [Google Scholar]
- 20.Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–9. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 21.Schulmann K, Sterian A, Berki A, et al. Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett’s-associated neoplastic progression and predicts progression risk. Oncogene. 2005;24:4138–48. doi: 10.1038/sj.onc.1208598. [DOI] [PubMed] [Google Scholar]
- 22.Bian YS, Osterheld MC, Fontolliet C, et al. p16 inactivation by methylation of the CDKN2A promoter occurs early during neoplastic progression in Barrett’s esophagus. Gastroenterology. 2002;122:1113–21. doi: 10.1053/gast.2002.32370. [DOI] [PubMed] [Google Scholar]
- 23.Wong DJ, Barrett MT, Stoger R, et al. p16INK4a promoter is hypermethylated at a high frequency in esophageal adenocarcinomas. Cancer Res. 1997;57:2619–22. [PubMed] [Google Scholar]
- 24.Kempster S, Phillips WA, Baindur-Hudson S, et al. Methylation of exon 2 of p16 is associated with late stage oesophageal cancer. Cancer Lett. 2000;150:57–62. doi: 10.1016/s0304-3835(99)00372-9. [DOI] [PubMed] [Google Scholar]
- 25.Klump B, Hsieh CJ, Holzmann K, et al. Hypermethylation of the CDKN2/p16 promoter during neoplastic progression in Barrett’s esophagus. Gastroenterology. 1998;115:1381–6. doi: 10.1016/s0016-5085(98)70016-2. [DOI] [PubMed] [Google Scholar]
- 26.Wong DJ, Paulson TG, Prevo LJ, et al. p16(INK4a) lesions are common, early abnormalities that undergo clonal expansion in Barrett’s metaplastic epithelium. Cancer Res. 2001;61:8284–9. [PubMed] [Google Scholar]
- 27.Kawakami K, Brabender J, Lord RV, et al. Hypermethylated APC DNA in plasma and prognosis of patients with esophageal adenocarcinoma. J Natl Cancer Inst. 2000;92:1805–11. doi: 10.1093/jnci/92.22.1805. [DOI] [PubMed] [Google Scholar]
- 28.Levine DS, Haggitt RC, Blount PL, et al. An endoscopic biopsy protocol can differentiate high-grade dysplasia from early adenocarcinoma in Barrett’s esophagus. Gastroenterology. 1993;105:40–50. doi: 10.1016/0016-5085(93)90008-z. [DOI] [PubMed] [Google Scholar]
- 29.Reid BJ, Blount PL, Feng Z, et al. Optimizing endoscopic biopsy detection of early cancers in Barrett’s high-grade dysplasia. Am J Gastroenterol. 2000;95:3089–96. doi: 10.1111/j.1572-0241.2000.03182.x. [DOI] [PubMed] [Google Scholar]
- 30.Schlemper RJ, Riddell RH, Kato Y, et al. The Vienna classification of gastrointestinal epithelial neoplasia. Gut. 2000;47:251–5. doi: 10.1136/gut.47.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmisano WA, Divine KK, Saccomanno G, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–8. [PubMed] [Google Scholar]
- 33.House MG, Guo M, Iacobuzio-Donahue C, et al. Molecular progression of promoter methylation in intraductal papillary mucinous neoplasms (IPMN) of the pancreas. Carcinogenesis. 2003;24:193–8. doi: 10.1093/carcin/24.2.193. [DOI] [PubMed] [Google Scholar]
- 34.House MG, Herman JG, Guo MZ, et al. Aberrant hypermethylation of tumor suppressor genes in pancreatic endocrine neoplasms. Ann Surg. 2003;238:423–32. doi: 10.1097/01.sla.0000086659.49569.9e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conio M, Filiberti R, Blanchi S, et al. Risk factors for Barrett’s esophagus: a case-control study. Int J Cancer. 2002;97:225–9. doi: 10.1002/ijc.1583. [DOI] [PubMed] [Google Scholar]
- 36.Eloubeidi MA, Provenzale D. Clinical and demographic predictors of Barrett’s esophagus among patients with gastroesophageal reflux disease: a multivariable analysis in veterans. Journal of clinical gastroenterology. 2001;33:306–9. doi: 10.1097/00004836-200110000-00010. [DOI] [PubMed] [Google Scholar]
- 37.Gerson LB, Edson R, Lavori PW, et al. Use of a simple symptom questionnaire to predict Barrett’s esophagus in patients with symptoms of gastroesophageal reflux. Am J Gastroenterol. 2001;96:2005–12. doi: 10.1111/j.1572-0241.2001.03933.x. [DOI] [PubMed] [Google Scholar]
- 38.O’Connor JB, Falk GW, Richter JE. The incidence of adenocarcinoma and dysplasia in Barrett’s esophagus: report on the Cleveland Clinic Barrett’s Esophagus Registry. Am J Gastroenterol. 1999;94:2037–42. doi: 10.1111/j.1572-0241.1999.01275.x. [DOI] [PubMed] [Google Scholar]
- 39.Weston AP, Badr AS, Hassanein RS. Prospective multivariate analysis of clinical, endoscopic, and histological factors predictive of the development of Barrett’s multifocal high-grade dysplasia or adenocarcinoma. Am J Gastroenterol. 1999;94:3413–9. doi: 10.1111/j.1572-0241.1999.01602.x. [DOI] [PubMed] [Google Scholar]
- 40.Reid BJ, Levine DS, Longton G, et al. Predictors of progression to cancer in Barrett’s esophagus: baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol. 2000;95:1669–76. doi: 10.1111/j.1572-0241.2000.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnell TG, Sontag SJ, Chejfec G, et al. Long-term nonsurgical management of Barrett’s esophagus with high-grade dysplasia. Gastroenterology. 2001;120:1607–19. doi: 10.1053/gast.2001.25065. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Cosby H, Hicks L, et al. No occult cancer at esophagectomy in patients with Barrett’s esophagus with high-grade dysplasia who have undergone surveillance with the Seattle biopsy protocol. Gastroenterology. 2007;132:A64. [Google Scholar]
- 43.Clement G, Braunschweig R, Pasquier N, et al. Methylation of APC, TIMP3, and TERT: a new predictive marker to distinguish Barrett’s oesophagus patients at risk for malignant transformation. J Pathol. 2006;208:100–7. doi: 10.1002/path.1884. [DOI] [PubMed] [Google Scholar]
- 44.Reid BJ. p53 and neoplastic progression in Barrett’s esophagus. Am J Gastroenterol. 2001;96:1321–3. doi: 10.1111/j.1572-0241.2001.03844.x. [DOI] [PubMed] [Google Scholar]
- 45.Weston AP, Banerjee SK, Sharma P, et al. p53 protein overexpression in low grade dysplasia (LGD) in Barrett’s esophagus: immunohistochemical marker predictive of progression. Am J Gastroenterol. 2001;96:1355–62. doi: 10.1111/j.1572-0241.2001.03851.x. [DOI] [PubMed] [Google Scholar]
- 46.Rabinovitch PS, Longton G, Blount PL, et al. Predictors of progression in Barrett’s esophagus III: baseline flow cytometric variables. The American journal of gastroenterology. 2001;96:3071–83. doi: 10.1111/j.1572-0241.2001.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tselepis C, Morris CD, Wakelin D, et al. Upregulation of the oncogene c-myc in Barrett’s adenocarcinoma: induction of c-myc by acidified bile acid in vitro. Gut. 2003;52:174–80. doi: 10.1136/gut.52.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edelstein ZR, Farrow DC, Bronner MP, et al. Central adiposity and risk of Barrett’s esophagus. Gastroenterology. 2007;133:403–11. doi: 10.1053/j.gastro.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 49.Dong LM, Kristal AR, Peters U, et al. Dietary supplement use and risk of neoplastic progression in esophageal adenocarcinoma: a prospective study. Nutrition and cancer. 2008;60:39–48. doi: 10.1080/01635580701586762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Funkhouser EM, Sharp GB. Aspirin and reduced risk of esophageal carcinoma. Cancer. 1995;76:1116–9. doi: 10.1002/1097-0142(19951001)76:7<1116::aid-cncr2820760703>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]