Abstract
Carbon catabolite repression (CCR) by transcriptional regulators follows different mechanisms in gram-positive and gram-negative bacteria. In gram-positive bacteria, CcpA-dependent CCR is mediated by phosphorylation of the phosphoenolpyruvate:sugar phosphotransferase system intermediate HPr at a serine residue at the expense of ATP. The reaction is catalyzed by HPr kinase, which is activated by glycolytic intermediates. In this review, the distribution of CcpA-dependent CCR among bacteria is investigated by searching the public databases for homologues of HPr kinase and HPr-like proteins throughout the bacterial kingdom and by analyzing their properties. Homologues of HPr kinase are commonly observed in the phylum Firmicutes but are also found in the phyla Proteobacteria, Fusobacteria, Spirochaetes, and Chlorobi, suggesting that CcpA-dependent CCR is not restricted to gram-positive bacteria. In the α and β subdivisions of the Proteobacteria, the presence of HPr kinase appears to be common, while in the γ subdivision it is more of an exception. The genes coding for the HPr kinase homologues of the Proteobacteria are in a gene cluster together with an HPr-like protein, termed XPr, suggesting a functional relationship. Moreover, the XPr proteins contain the serine phosphorylation sequence motif. Remarkably, the analysis suggests a possible relation between CcpA-dependent gene regulation and the nitrogen regulation system (Ntr) found in the γ subdivision of the Proteobacteria. The relation is suggested by the clustering of CCR and Ntr components on the genome of members of the Proteobacteria and by the close phylogenetic relationship between XPr and NPr, the HPr-like protein in the Ntr system. In bacteria in the phylum Proteobacteria that contain HPr kinase and XPr, the latter may be at the center of a complex regulatory network involving both CCR and the Ntr system.
INTRODUCTION
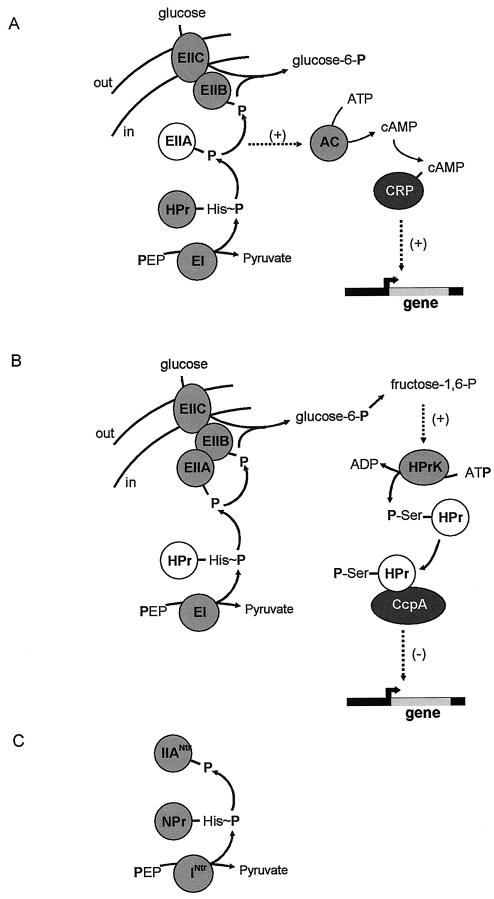
In bacteria, the phosphoenolpyruvate-dependent phosphotransferase system (PTS) is the main carbohydrate uptake system and, in addition, plays an important role in the regulation of expression of catabolic genes and operons (25, 36). The system is typical of bacteria and not found in the other kingdoms of life, Archaea and Eucarya. PTS-mediated uptake involves the transfer of the phosphoryl group of the high-energy metabolite phosphoenolpyruvate to the carbohydrate through a cascade of phosphotransfer proteins including EI, HPr, and EIIABC (Fig. 1A and B, left). In PTS-mediated gene regulation, the phosphorylation state of the intermediate proteins controls the expression of genes coding for sugar-specific components of the PTS (PRD-mediated induction [35]) and for transporters and enzymes needed for the catabolism of less favored carbon sources (carbon catabolite repression [CCR]). CCR is achieved by allosteric inhibition of transporters or cytoplasmic enzymes, which prevents the uptake or synthesis of inducers, respectively (inducer exclusion), or by interaction with transcriptional regulators (Fig. 1A and B, right). The latter mechanism is the topic of this review. CCR via transcriptional regulators follows distinct mechanisms in gram-negative and gram-positive bacteria, using different, unrelated transcriptional regulators, CRP and CcpA, respectively. In gram-negative bacteria, the phosphorylation state of the glucose-specific EIIAGlc regulates the activity of adenylate cyclase and, consequently, the level of cAMP in the cell (Fig. 1A). At sufficiently high levels, cAMP binds to the transcriptional regulator CRP, which induces binding to specific DNA sequences in the promoter region of the target genes, where it activates the initiation of transcription through interaction with the polymerase (25). In gram-positive bacteria, the signaling intermediate is HPr and not EIIAGlc (Fig. 1B). HPr in gram-positive bacteria is phosphorylated at two sites, a histidine residue and a serine residue. The histidine residue is phosphorylated by EI at the expense of phosphoenolpyruvate, while the serine is phosphorylated by HPr kinase (HPrK) at the expense of ATP or PPi (4, 24). HPr(His∼P) is involved in sugar transport and PRD-mediated regulation, and HPr(Ser-P) is involved in CCR. The primary sensor in the regulatory pathway is HPrK, which is activated by glycolytic intermediates. HPr(Ser-P) binds to the transcriptional regulator CcpA, thereby inducing the binding of the complex to so-called cre sites (for “catabolite responsive element”) in the promoter region of the target genes, which prevents transcription of the genes (13).
FIG. 1.
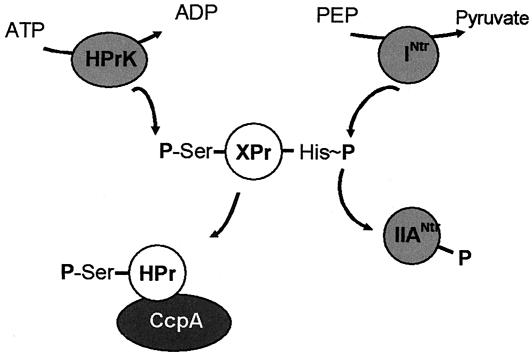
Schematic representation of CRP-dependent (A) and CcpA-dependent (B) carbon catabolite repression pathways and the Ntr regulatory pathway (C). (A and B) Shown on the left-hand side is PTS-mediated glucose uptake in the model organisms E. coli and B. subtilis, respectively. The phospho-carrier protein HPr is phosphorylated at the catalytic histidine residue by enzyme EI at the expense of phosphoenolpyruvate (PEP). The phosphoryl group is then transferred to EIIA, which is a cytoplasmic protein in E. coli (A) or part of the multidomain complex EIIABC in B. subtilis (B). From EIIA, the phosphoryl group is transferred to EIIB, a soluble domain attached to the integral membrane transporter domain EIIC. The glucose molecule is transported into the cell and at the same time phosphorylated by EIIB, yielding glucose-6-phosphate in the cell. Shown on the right-hand side is transcriptional regulator-mediated CCR. The transcriptional regulators CRP (A) and CcpA (B) are indicated by a dark background, and the PTS components involved, EIIA and HPr in panels A and B, respectively, are indicated by a white background. In E. coli (A), the degree of phosphorylation of EIIA determines the activity of adenylate cyclase (AC) and, consequently, the concentration of cAMP in the cell. Binding of cAMP to CRP results in a complex that stimulates transcription of target genes (positive regulation). In B. subtilis (B), fructose-1,6-phosphate produced from glucose-6-phosphate in glycolysis activates HPrK that phosphorylates HPr at the regulatory-site serine at the expense of ATP or PPi. Binding of HPr-Ser-P to CcpA results in acomplex that inhibits the transcription of target genes (negative regulation). The HPr molecule in HPr-mediated signal transduction is the same as the HPr involved in glucose uptake. In Crh-mediated signal transduction, the HPr molecule is not part of the uptake system and is termed Crh. (C) Phosphoryl group transfer chain in the nitrogen regulation pathway Ntr. The HPr-like protein NPr is phosphorylated by enzyme INtr at the expense of phosphoenolpyruvate, after which the phosphoryl group is transferred to EIIANtr. The pathway is thought to operate independently of the PTS sugar uptake pathway.
The above gives a generalized scheme for the CcpA-dependent transcriptional control in gram-positive bacteria and does not take into account other functions of CcpA (e.g., activation of glycolytic enzymes), mechanistic details (e.g., the location of the cre site), or the diversification reported in different organisms (for reviews, see references 12, 37, and 41). The scheme conforms to the CCR system in Bacillus subtilis, which has been studied extensively and is the prototype of CCR-regulated gene expression in gram-positives bacteria. The “prototype” contains one important variation not observed in most other gram-positive bacteria. During the B. subtilis genome-sequencing project, a second gene coding for an HPr-like protein was discovered (19). The protein, Crh (for “catabolite repression HPr”) has 45% sequence identity to HPr and contains the regulatory-site serine but not the active-site histidine (9). Accordingly, it was demonstrated that Crh was inactive in the PTS transport function but functional in CCR; Crh could be phosphorylated at the serine residue by HPrK and then act as a corepressor of CcpA. In B. subtilis the Crh- and the HPr-mediated regulatory pathways seem to operate in parallel. Studies using mutant strains containing HPr in which the regulatory-site serine was mutated to alanine showed that in vivo, Crh could (partly) take over the regulatory function of HPr in glucose-induced repression depending on the target gene(s). The regulation was not affected in a Crh knockout strain (2, 9). Apparently, Crh is redundant in glucose-potentiated catabolite repression in B. subtilis, leaving its function obscure. Recently, we demonstrated that repression of expression of the Mg2+ citrate transporter of B. subtilis grown in a medium containing succinate and glutamate was specifically mediated by Crh and not by HPr. It was suggested that Crh might be specifically involved in repression of metabolic genes by nonsugars (45).
A difference between CRP-dependent CCR found in gram-negative bacteria and CcpA-dependent CCR found in gram-positive bacteria is the strictness of the coupling between the PTS transport and regulatory functions. In the CRP-dependent mechanism (Fig. 1A), regulation of expression is directly coupled to turnover of IIAGlc. The level of phosphorylation of IIAGlc is determined by the uptake rate of glucose from the medium and by the uptake of other PTS sugars which compete for P-HPr and, thereby, limit the rate of phosphorylation of IIAGlc. In the CcpA-dependent mechanism (Fig. 1B), the primary sensor is HPrK that is activated by glycolytic intermediates (15, 17, 28) but, in principle, may be activated by other metabolites as well, making the mechanism more versatile. Our studies of the regulation of expression of the Mg2+ citrate transporter of B. subtilis demonstrate that, in addition to glucose, the pathway is potentiated by the non-PTS sugar inositol and by the nonsugars succinate and glutamate (44). Moreover, the signal transduction pathway in the HPr molecule is physically separated from the phosphoryl group transfer chain of the PTS transport function, i.e., via the regulatory-site Ser residue and active-site His residue, respectively. The regulatory state is modulated rather than being determined by the uptake system by virtue of the phosphorylation state of the active-site histidine (3, 27). Crh-mediated regulation may be a manifestation of this loose coupling between regulatory and transport function of the PTS; a single mutation in HPr (His to Gln) results in a regulatory pathway that is independent of the uptake system. The mechanism found in gram-positive bacteria, involving HPr and HPrK, may be a more general gene regulation system. Gram-negative bacteria may have compensated for their specialized but inflexible mechanism by developing the Ntr (nitrogen regulation) system, a PTS-based regulatory mechanism composed of a complete phosphotransfer chain that operates independently of the uptake system and is not found in gram-positive bacteria (Fig. 1C) (39). The Ntr system is involved in the regulation of nitrogen metabolism, but its influence may be much broader (23), including a role in virulence (33, 38).
In the present study, we investigated the distribution of CcpA-dependent CCR in the bacterial kingdom by searching the available sequence databases for HPrK homologues and HPr-like proteins and we investigated the evolutionary origin of the pathway by analyzing the relationship between the proteins and searching the genome databases for evolutionary links between regulatory systems. It follows that homologues of HPrK are found in many gram-negative bacteria; more importantly, the results suggest an evolutionary link between CcpA-dependent CCR and the Ntr type of regulation found in gram-negative bacteria.
STRATEGY
CcpA-dependent CCR is defined as the HPrK-catalyzed, ATP-dependent phosphorylation of an HPr-like molecule that in the seryl-phosphorylated state interacts with the transcriptional regulator CcpA to induce the binding of the latter to its cognate recognition site on the DNA. The signal transduction pathway may or may not be coupled to the PTS uptake system, i.e., HPr-mediated CCR or Crh-mediated CCR, respectively. The two modes differ in that in the latter, the HPr-like molecule Crh is not functional in carbohydrate uptake (21). B. subtilis Crh is the prototype of an HPr-like molecule that functions in Crh-mediated signal transduction. It is easily distinguished from B. subtilis HPr since it lacks the active-site histidine that, in the latter, is essential for PTS-mediated uptake. The HPrK proteins (HPrK/P) are bifunctional enzymes that are both kinases and phosphatases (8, 10, 17). They are thought to represent a new family of bacterial ATP-dependent protein kinases that are functional only in the phosphorylation of HPr-like molecules at the regulatory serine residue. Therefore, they are a pivotal component of CcpA-dependent CCR. CcpA is a member of the LacI-GalR family of DNA binding proteins that serve diverse functions in transcriptional regulation (13); therefore, the presence of a CcpA homologue in an organism may not be meaningful in a search for CcpA-dependent CCR in that organism. Summarizing, indicators for the identification of CcpA-dependent CCR in an organism are (i) the presence of HPrK/P, (ii) the presence of an HPr-like molecule containing the regulatory-site serine residue, and (iii) the presence of an additional HPr-like molecule missing the active-site histidine residue (Crh). Furthermore, the clustering of the genes coding for HPr-like proteins and HPrK/P on the genomes will play an important role in the analysis of the evolutionary context of the pathway.
PROTEIN FAMILIES
HPr Kinase
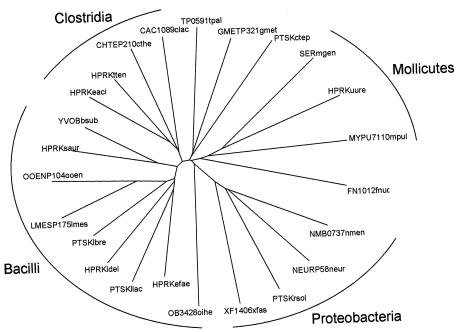
A BLAST search of the protein database at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/entrez/) (1) revealed the presence of 44 unique HPrK/P sequences (Table 1) that vary in length between 304 and 342 amino acid residues. The Fusobacterium nucleatum sequence FN1012 is twice the consensus length (615 residues) but represents a gene duplication with 23% sequence identity between the N- and C-terminal halves. Both halves have higher sequence identity to orthologues from other bacteria. The N-terminal half is 32% identical to HPRK of Staphylococcus aureus, and the C-terminal half is 34% identical to HPRK of Eubacterium acidaminophilum. All sequences are of bacterial origin. The majority originate from the phylum Firmicutes, containing the typical low-G+C gram-positive organisms, but homologues were also found in typical gram-negative bacteria in the β, γ, and δ subdivisions of the Proteobacteria and in the phyla Fusobacteria, Spirochaetes, and Chlorobi (see also references 8, 10, and 14) (Table 1). The different groups of bacteria are indicated in the phylogenetic tree of a subset of 24 typical sequences (Fig. 2). A small number of hits from the BLAST search represent proteins that are considerably smaller than the consensus length of HPrK/P (14). They contain between 141 and 159 residues, and, remarkably, all are found in bacteria belonging to the α subdivision of the Proteobacteria. Multiple sequence alignments of the full-length and short versions show that the latter corresponds to an internal fragment of the former, which corresponds to part of the catalytic domain. This is discussed in more detail later in this review.
TABLE 1.
The HPrK/P familya
| Phylum | Organism | HPrK/Pb | GIc |
|---|---|---|---|
| Firmicutes | |||
| Bacilli | Bacillus subtilis | YVOB | 16080553 |
| Bacillus halodurans | BH3590 | 15616152 | |
| Bacillus anthracis | BA0250 | 21397625 | |
| Listeria innocua | LIN2626 | 16801688 | |
| Listeria monocytogenes | LMO2483 | 16804521 | |
| Oceanobacillus iheyensis | OB2482 | 23099937 | |
| Staphylococcus aureus | HPRK | 15923750 | |
| Oenococcus oeni | OOENP104 | 23037825 | |
| Oceanobacillus iheyensis | OB3428 | 23100883 | |
| Enterococcus faecalis | HPRK | 6016253 | |
| Enterococcus faecium | EFAEP969 | 22991374 | |
| Lactobacillus casei | HPRK | 6688477 | |
| Lactobacillus brevis | PTSK | 14133562 | |
| Lactobacillus delbrueckii | HPRK | 16124246 | |
| Lactobacillus gasseri | LGASP106 | 23003221 | |
| Lactobacillus lactis | PTSK | 15672600 | |
| Streptococcus bovis | BAA77782 | 4884536 | |
| Streptococcus mutans | HPRK | 6647569 | |
| Streptococcus salivarius | HPRK | 6647570 | |
| Streptococcus pneumoniae | HPRK | 15903313 | |
| Streptococcus pyogenes | PTSK | 19745698 | |
| Streptococcus agalactiae | HPRK | 22536900 | |
| Leuconostoc mesenteroides | LMESP1751 | 23025086 | |
| Clostridia | Clostridium acetobutylicum | CAC1089 | 15894374 |
| Clostridium perfringens | HPRK | 18309986 | |
| Clostridium thermocellum | CHTEP210 | 23020136 | |
| Thermoanaerobacter tengcongensis | HPRK | 20808368 | |
| Eubacterium acidaminophilum | HPRK | 14250932 | |
| Mollicutes | Mycoplasma genitalium | SER | 12044937 |
| Mycoplasma pneumoniae | MPN223 | 13507962 | |
| Mycoplasma pulmonis | MYPU7110 | 15829182 | |
| Ureaplasma urealyticum | HPRK | 13357632 | |
| Proteobacteria | |||
| α subdivision | Mesorhizobium loti | MLL5093d | 13474247 |
| Sinorhizobium meliloti | SMC02752d | 15963795 | |
| Agrobacterium tumefaciens | AGRC51d | 15887390 | |
| Brucella melitensis | BMEI2034d | 17988317 | |
| Caulobacter crescentus | CC0239d | 16124494 | |
| Rhodobacter sphaeroides | RSPHP156d | 22957981 | |
| Rhodospirillum rubrum | RRUBP472d,e | 22965877 | |
| Magnetospirillum magnetotacticum | MAGNP764d | 23015048 | |
| Novosphingobium aromaticivorans | SAROP340d | 23110202 | |
| β subdivision | Neisseria meningitidis | NMB0737 | 15676635 |
| Ralstonia solanacearum | PTSK | 17545124 | |
| Ralstonia metallidurans | REUTP341 | 22978701 | |
| Burkholderio fungorum | BCEPP707 | 22989124 | |
| Nitrosomonas europaea | NEURP58 | 22954108 | |
| γ subdivision | Xylella fastidiosa | XF1406 | 15838007 |
| Xanthomonas campestris | PTSK | 21232236 | |
| Xanthomonas axonopodis | PTSK | 21243702 | |
| δ subdivision | Geobacter metallireducens | GMETP321 | 23056418 |
| Fusobacteria | Fusobacterium nucleatum | FN1012 | 19704347 |
| Spirochaetes | Treponema pallidum | TP0591 | 15639579 |
| Chlorobi | Chlorobium tepidum | PTSK | 21674451 |
the sequences were extracted from the NCBI protein database by a BLAST search using the B. subtilis sequence encoded by the translated product of the yvoB gene as the query. All unique HPrK/P sequences in the database are reported.
Typical and similar relationships. A typical sequence (bold) represents a group of similar sequences (indented) with at least 60% sequence identity.
GI indicates the unique GI number of the entry in the database.
Short version (see the text).
Contains an N-terminal extension that is not homologous to the N-terminal region of “normal” size homologues. The true translational start is probably at Met128.
FIG. 2.
Phylogenetic tree of HPrK/P. A subset of 24 typical HPrK/P sequences (the bold sequences in Table 1) was aligned using the Clustal X program (40), and then the tree was constructed using the DrawTree program in the Phylip package (6). The typical sequences represent groups of sequences with pairwise sequence identities of at least 60%. Organisms are indicated by a shorthand following the protein that consists of the first letter of the genus followed by the first three letters of the species. The short-version HPrKs that form a separate branch in the tree were left out of the analysis.
One hit in the BLAST search represented a protein outside the bacterial kingdom: MK1512 of the archaeon Methanopyrus kandleri. It resembles the short-version HPrKs in that it misses the N-terminal part of the full-length versions. The protein corresponds to the catalytic domain of full-length HPrKs (see below). This sequence is not considered further here.
HPr-Like Proteins
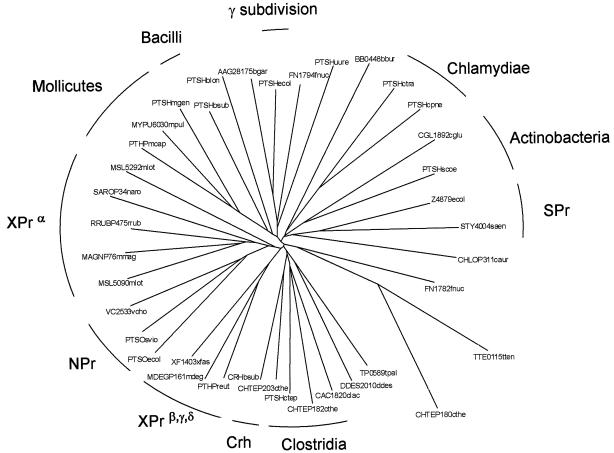
The NCBI protein database contained 105 unique HPr-like proteins when HPr entities that are part of multidomain proteins (the MTP, FruB, and FPr families) were left out (Table 2). The multidomain proteins do not seem to add much to the present discussion and have been reviewed before (see, for example, references 31 and 39). The HPr sequences vary in length between 82 and 112 residues. Eight sequences miss the active-site histidine, while no more than three (other) sequences do not contain the regulatory site serine. The phosphoenolpyruvate-dependent PTS is a typical bacterial uptake and signal transduction system; accordingly, all HPr-like proteins were of bacterial origin. The 105 HPr proteins were distributed over 87 different organisms, indicating that some bacteria contain more than one HPr-like molecule. The set of 87 organisms contains more or less equal numbers from the phyla Firmicutes (n = 34) and Proteobacteria (n = 41) and fewer from the other phyla (n = 12). Figure 3 shows a phylogenetic tree of a subset of the HPr-like molecules.
TABLE 2.
The family of HPr-like proteinsa
| Phylum | Organism | Gb | HPre | GI | Clusterc | HPrKd |
|---|---|---|---|---|---|---|
| Firmicutes | ||||||
| Bacilli | Bacillus subtilis | • | PTSH | 16078454 | IIABC-H-I | • |
| Bacillus anthracis | • | BA4727 | 21402102 | IIABC-H-I | • | |
| Bacillus megaterium | PTHP | 13633375 | ||||
| Bacillus halodurans | • | PTSH | 15615636 | H-I | • | |
| Geobacillus stearothermophilus | PTHP | 1172717 | H-I | |||
| Staphylococcus carnosus | PTHP | 131538 | ||||
| Staphylococcus xylosus | PTSH | 11596363 | • | |||
| Staphylococcus aureus | • | PTSH | 15924073 | H-I | • | |
| Lactobacillus casei | PTHP | 13633570 | • | |||
| Lactobacillus sakei | PTHP | 3122641 | ||||
| Lactobacillus gasseri | LGASP108 | 23003237 | H-I | • | ||
| Lactobacillus brevis | PTSH | 14133565 | • | |||
| Leuconostoc mesenteroides | LMESP115 | 23024451 | H-I | • | ||
| Lactococcus lactis | • | PTHP | 13633652 | H-I | • | |
| Enterococcus faecalis | PTHP | 1172718 | • | |||
| Streptococcus mutans | PTHP | 1172720 | • | |||
| Streptococcus salivarius | PTHP | 1172721 | • | |||
| Streptococcus bovis | PTHP | 13633635 | • | |||
| Streptococcus pyogenes | • | PTSH | 15675305 | H-I | • | |
| Streptococcus pneumoniae | • | SP1177 | 15901042 | H-I | • | |
| Streptococcus thermophilus | PTSH | 17978653 | ||||
| Streptococcus agalactiae | • | PTSH | 22536985 | H-I | • | |
| Oceanobacillus iheyensis | • | OB2344 | 23099799 | H | • | |
| Oenococcus oeni | OOENP127 | 23038069 | • | |||
| Listeria innocua | • | PTSH | 16800070 | H-I | • | |
| Bacillus subtilis | • | CRH | 16080527 | j-k-l-H | • | |
| Bacillus halodurans | • | CHR | 15616128 | j-k-l-H | • | |
| Bacillus anthracis | • | BA0239 | 21397614 | j-k-l-H | • | |
| Oceanobacillus iheyensis | • | OB2465 | 23099920 | j-k-l-H | • | |
| Clostridia | Thermoanaerobacter tengcongensis | • | FRUB | 20808237 | j-k-x10-l-H | • |
| Clostridium thermocellum | CHTEP203 | 23020129 | j-k-x-l-H | • | ||
| Thermoanaerobacter tengcongensis | • | TTE0115 | 20806644 | H | • | |
| Clostridium thermocellum | CHTEP180 | 23020106 | H | • | ||
| Clostridium thermocellum | CHTEP182 | 23021750 | H-I | • | ||
| Clostridium acetobutylicum | • | CAC1820 | 15895096 | H | • | |
| Clostridium perfringens | • | HPR | 18310651 | H | • | |
| Mollicutes | Mycoplasma capricolum | PTHP | 1172719 | |||
| Mycoplasma genitalium | • | PTSH | 12044891 | H | • | |
| Mycoplasma pneumoniae | • | PTSH | 13507792 | H | • | |
| Mycoplasma pulmonis | • | MYPU6030 | 15829074 | H-I | • | |
| Ureaplasma urealyticum | • | PTSH | 13358152 | H | • | |
| Proteobacteria | ||||||
| α subdivision | Mesorhizobium loti | • | MSL5292 | 13474414 | H-I | • |
| Mesorhizobium loti | • | MSL5090 | 13474245 | K′-IIA-H | ▪ | |
| Sinorhizobium meliloti | • | SMC02754 | 15963793 | K′-IIA-H | ▪ | |
| Agrobacterium tumefaciens | • | AGRC49 | 15887388 | K′-IIA-H | ▪ | |
| Brucella melitensis | • | BMEI2031 | 17988314 | K′-?-IIA-H | ▪ | |
| Rhodopseudomonas palustris | RPALP163 | 22962348 | K′-IIA-H | ▪ | ||
| Caulobacter crescentus | • | CC0241 | 16124496 | K′-IIA-H | ▪ | |
| Rhodobacter sphaeroides | RSPHP157 | 22957984 | K′-j-IIA-H | ▪ | ||
| Magnetospirillum magnetotacticum | MAGNP479 | 23011831 | ▪ | |||
| Novosphingobium aromaticivorans | SAROP34 | 23110205 | K′-j-IIA-H | ▪ | ||
| Rhodospirillum rubrum | RRUBP475 | 22965880 | K′-j-IIA-H-I | ▪ | ||
| Magnetospirillum magnetotacticum | MAGNP76 | 23015045 | K′-j-IIA-H-I | ▪ | ||
| β subdivision | Ralstonia eutropha | PTHP | 131532 | |||
| Ralstonia solanacearum | • | PSTH | 17545066 | IIA-H-I | • | |
| Ralstonia metallidurans | REUTP495f | 22980242 | IIA-H-I | • | ||
| Nitrosomonas europaea | NEURP196 | 22955983 | - | • | ||
| Burkholderia fungorum | BCEPP510 | 22987169 | IIA-H-I | • | ||
| Neisseria meningitidis | • | NMB2045 | 15677867 | IIA-H-I | • | |
| γ subdivision | Pseudomonas putida | PTHP | 13633649 | |||
| Pseudomonas aeruginosa | • | PA4466 | 15599662 | N-h-IIA-j-H | ||
| Pseudomonas fluorescens | PFLUP387 | 23061779 | N-h-IIA-j-H | |||
| Pseudomonas stutzeri | PTSO | 22138783 | ||||
| Microbulbifer degradans | MDEGP161 | 23027798 | N-IIA-j-H | |||
| Azotobacter vinelandii | AVINP175 | 23103593 | N-h-IIA-j-H | |||
| Escherichia coli | • | PTSH | 15802948 | H-I-IIA | /PICK> | |
| Klebsiella pneumoniae | PTHP | 131536 | ||||
| Yersinia pestis | • | PTSH | 16123174 | H-I-IIA | ||
| Serratia marcescens | PTSH | 21039016 | ||||
| Haemophilus influenzae | • | HI1713 | 16273600 | H-I-IIA | ||
| Haemophilus somnus | HSOM0557 | 23466976 | H-I-IIA | |||
| Pasteurella multocida | • | PTSH | 15602763 | H-I-IIA | ||
| Buchnera sp. | • | PTSH | 15616690 | H-I-IIA | ||
| Buchnera aphidicola | • | PTSH | 21622959 | H-I-IIA | ||
| Vibrio cholerae | • | VC0966 | 15640982 | H-I-IIA | ||
| Salmonella enterica | • | PTSH | 16765751 | H-I-IIA | ||
| Escherichia coli | • | PTSO | 15803746 | N-h-IIA-j-H | ||
| Klebsiella pneumoniae | PTSO | 1709908 | ||||
| Salmonella enterica | • | PTSO | 16762086 | N-h-IIA-j-H | ||
| Yersinia pestis | • | PTSO | 16123729 | N-h-IIA-j-H | ||
| Proteus mirabilis | PTSO | 13633651 | ||||
| Shewanella violacea | PTSO | 13633605 | ||||
| Vibrio cholerae | • | VC2533 | 15642528 | N-h-IIA-j-H | ||
| Escherichia coli | • | Z4879 | 15804017 | IIA-IIB-IIC-sk-H | ||
| Salmonella enterica | • | STY4004 | 16762540 | IIB-IIC-sk-x-H | ||
| Xanthomonas campestris | • | PTSH | 21232239 | N-h-IIA-K-j-IIA-H-I | • | |
| Xanthomonas axonopodis | • | PTSH | 21243705 | N-h-IIA-K-j-IIA-H-I | • | |
| Xylella fastidiosa | • | XF1403 | 15838004 | N-h-K-j-IIA-H-I | • | |
| δ subdivision | Geobacter metallireducens | GMETP32 | 23056421 | N-h-K-j-IIA-H-I | • | |
| Desulfovibrio desulfuricans | DDES2010 | 23475093 | IID-H-I | |||
| Fusobacteria | Fusobacterium nucleatum | • | FN1782 | 19705087 | H | • |
| Fusobacterium nucleatum | • | FN1794 | 19705099 | H-I | • | |
| Actinobacteria | Corynebacterium glutamicum | • | CGL1892 | 19553142 | IIABC-H | |
| Streptomyces coelicolor | • | PTSH | 21224185 | H | ||
| Bifidobacterium longum | • | PTSH | 23465010 | H-I | ||
| Chlamydiae | Chlamydia trachomatis | • | PTSH | 15605060 | H-I | |
| Chlamydia muridarum | • | TC0614 | 15835231 | H-I | ||
| Chlamydophila pneumoniae | • | PTSH | 15617961 | H-I | ||
| Spirochaetes | Borrelia garinii | AAG28175 | 11055615 | |||
| Borrelia burgdorferi | • | BB0557 | 15594902 | H-I-IIA | ||
| Borrelia burgdorferi | • | BB0448 | 15594793 | N-h-H | ||
| Treponema pallidum | • | TP0589 | 15639577 | K-x-H | • | |
| Chlorobi | Chlorobium tepidum | • | PTSH | 21648255 | H | • |
| Chloroflexi | Chloroflexus aurantiacus | CHLOP311 | 22973407 |
the sequences were extracted from the NCBI protein database by a BLAST search using the B. subtilis ptsH sequence gene initially as the query. All unique HPr sequences in the database were reported together with the unique GI number of the entry in the database.
A bullet indicates that the whole genome sequence of an organism is available.
Clustering of genes, possibly in an operon-like structure, on the genome. H, HPr-like protein; N, σN transcription factor; I, PTS enzyme I; IIABC, PTS enzyme IIABC; IIA, IIB, IIC, IID, PTS enzymes IIA, IIB, IIC, and IID; K and K′, normal and short version of HPrK/P homologue, respectively; j, h unknown conserved genes; sk, putative sugar kinase.
A dot or square indicates the presence in the organism of a normal or short version of an HPrK/P homologue, respectively (taken from Table 1).
Typical and similar relationships. A typical sequence (bold) represents a group of similar sequences (indented) with at least 55% sequence identity.
REUTP495 is 140 residues long, which most probably is an annotation error. The true translational start is likely to correspond to the Met residue at position 52.
FIG. 3.
Phylogenetic tree of the HPr-like proteins. The tree containing a subset of 38 typical HPr-like sequences was constructed as described in the legend to Fig. 2. A typical sequence (the bold sequences in Table 2) represents a group of sequences with pairwise sequence identities of at least 55%. XPrα, NPr, XPrβ,γ,δ, SPr, and Crh represent HPr-like proteins that are explained in the text. Organisms are indicated by a shorthand following the protein that consists of the first letter of the genus followed by the first three letters of the species.
DISTRIBUTION AND GENOME ANALYSIS
Firmicutes
At the time of compilation of Table 2, the complete genome sequence was available for 17 of the 34 bacteria in the phylum Firmicutes. All these genomes contained a sequence coding for HPrK. Moreover, an HPrK homologue was found in 11 of the remaining 17 organisms, and it is to be expected that in the final 6 organisms an HPrK homologue will be found eventually. All HPr-like molecules from the phylum Firmicutes contained the regulatory-site serine, while all 34 bacteria, except for Ureaplasma urealyticum (see below), contained at least one HPr-like molecule with the active site histidine. Clearly, CcpA-dependent carbon catabolite repression is the mechanism in the phylum Firmicutes.
The eight HPr-like proteins in the complete set that do not contain the active-site histidine and, therefore, potential Crh molecules are all found in the Firmicutes (Table 2). Five of these proteins are found in bacteria (B. subtilis, Bacillus halodurans, Bacillus anthracis, Oceanobacillus iheyensis, and Thermoanaerobacter tengcongensis) that, in addition, contain an HPr-like protein with the active-site histidine. Clostridium thermocellum contains three HPr-like molecules, one with and two without the active-site histidine. Finally, Ureaplasma urealyticum is the only bacterium that contains a single HPr-like molecule without the active-site histidine. In B. subtilis Crh, the active-site histidine is replaced by a glutamine while the adjacent residues are still very similar as in HPr sequences of similar organisms (see also Table 5, region A). A multiple sequence alignment of the HPr-like molecules revealed the same sequence motif in the three HPr-like proteins of the other bacilli that therefore contain Crh in addition to HPr. An investigation of the unfinished genome sequence data of Geobacillus stearothermophilus also revealed a second HPr-like molecule in addition to HPr, with the same characteristics of B. subtilis Crh (www.genome.ou.edu/bstearo.html) (not in Table 2). T. tengcongensis, which belongs to the clostridia, contains two HPr-like molecules, one with the active-site histidine (FRUB) and one in which the histidine is deleted (TTE0115). Apart from the different active-site substitution, the molecule differs from the bacillus Crhs in that the conserved sequence motif around the site is not retained (see Table 5). Nevertheless, the missing histidine suggests a function other than in PTS-mediated transport, possibly in Crh-mediated signal transduction. C. thermocellum is the only bacterium in the phylum Firmicutes with three HPr-like molecules, one containing the active-site histidine (CHTEP203) and two without (CHTEP180 and CHTEP182). CHTEP180 is closely related to TTE0115 of T. tengcongensis and also shows a deletion at the position of the active-site histidine. These two sequences are the most distant from the other members of the family (Fig. 3). In the second sequence of C. thermocellum without the active-site histidine (CHTEP182), His is replaced by Glu and, also, the surrounding region is not conserved as in the bacillus Crhs. Remarkably, the two HPr-like proteins of T. tengcongensis and C. thermocellum, FRUB and CHTEP203, respectively, that contain the active-site histidine residues are the closest relatives of the Crh proteins of the Bacillus species (Fig. 3).
TABLE 5.
Consensus sequence around the active-site histidine (region A) and the regulatory-site serine (region B) in the HPr-like proteins
| Phylum | HPr-like proteina | Region Ab | Region Bb | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Firmicutes | HPr | G | (ILV) | H | (AT) | R | P | A | N | (AL) | K S | (IL) | (MIL) | G | (VIL) | M | S | L |
| Crh | G | L | Q | A | R | P | A | N | A | K S | I | M | G | L | M | S | L | |
| TTE0115 | I | E | S | V | D | A | K S | I | M | G | L | F | S | L | ||||
| CHTEP180 | I | T | D | V | D | A | K S | I | M | G | I | F | S | L | ||||
| CHTEP182 | G | W | E | A | K | L | A | N | A | K S | L | L | G | V | L | S | L | |
| PTSHuure | S | A | F | I | K | A | I | D | A | K S | I | I | N | I | M | A | L | |
| Proteobacteria | ||||||||||||||||||
| γ | HPr | G | L | H | T | R | P | A | (NS) | A | K S | L | F | K | (IL) | Q | T | L |
| γ | NPr | G | (LM) | H | A | R | (PA) | A | A | X S | V | X | (AG) | (ML) | L | M | L | |
| α | XPr | G | L | H | A | R | A | (AS) | (AG) | X S | I | M | G | L | M | M | L | |
| β | XPr | G | L | H | A | R | A | S | (ND) | (AG) | K S | I | M | G | (VIL) | M | (MT) | L |
| γ | XPr | G | L | H | A | R | A | (AT) | (ND) | (AG) | K S | I | M | G | (VL) | M | L | L |
XPr is defined in the text.
parentheses represent single positions where the indicated residues are found in different species. The active-site histidine and regulatory-site serine are shown in bold type.
The pathogen U. urealyticum contains a single HPr-like protein (PTSH) in which no active-site histidine is present. In agreement, the genome sequence does not contain a gene homologous to EI, the PTS enzyme responsible for the phosphorylation of HPr(His) from phosphoenolpyruvate, but does contain an HPrK/P homologue, that catalyzes the phosphorylation of HPr/Crh(Ser) from ATP. The analysis suggests that U. urealyticum has lost the PTS uptake system but has retained Crh-mediated signal transduction (11). The organism belongs to the class Mollicutes, which is considered to represent minimal life forms. Other parasites in this class, the mycoplasmas, do have a single HPr-like molecule containing the active-site histidine and PTS enzyme I.
The genes coding for HPr of the bacilli are organized in an operon together with PTS enzyme I and usually are polycistronically transcribed in a single mRNA. In B. subtilis and B. anthracis, the pair of genes is preceded by the gene coding for the sugar-specific PTS enzyme IIABC (Tables 2 and 3). The gene coding for Crh is clustered on the chromosome of the four Bacillus species with three genes, yvcJ, yvcK, and yvcL, in B. subtilis that code for proteins of unknown function. We will name the genes j, k, and l, respectively. Except for the Mollicutes, the three genes are also present as a cluster in the Firmicutes that do not contain a Crh protein. Then, the cluster is not associated with any of the PTS proteins. The close phylogenetic relationship mentioned above between the HPr proteins of T. tengcongensis and C. thermocellum, FRUB and CHTEP203, respectively, and the bacillus Crh proteins is also supported by the location of the proteins on the genome. Both are located close to the j, k, and l proteins. Remarkably, CHTEP182 of C. thermocellum that does not contain the active-site histidine is found in an operon together with PTS enzyme I, which is responsible for phosphorylation of the histidine residue.
TABLE 3.
Typical gene clusters containing HPr-like proteins in the phyla Firmicutes and Proteobacteriaa
| Phylum | Gene cluster containing:
|
||||
|---|---|---|---|---|---|
| HPr | NPr | Crh | SPr | XPrb | |
| Firmicutes | H-I, H | j-k-l-H | |||
| Proteobacteria | |||||
| α | H-I | N-h-IIA∥K′-(j)-IIA-H-(I) | |||
| β | (N)-(h)-IIA-K-j∥IIA-H-I | ||||
| γ | H-I-IIA | N-h-IIA-j-H | IIA-IIB-IIC-sk-H | N-h-(IIA)-K-j-IIA-H-I | |
| δ | N-h-K-j-IIA-H-I | ||||
SPr and XPr are defined in the text. Symbols used are as follows: H, HPr-like protein; N, σN transcription factor; I, PTS enzyme I; IIA, IIB, and IIC, PTS enzymes IIA, IIB, and IIC; K and K′, full-length and short-version HPrK/P homologues, respectively; j, k, l, and h, unknown conserved genes; sk, putative sugar kinase.
Subclusters left and right of ∥ are distantly located on the genomes. Parentheses indicate that the genes are not present in all species.
Proteobacteria
All HPr-like molecules found in the phylum Proteobacteria contain the active-site histidine. Most HPr-like molecules are found in the γ subdivision, which contains the typical gram-negative bacteria such as Escherichia coli (Table 2). A number of these, Klebsiella pneumoniae, E. coli, Yersinia pestis, Vibrio cholerae, and Salmonella enterica, possess more than one HPr-like molecule but no HPrK/P. These organisms contain, in addition to HPr, a homologue termed NPr, which is a component of the Ntr regulatory system involved in the regulation of nitrogen metabolism, among others (26). The NPrs form a separate cluster in the phylogenetic analysis of the HPr-like molecules (the ptsO genes [Fig. 3]). HPr and NPr molecules are distinguished by the clustering of the coding genes with other genes on the chromosome (Tables 2 and 3). The gene coding for HPr (ptsH) is organized together with genes coding for the PTS enzymes EI and glucose-specific IIAGlc in the pts operon in the order H-I-IIA. The three proteins are the PTS components involved in CRP-dependent carbon catabolite repression. The gene coding for NPr (ptsO) is located elsewhere on the chromosome in the Ntr cluster, together with four other genes that are involved in the Ntr regulatory pathway (N-h-IIA-j-H). The cluster starts with an RNA polymerase σ54 factor (N) followed by a putative σ54 modulation protein (h), a PTS IIA homologue termed IIANtr, a P-loop-containing protein (j), and, finally, the gene coding for NPr. The P-loop-containing protein j is homologous to the j protein in the j-k-l-Crh cluster found in the Firmicutes (Table 3). The IIANtr protein and NPr, together with a third protein termed INtr, form an independent phosphoryl group transfer chain that uses phosphoenolpyruvate as the donor (Fig. 1C). INtr is a multidomain protein consisting of a homologue of the PTS EI and a domain also found in NifA activator proteins (30).
The whole-genome sequences of Haemophilus influenzae, Pasteurella multocida, Buchnera sp., and Buchnera aphidicola in the γ subdivision contain a single HPr-like molecule embedded in an HPr-like gene cluster (Table 2), suggesting that these organisms do not use NPr-mediated regulation. For sequence similarity reasons, the same is likely to be true for Serratia marcescens and Haemophilus somnus. In contrast, the genome of Pseudomonas aeruginosa contains a single HPr-like molecule embedded in an NPr-like gene cluster that, therefore, is likely to be an NPr species. The strictly aerobic Pseudomonas and also the Azotobacter species were thought not to use the PTS for uptake of sugars, except for fructose (32). Analysis of the P. aeruginosa genome revealed two complete sugar-specific PTSs, the one for fructose and another one for N-acetylglucosamine, in which HPr moieties are present as components of multidomain proteins (29). These HPr domains are not likely to play a role in sugar uptake as general PTS components. The Pseudomonas and Azotobacter genera (and the same may be true for the Shewanella, Proteus, and Microbulbifer genera [Table 2]) use an HPr-like protein only for regulatory purposes.
E. coli and S. enterica (and also S. enterica serovar Typhimurium) both have, in addition to HPr and NPr, a third HPr-like molecule, Z4879 and STY4004, respectively. In E. coli, the protein is present only in the enterohemorrhagic strains OD157:H7 and OD157:H7 EDL933 and not in the K-12 strain. The proteins in the Escherichia and Salmonella strains are closely related (Fig. 3) and are located in a similar gene cluster on the genome (Table 2). Upstream of the gene coding for the HPr-like protein, a gene annotated as a sugar kinase (sk) and a complete set of PTS enzyme II proteins (IIA-IIB-IIC) are located. The HPr-like protein in the cluster may represent a sugar-specific HPr (SPr, for “sugar-specific HPr” [Table 3]).
Three bacteria in the γ-subdivision contain an HPrK homologue: Xanthomonas campestris, Xanthomonas axonopodis, and Xylella fastidiosa (Table 2). They also contain a single HPr-like protein; remarkably, both are in the same gene cluster that consists of seven or eight genes (N-h-IIA-K-j-IIA-H-I). We will term this cluster and the single HPr-like protein the X-cluster and XPr, respectively (for “Xanthomonas cluster” and “Xanthomonas HPr”). The first genes in the X-cluster form an Ntr gene cluster from which the gene coding for NPr is missing and in which the gene coding for HPrK is inserted upstream of the j gene (Table 3). The IIA protein encoded in this part of the cluster is of the IIANtr type and is not found in the X. fastidiosa cluster (Table 2). A second IIA gene and the genes coding for XPr and PTS enzyme I follow the Ntr-like cluster. The second IIA protein is of the IIAMan type, homologues of the IIA domain of the mannose-specific IIABMan that is part of the mannose uptake system in E. coli (34). The two Xanthomonas species and X. fastidiosa lack both NPr and INtr; i.e., it is not clear how the IIANtr protein encoded in the X-cluster is phosphorylated. An exact copy of the X-cluster found in X. fastidiosa, without IIANtr, is also found in Geobacter metallireducens in the δ subdivision of the Proteobacteria.
With the exceptions of Mesorhizobium loti and Magnetospirillum magnetotacticum in the α subdivision, the bacteria in the α and β subdivisions of the Proteobacteria contain a single HPr-like molecule (Table 2). The complete genome sequence and the data from unfinished genomes suggest that the presence of HPrK in both subdivisions is common. The HPrKs of the α subdivision are all of the short-version type (see above) (14), and the HPr-like proteins are in the same gene cluster that, in addition, contains at least a IIA molecule of the IIAMan type. The cluster resembles the last part of the X-cluster found in the γ and δ subdivisions, but the genes coding for protein j and enzyme I are not always present (Table 3). The Rhodospirillum rubrum and M. magnetotacticum clusters are an exact match of this part of the X-cluster (Table 2). The genes in the remaining part of the X-cluster, N-h-IINtr, are also found clustered on the genome of the bacteria in the α subdivision but are located distantly from the XPr/HPrK part of the cluster (Table 3). All bacteria in the α subdivision contain the gene coding for INtr, while only M. loti, M. magnetotacticum, and R. rubrum contain the classical enzyme I. The presence of enzyme I correlates with the presence of a second HPr-like protein in M. loti and M. magnetotacticum.
The situation observed in the β subdivision is similar, but the X-cluster is broken up in different parts (Table 3). The single gene coding for the HPr-like molecule (XPr) clusters with the genes coding for enzyme I and IIAMan in the same order as in the X-cluster of the γ and δ subdivisions. The gene coding for HPrK clusters elsewhere on the genome, together with IIANtr and protein j in the order IIA-K-j. In the complete genome sequences of Ralstonia solanacearum and Neisseria meningitidis, the three genes are preceded only in the latter organism by the N and h genes (Tables 2 and 3). A difference with the α subdivision is that in the β subdivision, all bacteria contain the PTS enzyme I but not INtr.
Summarizing, HPrK is found in all four subdivisions of the Proteobacteria and seems to cluster on the genomes together with the genes coding for components involved in Ntr-type of gene regulation. The X-cluster in the Xanthomonas species in the γ subdivision represents the most complete cluster, while fission of the cluster is observed in the α and β subdivisions.
Other Phyla
In the phyla other than Firmicutes and Proteobacteria, most organisms have a single HPr-like molecule that clusters with the gene coding for enzyme I (H-I), as found in the Firmicutes (Table 2). Three bacteria contain an HPrK homologue, Fusobacterium nucleatum, Treponema pallidum, and Chlorobium tepidum. F. nucleatum contains two HPr-like molecules, one of which, FN1794, is adjacent to the gene coding for PTS EI, indicating that FN1794 is HPr. The second gene, FN1782, is located a couple of genes upstream of this pair in no apparent operon structure. The genome contains genes coding for IIANtr and the j protein, but, like HPrK, these do not all cluster together on the genome. No homologues are found for IIAMan or INtr. On the genome of T. pallidum, the genes coding for the HPrK homologue and the HPr-like protein are separated by a single gene, which is not related to any of the genes found in the X-cluster in the Proteobacteria. Both T. pallidum and C. tepidum contain IIANtr encoded elsewhere on the genome, but they contain no IIAMan, j protein, or INtr. The HPr-like molecules of the bacteria in this category that do contain HPrK do not seem to be closely related to the HPr-like molecules found in the X-cluster of the Proteobacteria (Fig. 3).
SEQUENCE ANALYSES
Full-Length and Short-Version HPr Kinases
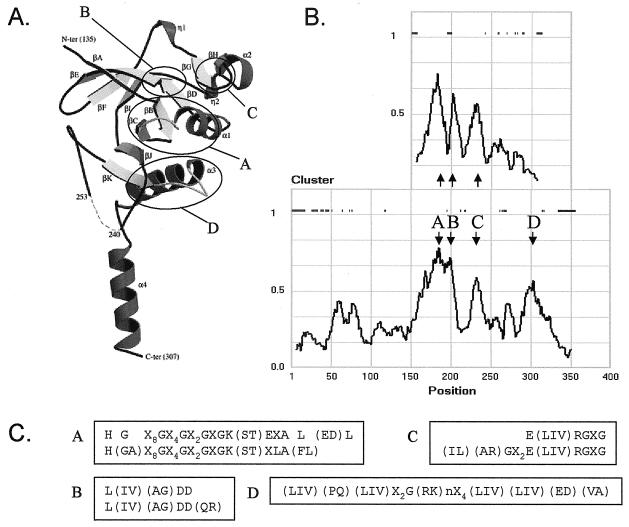
The genes coding for the short-version HPrK homologues in the α subdivision are organized, together with the HPr-like protein XPr, in a similar gene cluster (the X-cluster) to that of the full-length versions in the β, γ, and δ subdivisions of the Proteobacteria, strongly suggesting that they serve the same function, putatively as HPr kinases/phosphatases. HPrK is known to consist of two domains. The three-dimensional structure of HPrK of Staphylococcus xylosus resolved at 1.95 Å shows a hexameric arrangement (a dimer of trimers) in which the N-terminal and C-terminal domains are well separated, with no apparent intramolecular contacts (20). While the function of the N-terminal domain is not clear, the C-terminal domain is the catalytic domain. Deletion of the N-terminal 127 residues of Lactobacillus casei HPrK, more or less corresponding to the N-terminal domain, yielded an active entity whose three-dimensional structure was resolved separately at 2.8 Å (8). The structures of the C-terminal catalytic domains (Fig. 4A) from the two organisms closely matched each other. Multiple sequence alignment of the full-length and short-version HPrK homologues suggests that the latter corresponds to the catalytic domain; the beginning of the short versions correlates more or less with the beginning of the C-terminal domain of the full-length HPrKs. However, the length of the short versions and the C-terminal domains of the full-length proteins differ by roughly 50 residues; they are about 150 and 200 residues, respectively. The C-terminal domain of the full-length HPrK contains four conserved sequence motifs (A, B, C, and D in Fig. 4B), the first of which contains the so-called Walker A motif typical for the binding of the phosphate groups of ATP (43). Motifs A, B, and C are also found in the short-version homologues, but motif D is missing (Fig. 4B and C). In fact, the sequence similarity of the two versions covers only approximately the first 100 residues of the short version, the part that contains sequence motifs A, B, and C. The C-terminal 50 residues of the short versions do not seem to be related to the corresponding area in the full-length proteins. It follows that the short versions correspond to the top part in the structure depicted in Fig. 4A, up to strand βJ. The C-terminal part of the full-length proteins, consisting of βJ, βK, α3, and α4, would be missing. In the crystal structures (8, 20), the loop between βK and α3 in conserved domain D (the K3 loop) and the two α-helices α3 and α4 are involved in intimate contacts within two pairs of trimers that form the overall hexameric structure of the complex. The short versions may not form a multimeric structure and may instead exist as monomers.
FIG. 4.
Sequence signatures in full-length and short-version HPrK homologues. (A) Structural model of the C-terminal domain of the full-length HPrK/P. Ovals indicate conserved regions A, B, C, and D. (B) Pairwise sequence identity in the multiple sequence alignments of the short-version (top) and full-length (bottom) HPr kinases. The “cluster” function gives the fraction of identities at each position in the alignment in an all-against-all comparison. The scores were averaged over a sliding window of nine positions. The plot of the short-version HPrKs was “aligned” with the plot of the full-length versions based on the multiple sequence alignment. The set of full-length sequences used in the alignment is described in the legend to Fig. 2, and the set of short versions is taken from Table 2 (Proteobacteria, α subdivision). The bars in the top half of the plots indicate positions where a gap occurs in any of the sequences in the alignment. The four conserved regions in the full-length HPrK homologues are indicated as A to D. (C) Sequence motifs corresponding to regions A to D in panels A and B. The top and bottom sequences in the boxes correspond to the full-length and short-version HPr kinases, respectively. Conserved motif D is not present in the short-version HPr kinases. (Panel A reprinted from reference 8 with permission from the publisher.)
The structure of the catalytic domain of the L. casei HPrK/P in complex with B. subtilis HPr was resolved at 2.8 Å (7). The hexameric HPrK/P complex bound six HPr molecules, each at the interface of two monomers. The HPr molecules bound at two separate interfaces, one at each monomer. The catalytic interaction, positioning the regulatory-site serine of HPr close to the Walker A motif of HPrK, involves strand βA and conserved motifs A and B on one of the monomers (Fig. 4A). It follows that this interaction is still possible in the short-version HPrKs. The second interface involves C-terminal helix α4 on the neighboring monomer. This interaction is absent in the short versions. The analysis supports the conclusion of the genetic analysis, i.e., that the short-version HPrK homologues actually may function as HPrK/phosphatases.
Phylogenetic Relationships of HPr-Like Proteins
Phylogenetic analysis of the HPr-like proteins shows that the HPrs from the gram-positive (lacto)bacilli are closely related while those from the clostridia are more distant (Fig. 3). The four Crh and four HPr proteins of B. subtilis, B. halodurans, B. anthracis, and O. iheyensis cluster in different branches of the tree. The Crh proteins are more closely related to the HPr proteins of the clostridia than to the HPr proteins of the bacilli. Pairwise sequence analysis revealed a higher sequence identity between the HPr sequences of the different species and the Crh sequences of the different species than between HPr and Crh of the same species (Table 4). If Crh had originated from an HPr gene duplication during evolution, this must have happened before the primordial Bacillus diverged into the different species. A similar situation exists for the HPr and NPr proteins in gram-negative bacteria (Fig. 3).
TABLE 4.
Pairwise sequence identity of the HPr and Crh sequences of Bacillus speciesa
| Sequence | % Identity to:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| HPRbsub | HPRbhal | HPRbant | HPRoihe | CRHbsub | CRHbhal | CRHbant | CRHoihe | |
| HPRbsub | 64 | 72 | 61 | |||||
| HPRbhal | 61 | 70 | ||||||
| HPRbant | 55 | |||||||
| HPRoihe | ||||||||
| CRHbsub | 45 | 42 | 39 | 41 | 73 | 71 | 65 | |
| CRHbhal | 42 | 45 | 40 | 42 | 73 | 65 | ||
| CRHbant | 48 | 41 | 44 | 39 | 60 | |||
| CRHoihe | 42 | 43 | 38 | 36 | ||||
bsub, B. subtilis; bhal, B. halodurans; bant, B. anthracis; oihe, O. iheyensis.
The putative Crh proteins found in the clostridia, TTE0115 of the thermophile T. tengcongensis and CHTEP180 and CHTEP182 of C. thermocellum, and PTSH of the mollicute U. urealyticum are not in the same cluster as the bacillus Crhs. Closest is CHTEP182 in the clostridium cluster. TTE0115 and CHTEP180 are the most distant relatives in the HPr-like protein family, and PTSH of U. urealyticum clusters loosely with HPr-like proteins from the phyla Fusobacteria and Spirochaetes and HPrs from the γ subdivision of the Proteobacteria (Fig. 3). Consistent with the phylogenetic distance between Crh and HPr of the bacillus species, PTSH of U. urealyticum is quite distant from HPr of the related mollicutes and TTE0115 and CHTEP180 are distant from the clostridium branch. The conservation that is observed in the region around the mutated active-site histidine residue in the bacillus Crh proteins is completely absent in the four putative Crh proteins (see Table 5). Apart from the mutated active-site histidine, the latter do not seem to have much in common with the former. The gram-negative bacteria of the α, β, and δ subdivisions of the Proteobacteria are likely to have CcpA-dependent CCR since they possess HPrK. The HPr-like proteins of the α subdivision (XPrα) all cluster in one branch of the tree (Fig. 3). The HPr-like proteins of the β and δ subdivisions are on the same branch as the XPr proteins of the bacteria in the γ subdivision of the Proteobacteria that possess HPrK (Xanthomonas and Xylella species; XPrβ,γ,δ). The branch is distant from HPr of the gram-negative bacteria in the γ subdivision that do not contain HPrK (e.g., Escherichia and Klebsiella species). Both the XPrα and XPrβ,γ,δ proteins are distant from the gram-positive HPr proteins. Importantly, the XPrβ,γ,δ proteins and, especially, the XPrα proteins are loosely associated with the NPr proteins from the γ subdivision. Moreover, the XPr proteins of the β, γ, and δ subdivisions have >55% sequence identity to the HPr-like proteins of the Pseudomonas species in the γ subdivision that are likely to function in Ntr-type regulation (see above).
Sequence Motifs in HPr-Like Proteins
None of the residues in a multiple sequence alignment of the HPr-like family of proteins was completely conserved (data not shown). An analysis of an all-against-all pairwise sequence alignment reveals the presence of three well-conserved regions: region A around position 16 (B. subtilis Crh numbering), region B around position 46, and region C around position 70 in the C terminus (Fig. 5A). The solution structures of a number of HPr proteins originating from different species and of Crh of B. subtilis have been solved by nuclear magnetic resonance spectroscopy, and all of them show a similar overall folding (5, 16, 22, 42). The HPr polypeptide folds as an open-faced β-sandwich, with three α-helices (a, b, and c [Fig. 5B]) on top of a four-stranded β-sheet (strands β1 to β4 [Fig. 5B]). In a linear representation, the order of the secondary-structure elements would be β1aβ2β3bβ4c. Conserved region A contains the active-site histidine, which is positioned just in front of α-helix a. The region comprises the interface of the loop preceding helix a and helix a. The region is involved in the interaction of HPr and the PTS enzymes EI and IIA. Region B contains the regulatory-site serine, which is positioned at the interface of the loop between β3 and α-helix b. The region covers most of helix b, which is only two turns long. Region C comprises the loop between β4 and α-helix c plus the first turn at the N-terminal end of helix c. Region C is quite distant from the other two conserved regions on the surface of the protein. The HPr-like molecules from the Actinobacteria members Corynebacterium glutamicum and Streptomyces coelicolor, as well as from Chloroflexus aurantiacus, contain an insertion of 4 residues in the loop connecting strands β2 and β3, where they are not likely to disturb the folding of the protein (not shown).
FIG. 5.
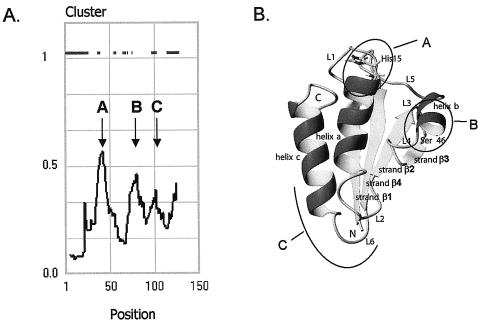
Conserved regions in HPr-like molecules. (A) Clustering of pairwise sequence identity. The plot is explained in the legend to Fig. 4. The “spike” at position 20 is an artifact caused by the presence of two sequences with an N-terminal extension. The bars in the top part of the plot indicate positions where a gap was found in any of the sequences. (B) Structural model of HPr-like proteins. Conserved regions A, B, and C are indicated. (C) Sequence analysis of conserved region C. The bars indicate the frequency of the negatively charged residues D and E at the indicated positions. The dotted line indicates the average frequency in the whole set. At positions with low frequencies, the dominant residue(s) is indicated. Position numbering is according to Crh of B. subtilis. (Panel B reprinted from reference 22 with permission from the publisher.)
With few exceptions, conserved region A in the HPr, Crh, NPr, and XPr proteins contains the consensus sequence G(LI)H(AT)R(PA)(AS), with the active-site histidine in bold (Table 5). It was noted before that in the bacillus Crhs, the motif is retained with the exception of the histidine-to-glutamine mutation, suggesting that Crh might still interact with the enzymes enzyme I and IIA (5). In contrast, in the putative Crh proteins from the clostridia and from U. urealyticum, TTE0115, CHTEP180, CHTEP182, and PTSH, the motif is completely lost during evolution, suggesting that these proteins have lost the interaction with the PTS uptake system.
The regulatory-site serine in conserved region B is the best-conserved residue in the whole family. Only three HPr-like molecules in the family do not contain a serine at this position: STY4004 of Salmonella enterica, CHLOP311 of Chloroflexus aurantiacus, and PTSH of Bifidobacterium longum (Table 2). None of these three organisms contains an HPrK homologue. The missing regulatory-site serine in the S. enterica protein is most probably an alignment artifact since the sequence SILG, which is similar to the consensus sequence (see below), is located 4 residues downstream. In contrast, the serine residue in B. longum is replaced by arginine while the surrounding residues are still conserved. In the C. aurantiacus protein, the serine residue is deleted and the adjacent residues are not conserved. The high conservation of the serine residue throughout the whole set makes it difficult to accept that the serine, or at least the conserved region, has no function in those bacteria that lack HPr kinase.
Conserved region B shows more clustering of the sequences following the phylogeny of the bacteria (Tables 1 and 2) than may be detected in the case of conserved region A (Table 5). Even though the HPrs and Crhs of (lacto)bacilli are in separate branches of the phylogenetic tree (Fig. 3), the regions around the regulatory-site serines are almost identical. The consensus sequence is VN(AL)KSIMG(VL)MSL(AG), in which the regulatory-site serine is shown in bold. The HPr and Crh sequences differ only at the third, ninth, and last positions, as indicated. The putative Crh proteins TTE0115 of T. tengcongensis and CHTEP180 of C. thermocellum have very similar sequences in region B, while CHTEP182 of C. thermocellum and PTSH of U. urealyticum are more divergent (Table 5). The HPr proteins of the gram-negative bacteria of the γ subdivision of the Proteobacteria are strongly conserved in region B, but the motif is quite different from that of HPr and Crh of the Firmicutes. Consistent with the presence of HPrK in these bacteria, the XPr proteins of the bacteria in the α, β, and γ subdivisions of the Proteobacteria contain B-motifs that resemble the motif observed in gram-positive bacteria. The triad IMG following the regulatory-site serine residue seems to be typical for the HPr-like proteins associated with HPrK. Remarkably, although XPr and NPr cluster on the same side of the phylogenetic tree, regions B are not conserved between the two types of HPr-like molecules. Region B of NPr is the least conserved among the HPr-like proteins (Table 5).
Conserved region C contains an unusually high fraction of the negatively charged residues aspartate and glutamate. Eight positions in this region contain 30% of all the D and E residues in the set. Figure 5C shows the frequency per position. In between the positions with high D-plus-E content are positions with conserved small or hydrophobic residues. Region C represents a negative patch on the surface of the protein. No specific function for the region is known.
CONCLUSIONS
Homologues of HPrK, the typical component of CcpA-dependent CCR, are commonly observed in the phylum Firmicutes but are also found in the phyla Proteobacteria, Fusobacteria, Spirochaetes, and Chlorobi, suggesting that CcpA-dependent CCR is not restricted to gram-positive bacteria. In the α and β subdivisions of the Proteobacteria, HPrK even appears to be common, while in the γ subdivision it is more of an exception and is found only in Xanthomonas and Xylella species.
HPrK/P homologues come in two versions: the full-length version, consisting of about 325 residues is the common one and is found in all phyla, whereas the short version, consisting of about 150 residues, is unique to the α subdivision of the Proteobacteria. The short version corresponds to an internal fragment of the full version. The full-length HPrKs may consist of three rather than two domains, an N-terminal domain of unknown function, a catalytic domain consisting of the next 100 residues, and a C-terminal domain that is responsible for the association of the protein into a multimeric structure. The short version corresponds to the internal catalytic domain.
Three lines of evidence suggest that the HPrK homologues in the phyla other than the Firmicutes function as HPrKs: (i) the interaction site with HPr on the catalytic domain is conserved (motifs A and B [Fig. 4]); (ii) the genes coding for the HPrK homologues and the HPr-like proteins of the same organisms (XPrs) are clustered on the genome (the X cluster [Table 3]); and (iii) the amino acid sequence around the regulatory-site serine in XPr (region B) is similar to that observed in the HPr-like proteins (HPr and Crh) of the Firmicutes that are phosphorylated by HPrK and different from the HPr-like proteins (HPr and NPr) from organisms that do not contain HPrK (Table 5). Overall, the XPr proteins do not cluster on the same branch of the phylogenetic tree as the HPr and Crh proteins of the Firmicutes (Fig. 3). A number of organisms in the α subdivision of the Proteobacteria do contain an HPrK homologue but no enzyme I, the PTS enzyme responsible for the phosphorylation of HPr at the active-site histidine (e.g., R. sphaeroides). XPr of these organisms seems to function only in CcpA-dependent gene regulation and not in PTS-mediated uptake, a situation similar to that observed in the strictly aerobic bacteria of the Pseudomonas species with respect to the Ntr regulatory system.
Crh-mediated signal transduction is observed only in the phylum Firmicutes. Uncoupling of the PTS transport function and the CCR function by mutation of the active-site histidine, yielding Crh proteins, allows the coexistence of independent uptake and regulatory functions in these bacteria. Again, a parallel may be drawn to the Ntr regulatory system involving NPr in the γ subdivision of the Proteobacteria, which is also thought to operate independently of the PTS uptake system. Close homologues of Crh of B. subtilis are found only in other bacilli. The putative Crh proteins of the Clostridia and U. urealyticum differ from the bacillius Crh in that the amino acid sequence of conserved region A containing the mutated active-site histidine is not conserved. Regions B of C. thermocellum CHTEP180 and of T. tengcongensis TTE0115 resemble the serine phosphorylation motif in the Firmicutes HPr and Crh proteins and therefore are the best candidates for a Crh function (Table 5). The presence of a second putative Crh protein in C. thermocellum with a divergent region B suggests a more differentiated regulatory system in this Clostridium species.
The most remarkable finding of the database searches presented here is a possible relationship between CcpA-dependent gene regulation and Ntr found in the γ subdivision of the Proteobacteria. The relationship is suggested by (i) the clustering of CCR and Ntr components on the genomes of members of the Proteobacteria (the X-cluster), (ii) the phylogenetic relationship between XPr and NPr (Fig. 3), and (iii) the presence of the j protein in the Ntr cluster and Crh cluster of the Firmicutes. The X-cluster in the Xanthomonas species in the γ subdivision represents the most complete cluster of genes. It contains the genes for IIANtr, HPrK, IIAMan, XPr, and enzyme I (Table 3). Since NPr and INtr are missing in the Xanthomonas species, their role may have been taken over by XPr and enzyme I, respectively. This would place XPr at the center of a complex regulatory network; it would be phosphorylated by HPrK in CcpA-dependent CCR and would be intermediate between IIANtr and enzyme I in the Ntr system. The X-clusters of X. fastidiosa and G. metallireducens in the δ subdivision lack the gene for IIANtr. If the N, h, and j genes are indicative of Ntr regulation, the function of IIANtr may have been taken over by IIAMan. In the α and β subdivision of the Proteobacteria, the genes of the X-cluster are organized in two subclusters. In the α subdivision, the gene coding for HPrK is found together with the genes for IIAMan and XPr, while in the β subdivision, the HPrK gene is clustered with the Ntr-associated genes. In both subdivisions, IIANtr and IIAMan are present. Most of the bacteria from the α subdivision do not possess enzyme I, but all of them contain INtr. The components in a bacterium like Caulobacter crescentus in the α subdivision provide the best evidence for XPr being an intermediate in both CcpA-dependent CCR and Ntr type of regulation. It contains the putative phosphotransfer chain INtr > XPr > IIANtr, and XPr contains the serine phosphorylation motif (region B), which allows phosphorylation by (the short version of) HPrK (Fig. 6). The mechanism could serve as a coupling between carbon and nitrogen metabolism of the cell.
FIG. 6.
Putative signal transduction network in the α subdivision of Proteobacteria. HPr-like protein XPr would connect two signal transduction pathways, CcpA-mediated gene regulation on the left and the Ntr-regulation system on the right.
PROSPECTS
Since the first description of the PTS in E. coli by Kundig and Roseman in 1964 (18), our knowledge of the physiological relevance of the system, which turned out to be a typical bacterial system, has grown enormously. The many sugar-specific components that were discovered defined the PTS as one of the transporter classes next to the primary, secondary, and ATP binding cassette-type transporters. The roles of the PTS in regulation of the expression of metabolic genes showed that the system was one of the important regulatory networks controlling metabolism in the bacterial cell. The many bacterial genome sequences that are available to date provide an inventory of the “hardware” of the PTS in different organism. The hardware comprises the genes coding for PTS proteins and their clustering on the genome. Only experiments can reveal the physiological function of the proteins by showing expression of the genes, control on other genes, interaction with other proteins, interaction with sites on the genome, phosphorylation states of the proteins, etc. Already it is clear that the hardware differs significantly among different bacteria. Only experiments will be able to tell how this translates to differences in the physiology of the organisms. Most of the analysis presented here is based purely on data mining, and none of the conclusions should be considered proven as such. For many organisms, we do not know if the open reading frames presented here as genes are actually expressed, nor do we know if the clusters of genes that play an important role in the analysis represent real operon structures that are transcribed in single messengers. Nevertheless, the analysis provides important clues that can and should be tested experimentally. If it shows one thing, it is that we cannot get a complete overview of a complex system like the PTS by studying one or two model organisms like E. coli and B. subtilis. To find the roots of the PTS (at least the regulatory part), more exotic bacteria in the α, β, γ, and δ subdivisions of the phylum Proteobacteria may have to be studied. For each question asked, we may have to find the best organism to study. With the ongoing genome-sequencing projects, this may soon become reality.
Acknowledgments
This work was supported by grants from the Ministry of Economic Affairs of The Netherlands, in the framework of “IOP Milieutechnologie/Zware Metalen,” project IZW97404.
REFERENCES
- 1.Altschul, S. F., and E. V. Koonin. 1998. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci. 23:444-447. [DOI] [PubMed] [Google Scholar]
- 2.Darbon, E., A. Galinier, D. Le Coq, and J. Deutscher. 2001. Phosphotransfer functions mutated Bacillus subtilis HPr-like protein Crh carrying a histidine in the active site. J. Mol. Microbiol. Biotechnol. 3:439-444. [PubMed] [Google Scholar]
- 3.Deutscher, J., E. Kuster, U. Bergstedt, V. Charrier, and W. Hillen. 1995. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol. Microbiol. 15:1049-1053. [DOI] [PubMed] [Google Scholar]
- 4.Deutscher, J., and M. H. Saier, Jr. 1983. ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 80:6790-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favier, A., B. Brutscher, M. Blackledge, A. Galinier, J. Deutscher, F. Penin, and D. Marion. 2002. Solution structure and dynamics of Crh, the Bacillus subtilis catabolite repression HPr. J. Mol. Biol. 317:131-144. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 2002. PHYLIP (Phylogeny Inference Package), version 3.6a3. Department of Genome Sciences, University of Washington, Seattle.
- 7.Fieulaine, S., S. Morera, S. Poncet, I. Mijakovic, A. Galinier, J. Janin, J. Deutscher, and S. Nessler. 2002. X-ray structure of a bifunctional protein kinase in complex with its protein substrate HPr. Proc. Natl. Acad. Sci. USA 99:13437-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fieulaine, S., S. Morera, S. Poncet, V. Monedero, V. Gueguen-Chaignon, A. Galinier, J. Janin, J. Deutscher, and S. Nessler. 2001. X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain. EMBO J. 20:3917-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galinier, A., J. Haiech, M. C. Kilhoffer, M. Jaquinod, J. Stülke, J. Deutscher, and I. Martin-Verstraete. 1997. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc. Natl. Acad. Sci. USA 94:8439-8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galinier, A., M. Kravanja, R. Engelmann, W. Hengstenberg, M. C. Kilhoffer, J. Deutscher, and J. Haiech. 1998. New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc. Natl. Acad. Sci. USA 95:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glass, J. I., E. J. Lefkowitz, J. S. Glass, C. R. Heiner, E. Y. Chen, and G. H. Cassell. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407:757-762. [DOI] [PubMed] [Google Scholar]
- 12.Guédon, E., E. Jamet, and P. Renault. 2002. Gene regulation in Lactococcus lactis: the gap between predicted and characterized regulators. Antonie Leeuwenhoek 82:93-112. [PubMed] [Google Scholar]
- 13.Henkin, T. M. 1996. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol. Lett. 135:9-15. [DOI] [PubMed] [Google Scholar]
- 14.Hu, K. Y., and M. H. Saier, Jr. 2002. Phylogeny of phosphoryl transfer proteins of the phosphoenolpyruvate-dependent sugar-transporting phosphotransferase system. Res. Microbiol. 153:405-415. [DOI] [PubMed] [Google Scholar]
- 15.Jault, J. M., S. Fieulaine, S. Nessler, P. Gonzalo, A. Di Pietro, J. Deutscher, and A. Galinier. 2000. The HPr kinase from Bacillus subtilis is a homo-oligomeric enzyme which exhibits strong positive cooperativity for nucleotide and fructose 1,6-bisphosphate binding. J. Biol. Chem. 275:1773-1780. [DOI] [PubMed] [Google Scholar]
- 16.Jia, Z., M. Vandonselaar, W. Hengstenberg, J. W. Quail, and L. T. Delbaere. 1994. The 1.6 Å structure of histidine-containing phosphotransfer protein HPr from Streptococcus faecalis. J. Mol. Biol. 236:1341-1355. [DOI] [PubMed] [Google Scholar]
- 17.Kravanja, M., R. Engelmann, V. Dossonnet, M. Blüggel, H. E. Meyer, R. Frank, A. Galinier, J. Deutscher, N. Schnell, and W. Hengstenberg. 1999. The hprK gene of Enterococcus faecalis encodes a novel bifunctional enzyme: the HPr kinase/phosphatase. Mol.Microbiol. 31:59-66. [DOI] [PubMed] [Google Scholar]
- 18.Kundig, W., S. Gosh, and S. Roseman. 1964. Phosphate bound to histidine in a protein as an intermediate in a novel phosphotransferase system. Proc. Natl. Acad Sci. USA 52:1067-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessières, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, and A. Danchin. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 20.Marquez, J. A., S. Hasenbein, B. Koch, S. Fieulaine, S. Nessler, R. B. Russell, W. Hengstenberg, and K. Scheffzek. 2002. Structure of the full-length HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution: mimicking the product/substrate of the phospho transfer reactions. Proc. Natl. Acad Sci. USA 99:3458-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin-Verstraete, I., A. Galinier, E. Darbon, Y. Quentin, M. C. Kilhoffer, V. Charrier, J. Haiech, G. Rapoport, and J. Deutscher. 1999. The Q15H mutation enables Crh, a Bacillus subtilis HPr-like protein, to carry out some regulatory HPr functions, but does not make it an effective phosphocarrier for sugar transport. Microbiology 145:3195-3204. [DOI] [PubMed] [Google Scholar]
- 22.Maurer, T., R. Doker, A. Gorler, W. Hengstenberg, and H. R. Kalbitzer. 2001. Three-dimensional structure of the histidine-containing phosphocarrier protein (HPr) from Enterococcus faecalis in solution. Eur. J. Biochem. 268:635-644. [DOI] [PubMed] [Google Scholar]
- 23.Michiels, J., T. Van Soom, I. D'hooghe, B. Dombrecht, T. Benhassine, P. de Wilde, and J. Vanderleyden. 1998. The Rhizobium etli rpoN locus: DNA sequence analysis and phenotypical characterization of rpoN, ptsN, and ptsA mutants. J. Bacteriol. 180:1729-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mijakovic, I., S. Poncet, A. Galinier, V. Monedero, S. Fieulaine, J. Janin, S. Nessler, J. A. Marquez, K. Scheffzek, S. Hasenbein, W. Hengstenberg, and J. Deutscher. 2002. Pyrophosphate-producing protein dephosphorylation by HPr kinase/phosphorylase: a relic of early life? Proc. Natl. Acad. Sci. USA 99:13442-13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powell, B. S., D. L. Court, T. Inada, Y. Nakamura, V. Michotey, X. Cui, A. Reizer, M. H. Saier, Jr., and J. Reizer. 1995. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIANtr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J. Biol. Chem. 270:4822-4839. [DOI] [PubMed] [Google Scholar]
- 27.Reizer, J., U. Bergstedt, A. Galinier, E. Küster, M. H. Saier, Jr., W. Hillen, M. Steinmetz, and J. Deutscher. 1996. Catabolite repression resistance of gnt operon expression in Bacillus subtilis conferred by mutation of His-15, the site of phosphoenolpyruvate-dependent phosphorylation of the phosphocarrier protein HPr. J. Bacteriol. 178:5480-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reizer, J., C. Hoischen, F. Titgemeyer, C. Rivolta, R. Rabus, J. Stülke, D. Karamata, M. H. Saier, Jr., and W. Hillen. 1998. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol. Microbiol. 27:1157-1169. [DOI] [PubMed] [Google Scholar]
- 29.Reizer, J., A. Reizer, M. J. Lagrou, K. R. Folger, C. K. Stover, and M. H. Saier, Jr. 1999. Novel phosphotransferase systems revealed by bacterial genome analysis: the complete repertoire of pts genes in Pseudomonas aeruginosa. J. Mol. Microbiol. Biotechnol. 1:289-293. [PubMed] [Google Scholar]
- 30.Reizer, J., A. Reizer, and M. H. Saier, Jr. 1995. Novel phosphotransferase system genes revealed by bacterial genome analysis—a gene cluster encoding a unique Enzyme I and the proteins of a fructose-like permease system. Microbiology 141:961-971. [DOI] [PubMed] [Google Scholar]
- 31.Reizer, J., and M. H. Saier, Jr. 1997. Modular multidomain phosphoryl transfer proteins of bacteria. Curr. Opin. Struct. Biol. 7:407-415. [DOI] [PubMed] [Google Scholar]
- 32.Romano, A. H., S. J. Eberhard, S. L. Dingle, and T. D. McDowell. 1970. Distribution of the phosphoenolpyruvate: glucose phosphotransferase system in bacteria. J. Bacteriol. 104:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segura, D., and G. Espín. 1998. Mutational inactivation of a gene homologous to Escherichia coli ptsP affects poly-β-hydroxybutyrate accumulation and nitrogen fixation in Azotobacter vinelandii. J. Bacteriol. 180:4790-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stolz, B., M. Huber, Z. Markovic-Housley, and B. Erni. 1993. The mannose transporter of Escherichia coli. Structure and function of the IIABMan subunit. J. Biol. Chem. 268:27094-27099. [PubMed] [Google Scholar]
- 35.Stülke, J., M. Arnaud, G. Rapoport, and I. Martin-Verstraete. 1998. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 28:865-874. [DOI] [PubMed] [Google Scholar]
- 36.Stülke, J., and W. Hillen. 1998. Coupling physiology and gene regulation in bacteria: the phosphotransferase sugar uptake system delivers the signals. Naturwissenschaften 85:583-592. [DOI] [PubMed] [Google Scholar]
- 37.Stülke, J., and W. Hillen. 2000. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 54:849-880. [DOI] [PubMed] [Google Scholar]
- 38.Tan, M. W., L. G. Rahme, J. A. Sternberg, R. G. Tompkins, and F. M. Ausubel. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad Sci. USA 96:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tchieu, J. H., V. Norris, J. S. Edwards, and M. H. Saier, Jr. 2001. The complete phosphotransferase system in Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:329-346. [PubMed] [Google Scholar]
- 40.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Titgemeyer, F., and W. Hillen. 2002. Global control of sugar metabolism: a Gram-positive solution. Antonie Leeuwenhoek 82:59-71. [PubMed] [Google Scholar]
- 42.van Nuland, N. A., J. Grötzinger, K. Dijkstra, R. M. Scheek, and G. T. Robillard. 1992. Determination of the three-dimensional solution structure of the histidine-containing phosphocarrier protein HPr from Escherichia coli using multidimensional NMR spectroscopy. Eur.J.Biochem. 210:881-891. [DOI] [PubMed] [Google Scholar]
- 43.Walker, J. E. 1994. The regulation of catalysis in ATP synthase. Curr.Opin.Struct.Biol. 4:912-918. [DOI] [PubMed] [Google Scholar]
- 44.Warner, J. B., B. P. Krom, C. Magni, W. N. Konings, and J.S. Lolkema. 2000. Catabolite repression and induction of the Mg2+-citrate transporter CitM of Bacillus subtilis. J. Bacteriol. 182:6099-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warner, J. B., and J. S. Lolkema. 2003. A Crh specific function in carbon catabolite repression in Bacillus subtilis. FEMS Microbiol. Lett. 220:277-280. [DOI] [PubMed] [Google Scholar]