Abstract
The adenomatous polyposis coli (APC) protein is crucial to homeostasis of normal intestinal epithelia because it suppresses the β-catenin/TCF pathway. Consequently, loss or mutation of the APC gene causes colorectal tumors in humans and mice. Here, we describe our use of Multidimensional Protein Identification Technology (MudPIT) to compare protein expression in colon tumors to that of adjacent healthy colon tissue from ApcMin/+ mice. Twenty-seven proteins were found to be up-regulated in colon tumors and twenty-five down-regulated. As an extension of the proteomic analysis, the differentially expressed proteins were used as “seeds” to search for co-expressed genes. This approach revealed a co-expression network of 45 genes that is up-regulated in colon tumors. Members of the network include the antibacterial peptide cathelicidin (CAMP), Toll-like receptors (TLRs), IL-8, and triggering receptor expressed on myeloid cells 1 (TREM1). The co-expression network is associated with innate immunity and inflammation, and there is significant concordance between its connectivity in humans versus mice (Friedman: p value = 0.0056). This study provides new insights into the proteins and networks that are likely to drive the onset and progression of colon cancer.
Keywords: proteomics, network, MudPIT, mass spectrometry, transcriptomics, colon cancer, inflammation, ApcMin/+
INTRODUCTION
Colorectal cancer (CRC) is the third leading cause of cancer-related death in the United States 1. Seventy-five percent of CRCs occur sporadically, and the rest are hereditary. About five percent of hereditary CRC results from a germ-line mutation in the adenomatous polyposis coli (APC) gene 2, 3. Even in sporadic CRC, the APC gene is usually inactivated by mutations 4, leading many to consider APC the colorectal cancer gene 5. Other than genetic testing for mutations in APC, there are few biomarkers that can guide clinical management of CRC 6. In addition, classic chemotherapies remain the mainstay of CRC treatment. Clearly then there is a pressing need for accurate diagnostic/ prognostic biomarkers, and for new drugs to help reduce the mortality of CRC.
The ApcMin/+ mouse is an accepted model of human CRC because it contains a germ-line mutation in the Apc gene 7. Tumors and body fluids from this strain of mice have been subjected to transcriptome profiling 8, 9. Proteomic analysis has also been used to search for insights into the onset and progression of CRC. Xu et al studied distal colons from wild-type mice and compared them to intestinal adenomas from ApcMin/+ mice 10; while Huttlin et al compared the proteome of colon tumors of ApcMin/+ mice to that of normal tissue of wild-type mice 11. Ang et al analyzed fecal samples from ApcMin/+ mice 12. Here, we perform the first comparison of the proteome of colon tumors to that of adjacent non-tumor tissue in the ApcMin/+ mouse. We identify twenty-seven proteins that are up-regulated and twenty-five down-regulated in colon tumors. The identity of these proteins provide new insights into molecular changes that give rise to tumors and reveal new avenues that can be targeted for therapeutic intervention.
As an extension of straightforward proteome comparisons between tumor and non-tumor tissue, we also sought to determine if a network of co-regulated proteins/genes is associated with colon tumors in the ApcMin/+ mouse. This approach was taken in response to the increasing interest in network and pathway-based target discovery, which largely stems from the notion that complex phenotypes, like that of cancer, must arise from coordinated changes to a network of interacting genes and/or proteins 13. With this idea in mind our criteria for such a co-expression network were: i) that it exist in human and mouse so that studies in animal models can be reasonably extrapolated to the clinical condition; ii) that the network be highly cohesive, meaning that connections between individual members of the network be proximal, and achieve high statistical significance; and iii) that we have high confidence that members of the network are expressed as proteins (not just as transcripts), so they can be detected as biomarkers and/or be targeted with drug therapy. To identify such a network, protein “seeds” identified by proteomics were used to search a database for co-expressed genes (COXPRESdb) 14. This search identified a highly cohesive network of 45 genes, mostly associated with inflammation. Based on the fact that the central nodes of the network are up-regulated at the level of protein, and that a representative set of additional nodes are up-regulated at the level of transcript, we conclude the network is up-regulated in CRC and that it might represent a viable therapeutic target.
MATERIAL AND METHODS
Animal Husbandry and Tissue Collection
All animal procedures were approved by the IACUC of the Sanford-Burnham Medical Research Institute. C57BL6/J-ApcMin/+ mice were purchased from The Jackson Laboratory and bred in the Institute’s animal facility. Genotyping was conducted using the protocol provided by The Jackson Laboratory. Mice were housed in conventional cages under a 12 h light/12 h dark cycle with free access to Teklad Global 18% Protein Rodent Diet and tap water. Mice were sacrificed at ages 17, 21 and 25 weeks, and the colons were collected and rinsed with phosphate-buffered saline (PBS), pH 7.4. We found ~60% of the mice contained at least one visible tumor in the colon. The colon was cut vertically to expose the inner epithelia and then rinsed with PBS to remove fecal matter. Tumors in the distal colon were excised under a dissecting microscope. Non-tumor colon tissue was excised from a site at least 0.5 cm from the collected tumor. The samples collected at 25 weeks were used for proteomic profiling; those collected at 17 weeks and 21 weeks were used for quantitative PCR validation of the reconstructed co-expression network. The average body weights of these mice are 20.5 ± 2.7 g for 17 weeks (n=4, female), 21.3 ± 3.1 g for 21 weeks (n=4, female) and 21.2 ± 3.6 g for 25 weeks (two female and one male).
Protein Extraction and MudPIT
Freshly collected colon tumors, or adjacent non-tumor tissue, were placed directly into 200 μL of 6 M guanidine-HCl (Sigma) held at 100ºC, and homogenized. After centrifugation, the supernatant was collected and proteins in the sample were reduced with TCEP, alkylated with iodoacetamide, and digested overnight with trypsin. Trifluoroacetic acid was then added to a final concentration of 0.1%. The resulting peptides were purified with a Zip-Tip C18 resin and then re-suspended in 5% formic acid. Finally, the protein concentration was measured with a Nanodrop spectrophotometer (Thermo Scientific).
An automated 2D NanoLC-LTQ system was used for this study. The system consists of an Eksigent Nano-2D LC, an autosampler, a switch valve, a strong cation exchange (SCX) loading column, a C18 trap column (Agilent), a capillary separation column, and a LTQ ion trap mass spectrometer (Thermo Electron). A fused-silica column (250 μm i.d.) with 3 cm of SCX resin (Whatman) and a microfilter frit (Upchurch) was used as a loading column. The loading column was connected between the exit of Channel 1 and the C18 trap column of the Eksigent Nano-2D system. The 15 cm capillary separation column (100 μm i.d.) packed with Magic C18 resin (Michrom Bioresources) was mounted on the Michrom ADVANCE electrospray source.
The sample (25 μg) of tryptic peptides was loaded by the autosampler onto a SCX column in buffer A (2% acetonitrile, 0.1% formic acid) at a flow rate of 10 μL/min. After sample loading and washing, seven sequential elution cycles were applied to elute proteins from the separation column. The flow rate was maintained at 500 nL/min. The first elution cycle (45 min) was run with a linear gradient of 0 to 100% buffer B (98% acetonitrile, 0.1% formic acid). The following cycle was initiated by loading 5 μL of salt solution followed by a desalting wash, and then a 90 min linear gradient of 0 to 100% buffer B. In each successive cycle, the isocratic salt concentration increased from 10% to 100% buffer C (500 mM ammonium acetate, 2% acetonitrile, 0.1% formic acid). Peptides from each cycle were directly eluted into the LTQ, which was controlled by Xcalibur software. Tandem mass spectrometry (MS/MS) spectra were collected during the LC/MS runs. Each scan was set to acquire a full MS scan, followed by MS/MS scans on the four most intense ions from the preceding MS scan. Relative collision energy for collision-induced dissociation was set at 35%. Three replicate runs were performed for each sample, and all peptides identified in separate runs were pooled into one list for further analysis.
Protein Identification and Quantification
A database of semi-tryptic peptides, generated from the NCBI mouse database, was imported into SorcererTM 2 (SageN Research). The MS/MS spectra obtained from MudPIT were automatically extracted and searched against the database using Sorcerer SEQUEST. For the search, 57 Da was added to all cysteines to account for carboxyamidomethylation. An additional 42 Da was permitted on N-terminal residues to account for potential acetylation, and 16 Da was permitted on methionines to account for potential oxidation.
The results from the SEQUEST searches were automatically filtered, organized, and displayed by PeptideProphet and ProteinProphet (Institute for Systems Biology). ProteinProphet computed a probability score from 0 to 1 for each protein, based on peptides assigned to MS/MS spectra and analyzed by PeptideProphet. To minimize false positive identifications, the following criteria were used: i) proteins were considered present only if two peptides from its sequence were observed, ii) the threshold of adjusted probability score for each peptide was set at ≥ 0.85, and iii) the threshold of probability score for protein identification was set at ≥ 0.85. The false positive rate using these criteria was below 1%, based on decoy database analysis.
To quantify the relative differences in protein expression we used label-free methods based on spectrum counts 15, 16. Because comparisons were made between multiple MS runs on replicate samples, the spectral counts from all runs were first normalized using the normalize.quantiles routine 17. This step minimized variations between MudPIT runs and stochastic differences in biological replicates. The normalized spectral counts were then imported into GeneSpring and statistically different protein levels were identified by t-tests.
Similarity of the Co-expression Network in Human Versus Mouse
In gene co-expression networks, each gene corresponds to a node. The neighbors of a node i are the nodes/genes that are connected to the node i. Two genes are connected by an edge with a weight indicating the distance (rank, ς). A gene co-expression network can be represented by a distance matrix D = [dij], where dij is the distance of a connection between two nodes i and j. All co-expression networks considered in this paper were represented in a form of connectivity graphs. Every co-expression network has been defined by Obayashi et al., 2008 14 so that every pair of connected gene-nodes is weighted by two asymmetric distances (ranks). Thus, every gene-node in the network can be characterized as an array of distance-pairs (vertex degree: {Vd}). To evaluate the conservation of the network between mouse and human, we investigated the topological overlap of the network using a mouse and human distance matrix (Dm ≠ Dh) at a given vertex degree (Vd=10). The distance matrix was compiled for nine orthologous central nodes (Ωij=9) and their 32 non-redundant secondary nodes as described under Results. From all overlapping pairs of nodes {Vmax}, defined here as mouse-human connectivity, only 10 pairs {Vd} that include the MS-identified gene products were assembled into the distance matrix (Dh,m). We formed a statistical null-hypothesis that postulates: the mouse and human networks (Nm ~ Nh) are similar, if the difference of distance (rank, dς) among orthologous nodes (dς[Ω9]) each represented by vertices (dς[Ω9] ∀ [V10]) in a distance-matrix (Dm=dς[Ω9]∀[V10] → Dh=dς[Ω,9]∀[V,10]) are similar. An analysis was performed to test networks of different size, and with different vertex degrees (Vd=10) using Friedman 18 and Wilcoxon 19 tests for the paired observations.
RNA Purification and Quantitative RT-PCR (qPCR)
RNA purification and RT-PCR were performed as described previously 20. Briefly, RNA was isolated from colon tissues with TRIZOL reagent (Invitrogen, CA) and the RNeasy Mini Kit (Qiagen, CA). cDNA was synthesized using SuperScript First-Strand Synthesis System (Invitrogen, CA). qPCR was carried out on a Mx 3000P Real-Time PCR System (Stratagene, CA) using SYBR Green PCR Master Mix (Applied Biosystems, CA), and diluted cDNA as template. The primers were designed using the Primer-Blast program at the NCBI website, and the primer sequences are listed in Supplemental Table 1. Values obtained from qPCR were normalized to the expression of GAPDH. Differences in RNA expression between colon tumor and matched adjacent non-tumor colon tissue were expressed as fold change for cancer versus normal tissue.
RESULTS
Proteins Identified in Tumor and Non-tumor Tissue in the Colon of ApcMin/+ Mice
Proteins expressed in tumor and adjacent non-tumor tissue of colons of three ApcMin/+ mice were identified by MudPIT. More than 1000 proteins were identified in each tissue sample (Table 1, Supplemental Figure 1, Supplemental Table 2), and greater than 2,400 proteins were identified in the entire analysis. Of these, 993 proteins were expressed in tumors from all mice, and 865 proteins were expressed in the non-tumor tissue from all mice. The global functions of these two sets of protein were assessed with the DAVID Bioinformatics Resources from NCBI (http://david.abcc.ncifcrf.gov) (Supplemental Fig. 2), but no major distinctions in the global functions of the two sets of proteins were evident.
Table 1.
Number of proteins identified by MudPIT.
| Tumor | Non-tumor | Common in T/N pair | |
|---|---|---|---|
| Mouse # 1 | 1802 | 1496 | 1275 |
| Mouse # 2 | 1329 | 1237 | 1030 |
| Mouse # 3 | 1546 | 1411 | 1251 |
| Total | 2220 | 1970 | |
| Common in 3 mice | 993 | 865 |
Proteins Differentially Expressed in Tumor Versus Non-tumor Tissue
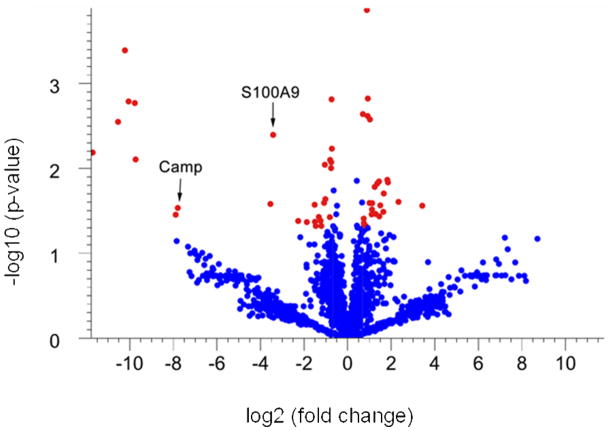
Proteins that are differentially expressed in tumor versus adjacent non-tumor tissue were identified based on statistical analyses of spectral counts. Spectral counts were first normalized with a statistically based quantile normalization method 17, 21, 22, and then proteins with significant differences in normalized expression were identified with a Welch t-test (parametric test, variances not assumed equal). The results of these analyses are shown in a “Volcano Plot” where statistical significance of each protein is plotted as a function of the difference in spectral count between samples (Fig. 1). To narrow our analysis to the proteins of highest significance we used a 1.6-fold change in expression, and a p-value of 0.05 as our thresholds. Using these criteria 52 proteins were differentially expressed: 27 were upregulated in tumors and 25 were downregulated. All of the 52 differentially expressed proteins that were identified from the Welch t-test also fit a beta-binomial distribution by maximum likelihood estimation 23 (Table 2 and Table 3).
Figure 1. Volcano plot of statistical analysis of identified proteins in non-tumor colon tissue versus colon tumor.
To identify differentially expressed proteins in colon tumors, the normalized spectrum counts of each protein were analyzed by Welch t-test using GeneSpring. Proteins that exhibit at least a 1.6-fold up (left) or down (right) expression in the tumor tissue compared to non-tumor tissue (spots that lie outside of the green lines) and that have a significant p-value (<0.05) (shown in red) are considered to be proteins that are different between tumor and non-tumor tissue.
Table 2.
Proteins over-expressed in colon tumors vs. non-tumor tissues.
| Protein ID | Symbol | Ave spec count in Non-tumor colon | Ave spec count in colon tumor | T/N ratio | p-value | Description |
|---|---|---|---|---|---|---|
| Antibiotic peptides and immune response | ||||||
| NP_032720 | Ngp | 0.00 | 11.67 | unique | 0.001 | neutrophilic granule protein |
| NP_032548 | Ltf | 0.00 | 17.56 | unique | 0.002 | lactotransferrin |
| NP_038678 | S100a8 | 0.00 | 46.97 | unique | 0.006 | S100 calcium binding protein A8 |
| NP_034022 | Chi3l3 | 0.00 | 12.22 | unique | 0.007 | Chitinase 3-like 3 |
| NP_034051 | Camp | 0.00 | 4.11 | unique | 0.029 | cathelicidin antimicrobial peptide |
| NP_034714 | Itln1 | 0.00 | 5.56 | unique | 0.035 | Intelectin 1 |
| NP_059068 | Lyz2 | 0.00 | 12.22 | unique | 0.000 | Lysozyme 2 |
| NP_598623 | Fgg | 2.58 | 30.78 | 11.91 | 0.026 | fibrinogen, gamma polypeptide |
| NP_033140 | S100a9 | 7.50 | 77.14 | 10.29 | 0.004 | S100 calcium binding protein A9 |
| NP_034860 | Anxa1 | 98.28 | 167.33 | 1.70 | 0.010 | annexin A1 |
| Cell proliferation, differentiation and death | ||||||
| NP_031492 | Api5 | 0.00 | 9.50 | unique | 0.001 | Apoptosis inhibitor 5 |
| NP_080390 | Eif2s1 | 4.78 | 15.92 | 3.33 | 0.043 | EIF2e subunit 1 alpha |
| NP_001074263 | G3bp2 | 2.06 | 5.67 | 2.76 | 0.026 | Ras-GTPase-activating protein (GAP120) SH3- |
| NP_032423 | Itga6 | 7.22 | 15.94 | 2.21 | 0.047 | integrin alpha 6 |
| NP_035010 | Ncl | 87.75 | 181.97 | 2.07 | 0.009 | Nucleolin |
| NP_067305 | Acp1 | 7.61 | 15.78 | 2.07 | 0.025 | acid phosphatase 1, soluble |
| NP_659207 | Eif4a1 | 58.14 | 98.22 | 1.69 | 0.008 | eukaryotic translation initiation factor 4A1 |
| Actin and cell migration | ||||||
| NP_663604 | Pls3 | 4.78 | 20.33 | 4.26 | 0.041 | T-plastin |
| NP_780660 | CKAP4 | 15.28 | 36.67 | 2.40 | 0.037 | cytoskeleton-associated protein 4 |
| NP_031713 | Cfl1 | 154.89 | 269.19 | 1.74 | 0.037 | cofilin 1, non-muscle |
| NP_032905 | Lcp1 | 39.72 | 65.39 | 1.65 | 0.005 | L-plastin |
| RNA and microRNA metabolism | ||||||
| NP_081219 | Snrpd2 | 6.72 | 17.67 | 2.63 | 0.047 | small nuclear ribonucleoprotein D2 |
| NP_062750 | Snd1 | 23.00 | 53.28 | 2.32 | 0.041 | staphylococcal nuclease domain containing 1 |
| XP_001479766 | LOC1000 | 21.28 | 36.94 | 1.74 | 0.008 | PREDICTED: similar to ribosomal protein L10a |
| NP_032800 | Pabpc1 | 58.58 | 97.50 | 1.66 | 0.001 | poly A binding protein, cytoplasmic 1 |
| Lysosome | ||||||
| NP_031534 | Atp6v1a | 2.56 | 6.78 | 2.65 | 0.042 | ATPase, H+ transporting, lysosomal V1 subunit |
| DNA assembly and gene expression | ||||||
| NP_031650 | Cbx3 | 17.61 | 35.17 | 2.00 | 0.023 | chromobox homolog 3 |
Table 3.
Proteins reduced in colon tumor vs. non-tumor tissue.
| Protein ID | Symbol | Ave spec | Ave spec | T/N | p-value | Description |
|---|---|---|---|---|---|---|
| Actinomyosin complex and cell-matrix interaction | ||||||
| NP_001074776 | Ppp1r12b | 12.78 | 3.44 | 0.27 | 0.0146 | protein phosphatase 1 regulatory subunit |
| NP_109617 | Sacm1l | 8.44 | 2.33 | 0.28 | 0.0137 | SAC1 (suppressor of actin mutations 1, |
| XP_619639 | Tns1 | 38.06 | 11.67 | 0.31 | 0.0198 | PREDICTED: tensin 1 |
| NP_964001 | Synm | 179.94 | 80.89 | 0.45 | 0.0258 | desmuslin isoform H and H |
| NP_034052 | Cnn1 | 777.56 | 381.33 | 0.49 | 0.0027 | calponin 1 |
| NP_647461 | Mylk | 230.11 | 121.00 | 0.53 | 0.0015 | myosin, light polypeptide kinase |
| XP_485171 | Myl9 | 223.92 | 127.78 | 0.57 | 0.0478 | PREDICTED: similar to Myosin, light |
| NP_663550 | Cald1 | 137.19 | 81.92 | 0.60 | 0.0476 | caldesmon 1 |
| NP_075891 | Mylc2b | 269.72 | 166.28 | 0.62 | 0.0023 | myosin light chain, regulatory B |
| NP_001030136 | Sorbs1 | 76.28 | 36.08 | 0.47 | 0.0305 | sorbin and SH3 domain containing 1 |
| NP_604443 | Dst | 39.53 | 14.25 | 0.36 | 0.0143 | dystonin isoforms a and b |
| NP_062733 | Dpt | 11.28 | 4.11 | 0.36 | 0.0274 | dermatopontin |
| Oxidative stress response, metabolism and detoxification | ||||||
| NP_598755 | Gstt3 | 9.61 | 3.67 | 0.38 | 0.0151 | glutathione S-transferase, theta 3 |
| NP_031646 | Cbr1 | 119.03 | 71.97 | 0.60 | 0.0441 | carbonyl reductase 1 |
| NP_964004 | Ugt1a7c | 31.53 | 3.17 | 0.10 | 0.0276 | UDP glucuronosyltransferase 1 family, |
| NP_080979 | Ndufa8 | 15.53 | 6.44 | 0.42 | 0.0165 | NADH dehydrogenase (ubiquinone) 1 |
| NP_033463 | Tst | 25.78 | 12.67 | 0.49 | 0.0256 | thiosulfate sulfotransferase, |
| NP_084501 | Dlst | 23.33 | 12.67 | 0.54 | 0.0001 | dihydrolipoamide S-succinyltransferase |
| NP_663533 | Hadhb | 66.14 | 40.06 | 0.61 | 0.0390 | mitochondrial trifunctional protein, beta |
| NP_031520 | Ass1 | 30.61 | 16.17 | 0.53 | 0.0024 | argininosuccinate synthetase |
| NP_084280 | Cryl1 | 22.89 | 8.72 | 0.38 | 0.0369 | crystallin, lambda 1 |
| NP_083907 | Abhd14b | 12.89 | 6.11 | 0.47 | 0.0350 | abhydrolase domain containing 14b |
| Signal transduction and regulatory pathways | ||||||
| NP_062650 | Plcd1 | 6.00 | 2.56 | 0.43 | 0.0345 | phospholipase C, delta 1 |
| NP_033176 | Selenbp1 | 223.50 | 75.00 | 0.34 | 0.0326 | selenium binding protein 1 |
| NP_031720 | Chgb | 10.89 | 2.22 | 0.20 | 0.0248 | chromogranin B |
Computational Reconstruction of a Co-expression Network Linked to Colon Cancer
The 52 differentially expressed proteins were used to search for networks of genes linked by co-expression. Two primary criteria were used to identify such networks. First, they must be highly cohesive, a measure of the statistical significance of their connectivity based on gene co-expression. Second, the network must have similar features in human and mouse because we reason that similarity across species would provide another measure of the significance of the network, and that it could guide the use of the ApcMin/+ mouse as a model of the human disease.
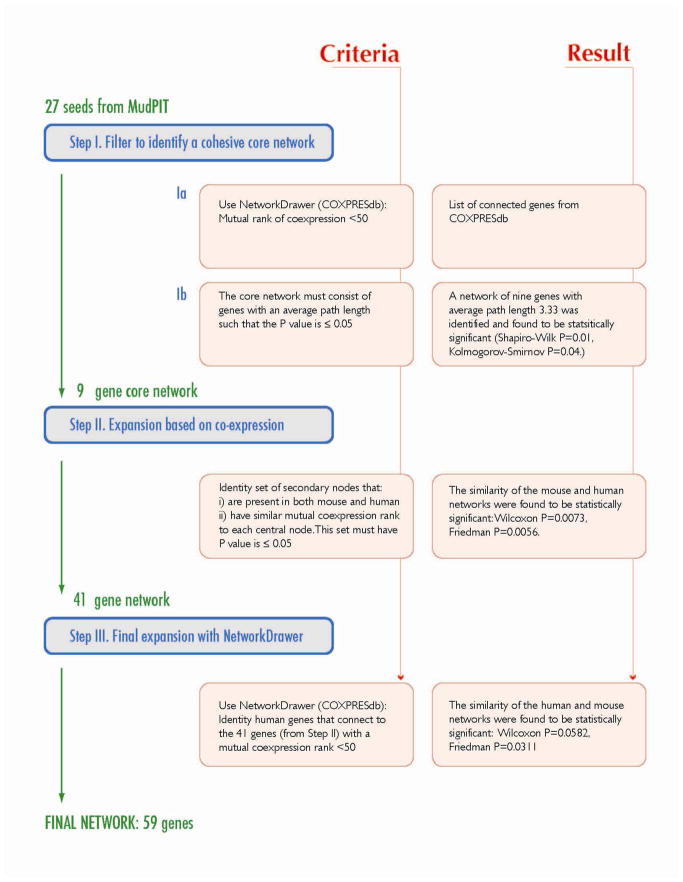
A straightforward three-step process was used to identify co-expression networks (Fig. 2). In Step I, we used the proteins identified from MudPIT as “seeds” to identify a core co-expression network. This was accomplished by submitting the gene IDs of all upregulated and downregulated proteins to the NetworkDrawer tool (within COXPRESdb; http://coxpresdb.hgc.jp/top_tool.shtml). This tool returns the set of seeds that are also linked by gene co-expression calculated for human and mouse genes across 63 different tissues including colon 14. In our case, 9 of the 27 upregulated proteins were found to be part of a statistically significant co-expression network, but none of the downregulated proteins were linked by co-expression.
Figure 2. Schema of Network Reconstruction.
The three steps used to generate CRC-INF-45 are outlined.
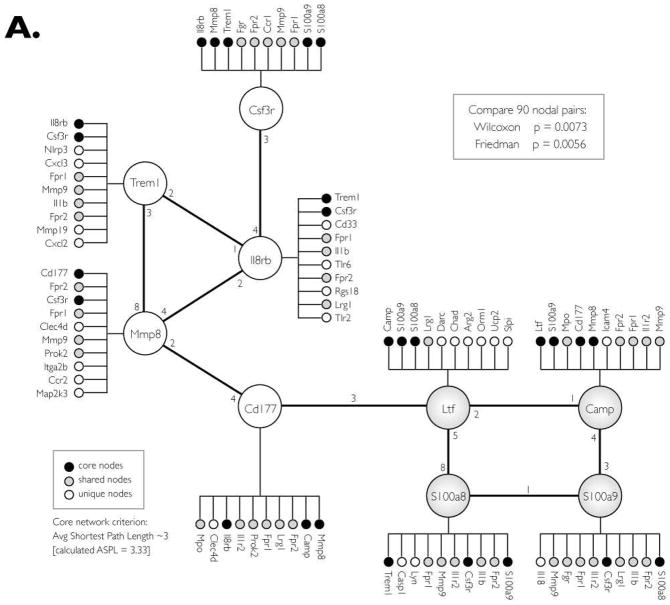
In Step II, the nine-gene network of up-regulated genes/proteins was expanded by searching for additional co-expressed genes. This was accomplished by assembling a list (from COXPRESdb) of the top 10 co-expressed genes for each of the 9 core nodes (Fig. 3A). The list was manually filtered to remove genes that are absent in either the mouse or human, and genes for which there is no co-expression data. The list was consolidated by removing redundant secondary nodes; genes that are co-expressed with more than one of the nine core nodes. This expansion yielded a network of 41 co-expressed genes whose co-expression patterns are statistically similar in human and mouse (Wilcoxon P=0.0073, Friedman P=0.0056).
Figure 3. Co-expression CRC-INF-45 network in colon tumors of ApcMin/+ mice.
(A) Proteins that are upregulated in tumor tissue were used to search COXPRESSdb for co-regulated genes within an average path length (rank) of three. In addition to the four nodes identified by MudPIT (large shaded nodes; Ltf, Camp, S100a8 and S100a9), this search yielded five additional core genes (open nodes; Cd177, Mmp8, Trem1, IL8rb and Csf3r). This nine-member core network was subsequently expanded to 41 genes (small circles) by identifying the top ten co-regulated genes for each core gene that is also co-expressed in humans. Black small circles indicate genes that are members of the core network, shaded small circles show genes that are shared among the core genes, and white circles are unique to the specific core member.
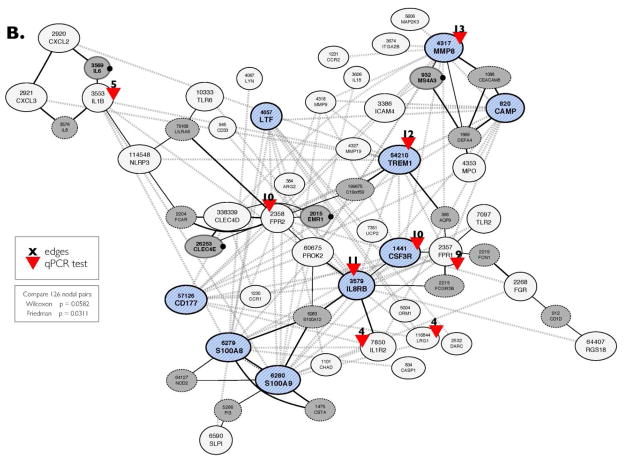
(B) Topology of the co-expression CRC-INF-45 network. To view the topology of connectivity, the 41 genes co-expressed in the mouse and human were submitted to NetworkDrawer tool from COXPRESSdb. The nine core genes are blue hatched nodes. All the other genes in the 41 member network are shown as white nodes. The automated NetworkDrawer tool included 18 additional linked genes (grey nodes). The four genes that contribute to the conservation of the network have a black dot to the right of the node. Nine representative genes with high connectivity were selected for quantification by quantitative PCR (red arrowheads); the number of nodes (edges) that each of these genes connect, based on co-expression data, is shown in black. The dotted grey lines represent co-expression linkages; the solid black lines represent protein-protein interactions identified using NetworkDrawer.
In Step III, to visualize the topology of the network, and to allow for limited automated expansion, we submitted the gene IDs of the 41 member network to the NetworkDrawer tool, which automatically displays the connectivity between nodes in the network (Fig. 3B). However, NetworkDrawer also has an automated feature that adds other nodes based on data from human genes and tissues that fit criteria as defined by the algorithm embedded in the tool. Consequently, 18 additional human genes were added to yield a 59 member human network. We present the entire 59 member human network for the sake of completeness (Fig. 3B), but for statistical comparisons 11 of the automatically added nodes were excluded because they lack counterparts in mice or because there is no data available in COXPRESdb. Three other nodes (DEFA4, CD1D, and APQ9) were excluded because their inclusion interfered with statistical similarity. Therefore, only four of the nodes (Fig. 3B, grey nodes with bold names and black dot on the right) that were added automatically by the NetworkDrawer tool, could be preserved in the network while still maintaining statistical similarity with mouse (Friedman: p = 0.0311, Wilcoxon: p = 0.0582). Thus, the co-expression network linked to colorectal cancer has 45 members.
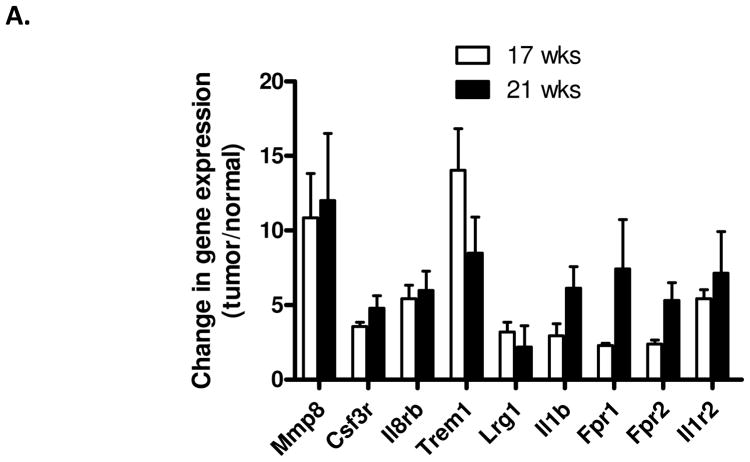
Quantitative RT-PCR was used to verify that members of the co-expression network are upregulated in colon tumors compared to adjacent non-tumor tissue. Nine representative genes were selected because their connectivity to other genes in the network ranged from low (connected to less than five other nodes) to high (connected to more than 10 other nodes). All nine representative genes were found to be upregulated in tumors compared to adjacent tissue, and some of these were upregulated by more than five-fold (Fig. 4). In addition, similar patterns of expression of the nine genes were observed between mice that were 17 weeks of age and 21 weeks of age, showing that the co-expression network is not simply a result of the onset of morbidity.
Figure 4. Members of the co-expression CRC-INF-45 network are upregulated in colon tumors compared to adjacent non-tumor tissue.
Tissue RNA was isolated from four sets of matched colon tumor and adjacent colon tissue of 17 and 21 week old ApcMin/+ mice and used as template in qPCR assays as described in Materials and Methods section. Specific primer sets (Supplemental Table 1) were used to quantify mRNA encoding Mmp8, Csf3r, Il8rb, Trem1, Lrg1, Il1b, Fpr1, Fpr2, and Il1r2; red arrowheads in Figure 3B. The qPCR was performed in duplicate, and values were normalized to expression of GAPDH prior to calculating the difference in the level of expression in tumor vs. normal tissue. All results are presented as mean ± SEM.
The Co-expression Network is Linked to Inflammation
To determine if the network is similar to any known cellular pathways, we tested its overlap with an extensive list of pathway ontologies listed on the PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System (www.pantherdb.org/). Based on the binomial distribution function 24 (p<0.001), the network is similar to three known pathways: 1) the toll-like receptor (TLR) signaling pathway (p<0.00005), the interleukin (IL) signaling pathway (p<0.002), and 3) the inflammation pathway, which showed the strongest positive coordinated association with the gene network (p= 0.00000009; 9 e-8). Thus, this network is named CRC-INF-45.
DISCUSSION
The primary outcomes of this study are: 1) the identification of 27 proteins with elevated, and 25 protein with reduced expression in colon tumors compared to adjacent non-tumor tissue of the ApcMin/+ mice; 2) the development of a straightforward method for reconstructing co-expression networks using proteins identified by proteomics as seeds to search for co-expressed genes; 3) the identification of a network of 45 gene/proteins that are linked by co-expression, associated with inflammation, and that are up-regulated in colon tumors of the ApcMin/+ mice. Together these observations provide the new molecular targets for follow-up research on progression of colon tumors.
Some of the proteins we found to be over-expressed in colon tumors were previously linked to other types of cancer. One of these is the nuclear protein apoptosis inhibitor 5 (Api5; also called AAC-11). It is upregulated in B cell leukemia 25, in non-small cell lung cancer 26, and in cervical cancer 27, but this study is the first evidence that it could be linked to CRC. Api5 is apparently necessary for E2F-induced apoptosis 28, probably because it binds to Acinus and prevents Acinus mediated DNA fragmentation 29. The fact that knock down of Api5 sensitizes tumor cells to chemotherapeutic drugs29 is also consistent with this mechanism of action. Api5 expression has also been suggested to be a prognostic marker. Taken together, these findings indicate that Api5 is likely to be reasonable therapeutic target for colorectal cancer.
Ras-GTPase activating protein-SH3-domain-binding proteins 2 (G3bp2) is another protein up-regulated in tumor tissue. This protein interacts with IκBα and IκBα/NFκB complexes, and has been shown to be over-expressed in human breast cancer tissues 30. As far as we are aware, over expression of G3bp2 has not previously been described in colorectal cancer, and it is possible that its increased expression in tumors may indicate modulation of NFκB activity. There is also evidence that Api5 is regulated by NFκB 31, so it is conceivable that up-regulation of G3bp2 is functionally linked to the observed up-regulation of Api5.
Interestingly no proteins were found to be absent in tumors compared to non-tumor tissue. However, a single protein, UDP glucuronosyltransferase 1A7, is expressed at levels 10-fold lower in tumor compared to non-tumor tissue. This protein acts as a detoxifying enzyme by adding glucuronidate to polycyclic aromatic hydrocarbons and heterocyclic amines, both of which are known environmental carcinogens that can give rise to CRC. In fact, polymorphisms in the gene encoding UDP glucuronosyltransferase 1A7 are risk factors to CRC, and these polymorphisms give rise to truncated form of the enzyme 32. In conjunction with these studies, the fact that we observe UDP glucuronosyltransferase 1A7 to be substantially lower in tumors suggests that its down-regulation may contribute to tumor onset in the ApcMin/+ mice.
In an effort to expand the scope of our proteomic study we established a strategy to identify gene co-expression networks that encompass some of the differentially expressed proteins identified by proteomics. The strategy is straightforward in approach, and simply uses all proteins that are identified as differentially expressed to search the COXPRESSdb to identify the proteins that are linked together through a gene co-expression network. With this strategy we identified a single cohesive co-expression network (called CRC-INF-45) that encompasses nine of the 27 proteins up-regulated in tumors of the ApcMin/+ mice. Inspection of the genes within this network shows them to be mostly involved in inflammatory response. In fact, four of the genes within CRC-INF-45 are also members of a network of interacting genes involved in the systemic inflammatory response to bacterial endotoxin 33. Since mechanistic links between inflammation and cancer are now becoming widely accepted 34–36, inflammatory networks are of increasing interest, especially in CRC where clinical experience has shown that patients with inflammatory bowel disease (IBD), particularly ulcerative colitis (UC), have increased risk of developing CRC (see review 34–36). Several interleukins and their receptors that have been previously associated with CRC 35, are also prominent in CRC-INF-45. These include IL-1B, a gene with polymorphisms associated with colon cancer recurrence 37 and IL-6, which is high in the serum of patients with CRC and is associated with metastasis and decreased survival 38. IL-8 is also encompassed within CRC-INF-45, and its mRNA levels are increased in precancerous adenomas, and rise even further in later stages of CRC 39. However, interleukins and their receptors are probably not optimal therapeutic targets because they can either stimulate or inhibit tumor growth and progression 35. For example, IL-17 promotes angiogenesis and tumor growth 40, but can also inhibit tumor growth through T-cell activation 41.
More viable targets within CRC-INF-45 include the triggering receptor expressed on myeloid cells-1 (TREM1), which is expressed approximately ten-fold higher in tumor than in normal adjacent tissue (Table 2). TREM1 is a transmembrane receptor expressed on monocytes and neutrophils, and it contains a single extracellular Ig-like ligand binding domain a transmembrane region, and a cytoplasmic tail 42. While the ligand for TREM-1 is not yet known, occupancy of this receptor induces signaling through the transmembrane adapter protein DNAX activation protein 12 (DAP12). The end-result is amplification in secretion of pro-inflammatory cytokines 43. TREM-1 is already receiving attention as a drug target because inhibition of its function attenuates inflammation in inflammatory bowel disease 44.
The topology of CRC-INF-45 also provides support for the Toll-like receptors (TLRs) as potential therapeutic targets. TLRs are pattern recognition receptors that play critical roles in initiating both innate and adaptive immune responses against microbial pathogens. In the gut, they also help maintaining intestinal homeostasis 45. As with many immune receptors, ligand binding to TLRs initiates a signaling cascade that activates NF-κB, a pro-inflammatory transcription factor, and numerous studies have suggested TLRs promote tumorigenesis through pro-inflammatory effects. Consistent with this view, TLRs are over-expressed in many tumor types, and in one small study with CRC patients TLR2 mRNA levels correlated with advanced CRC 46. In addition, a microsatellite polymorphism in TLR2 has been associated with colorectal cancer 47. Although not as extensively studied, TLR6 has been shown to be over-expressed in mucosa-associated lymphoid tissue (MALT) lymphoma 48, which is associated with Helicobacter pylori infection 49. These published studies, in conjunction with the fact that both TLR2 and TLR6 are members of CRC-INF-45, suggest that antagonists of TLR2 and/or TLR6 may have anti-tumor effects in CRC.
One central node of CRC-INF-45 that has not previously been associated with CRC is the anti-microbial peptide cathelicidin (CAMP; also known as LL-37). Cathelicidin is over-expressed in lung 50, breast 51, prostate 52 and ovarian 53 cancers; it appears to have pro-invasive effects 54, to suppress apoptosis 55 and to be mitogenic 56. At least some of the pro-tumorigenic effects of cathelicidin probably arise from its ability to stimulate recruitment of mesenchymal stem cells to tumors 25. Our finding that cathelicidin is a central node of CRC-INF-45, in addition to the findings by others, suggest that cathelicidin has an importance role in tumorigenesis and illustrate its potential as a therapeutic target in CRC.
Proteomic analysis requires a substantial amount of starting material, so to obtain colon tumors of sufficient size, we allowed the mice to live to 25 weeks of age. At this time point the animals have a considerable tumor burden in the small intestine, so it conceivable that general morbidity could potentially contribute to expression of CRC-INF-45. However, the study was performed by comparing tumor tissue to non-tumor adjacent tissue, so any systemic illness or morbidity should be eliminated from the MudPIT analysis. The PCR studies that confirm the expression of CRC-INF-45 were performed on animals at 17 weeks of age, which also have a high tumor burden, but these comparisons also involved the comparison of tumor to adjacent non-tumor tissue. So, while we cannot exclude the possibility that systemic illness, as a result of tumor burden, contributes to expression of CRC-INF-45, the simplest interpretation of our findings is that this network is up-regulated because of the formation of a microenvironment of inflammatory cells. Future work will dissect the time frame in which CRC-INF-45 becomes apparent.
The present study is not the first to use proteomics to search for differentially expressed proteins in ApcMin/+ mice, but there are substantial differences between the prior studies and the comparison reported here. With the intent of identifying potential biomarkers for CRC, Caprioli’s group used laser capture microdissection and MS to identify proteins that are over-expressed in the top vs. the bottom of crypts from adenomas from ApcMin/+ mice 10. In another study, Huttlin et al compared the proteome of colon tumors of ApcMin/+ mice to that of normal tissue of wild-type mice 11. While we identified nine of the ten proteins that were observed to be differentially regulated by Huttlin et al in at least one mouse pair in our study (Supplemental Table 2), none of these reached our threshold for significance.
Other investigators have also proposed a “seeding” strategy for identifying larger networks based on proteins identified from proteomics 57. However, an important distinction between the networks reconstructed by Nibbe et al, and that reported here is that Nibbe et al reconstructed a protein-protein interaction network, as opposed to a co-expression network of genes, as is reported here. That protein-protein interaction network is largely based on protein-protein interactions documented in the Human Protein Reference Database (HPRD), all of which are based on information in the literature which is manually extracted, interpreted and curated. As stated by the developer of HPRD, there is no established gold standard for determining the existence of a protein-protein interaction, so it can be difficult to evaluate the significance of such networks. Given the possible uncertainty of that strategy, we elected to limit our study to reconstruction of a co-expression network that is not biased by human interpretation, because the transcriptomic data used for the search is obtained from transcriptome profiling data in NCBI GEO 14.
In conclusion, we have used a proteomics-driven approach to reconstruct a co-expression network implicated in CRC: the CRC-INF-45 network. This network provides a comprehensive view, which may lead to the direct identification of genes underlying CRC, and provides a bridge to link individual genes to the complex traits of CRC. Importantly, we noted that up-regulated genes are associated with inflammation, which is known to play a significant role in cancer. Although more research is required to detail the interactions between the immune and gastrointestinal systems at the molecular level, this study has revealed a unique inflammatory network involved in CRC, and provides critical information for development of therapeutic strategies against colorectal cancer.
Supplementary Material
Acknowledgments
The authors thank Dr. Roy Williams and Dr. Yoshinobu Igarashi for their help with data analysis. This work was supported by NIH grant # RR020843.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hedge P, McKechnie D, et al. Identification of FAP locus genes from chromosome 5q21. Science. 1991;253:661–5. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 4.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 5.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 6.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–99. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 7.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–4. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 8.Leclerc D, Deng L, Trasler J, Rozen R. ApcMin/+ mouse model of colon cancer: gene expression profiling in tumors. J Cell Biochem. 2004;93:1242–54. doi: 10.1002/jcb.20236. [DOI] [PubMed] [Google Scholar]
- 9.Paoni NF, Feldman MW, Gutierrez LS, Ploplis VA, Castellino FJ. Transcriptional profiling of the transition from normal intestinal epithelia to adenomas and carcinomas in the APCMin/+ mouse. Physiol Genomics. 2003;15:228–35. doi: 10.1152/physiolgenomics.00078.2003. [DOI] [PubMed] [Google Scholar]
- 10.Xu BJ, Li J, Beauchamp RD, Shyr Y, Li M, Washington MK, Yeatman TJ, Whitehead RH, Coffey RJ, Caprioli RM. Identification of early intestinal neoplasia protein biomarkers using laser capture microdissection and MALDI MS. Mol Cell Proteomics. 2009;8:936–45. doi: 10.1074/mcp.M800345-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huttlin EL, Chen X, Barrett-Wilt GA, Hegeman AD, Halberg RB, Harms AC, Newton MA, Dove WF, Sussman MR. Discovery and validation of colonic tumor-associated proteins via metabolic labeling and stable isotopic dilution. Proc Natl Acad Sci U S A. 2009;106:17235–40. doi: 10.1073/pnas.0909282106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ang CS, Rothacker J, Patsiouras H, Burgess AW, Nice EC. Murine fecal proteomics: A model system for the detection of potential biomarkers for colorectal cancer. J Chromatogr A. 2009 doi: 10.1016/j.chroma.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–13. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 14.Obayashi T, Hayashi S, Shibaoka M, Saeki M, Ohta H, Kinoshita K. COXPRESdb: a database of coexpressed gene networks in mammals. Nucleic Acids Res. 2008;36:D77–82. doi: 10.1093/nar/gkm840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR., 3rd Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–3. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 17.Huang SK, Darfler MM, Nicholl MB, You J, Bemis KG, Tegeler TJ, Wang M, Wery JP, Chong KK, Nguyen L, Scolyer RA, Hoon DS. LC/MS-based quantitative proteomic analysis of paraffin-embedded archival melanomas reveals potential proteomic biomarkers associated with metastasis. PLoS ONE. 2009;4:e4430. doi: 10.1371/journal.pone.0004430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman M. The use of ranks to avoid the assumption of normality implicit in the analysis of variance. J Am Stat Assoc. 1937;32:6750701. [Google Scholar]
- 19.Siegel S. Non-parametric statistics for the behavioral sciences. McGraw-Hill, Ed; New York: 1956. pp. 75–83. [Google Scholar]
- 20.Fang C, Dean J, Smith JW. A novel variant of ileal bile acid binding protein is up-regulated through nuclear factor-kappaB activation in colorectal adenocarcinoma. Cancer Res. 2007;67:9039–46. doi: 10.1158/0008-5472.CAN-06-3690. [DOI] [PubMed] [Google Scholar]
- 21.Callister SJ, Barry RC, Adkins JN, Johnson ET, Qian WJ, Webb-Robertson BJ, Smith RD, Lipton MS. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J Proteome Res. 2006;5:277–86. doi: 10.1021/pr050300l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgs RE, Knierman MD, Gelfanova V, Butler JP, Hale JE. Comprehensive label-free method for the relative quantification of proteins from biological samples. J Proteome Res. 2005;4:1442–50. doi: 10.1021/pr050109b. [DOI] [PubMed] [Google Scholar]
- 23.Pham TV, Piersma SR, Warmoes M, Jimenez CR. On the beta-binomial model for analysis of spectral count data in label-free tandem mass spectrometry-based proteomics. Bioinformatics. 26:363–9. doi: 10.1093/bioinformatics/btp677. [DOI] [PubMed] [Google Scholar]
- 24.Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–5. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 25.Coffelt SB, Marini FC, Watson K, Zwezdaryk KJ, Dembinski JL, LaMarca HL, Tomchuck SL, Honer zu Bentrup K, Danka ES, Henkle SL, Scandurro AB. The pro-inflammatory peptide LL-37 promotes ovarian tumor progression through recruitment of multipotent mesenchymal stromal cells. Proc Natl Acad Sci U S A. 2009;106:3806–11. doi: 10.1073/pnas.0900244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki H, Moriyama S, Yukiue H, Kobayashi Y, Nakashima Y, Kaji M, Fukai I, Kiriyama M, Yamakawa Y, Fujii Y. Expression of the antiapoptosis gene, AAC-11, as a prognosis marker in non-small cell lung cancer. Lung Cancer. 2001;34:53–7. doi: 10.1016/s0169-5002(01)00213-6. [DOI] [PubMed] [Google Scholar]
- 27.Kim JW, Cho HS, Kim JH, Hur SY, Kim TE, Lee JM, Kim IK, Namkoong SE. AAC-11 overexpression induces invasion and protects cervical cancer cells from apoptosis. Lab Invest. 2000;80:587–94. doi: 10.1038/labinvest.3780063. [DOI] [PubMed] [Google Scholar]
- 28.Morris EJ, Michaud WA, Ji JY, Moon NS, Rocco JW, Dyson NJ. Functional Identification of Api5 as a Suppressor of E2F-Dependent Apoptosis In Vivo. PLoS Genet. 2006;2:e196. doi: 10.1371/journal.pgen.0020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rigou P, Piddubnyak V, Faye A, Rain JC, Michel L, Calvo F, Poyet JL. The antiapoptotic protein AAC-11 interacts with and regulates Acinus-mediated DNA fragmentation. Embo J. 2009;28:1576–88. doi: 10.1038/emboj.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.French J, Stirling R, Walsh M, Kennedy HD. The expression of Ras-GTPase activating protein SH3 domain-binding proteins, G3BPs, in human breast cancers. Histochem J. 2002;34:223–31. doi: 10.1023/a:1021737413055. [DOI] [PubMed] [Google Scholar]
- 31.Ren K, Zhang W, Shi Y, Gong J. Pim-2 activates API-5 to inhibit the apoptosis of hepatocellular carcinoma cells through NF-kappaB pathway. Pathol Oncol Res. 2010;16:229–37. doi: 10.1007/s12253-009-9215-4. [DOI] [PubMed] [Google Scholar]
- 32.Strassburg CP, Vogel A, Kneip S, Tukey RH, Manns MP. Polymorphisms of the human UDP-glucuronosyltransferase (UGT) 1A7 gene in colorectal cancer. Gut. 2002;50:851–6. doi: 10.1136/gut.50.6.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen BS, Yang SK, Lan CY, Chuang YJ. A systems biology approach to construct the gene regulatory network of systemic inflammation via microarray and databases mining. BMC Med Genomics. 2008;1:46. doi: 10.1186/1755-8794-1-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clevers H. At the crossroads of inflammation and cancer. Cell. 2004;118:671–4. doi: 10.1016/j.cell.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Itzkowitz SH, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol. 2004;287:G7–17. doi: 10.1152/ajpgi.00079.2004. [DOI] [PubMed] [Google Scholar]
- 37.Lurje G, Hendifar AE, Schultheis AM, Pohl A, Husain H, Yang D, Manegold PC, Ning Y, Zhang W, Lenz HJ. Polymorphisms in interleukin 1 beta and interleukin 1 receptor antagonist associated with tumor recurrence in stage II colon cancer. Pharmacogenet Genomics. 2009;19:95–102. doi: 10.1097/FPC.0b013e32831a9ad1. [DOI] [PubMed] [Google Scholar]
- 38.Knupfer H, Preiss R. Serum interleukin-6 levels in colorectal cancer patients--a summary of published results. Int J Colorectal Dis. 25:135–40. doi: 10.1007/s00384-009-0818-8. [DOI] [PubMed] [Google Scholar]
- 39.Cui G, Yuan A, Goll R, Vonen B, Florholmen J. Dynamic changes of interleukin-8 network along the colorectal adenoma-carcinoma sequence. Cancer Immunol Immunother. 2009;58:1897–905. doi: 10.1007/s00262-009-0702-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Numasaki M, Fukushi J, Ono M, Narula SK, Zavodny PJ, Kudo T, Robbins PD, Tahara H, Lotze MT. Interleukin-17 promotes angiogenesis and tumor growth. Blood. 2003;101:2620–7. doi: 10.1182/blood-2002-05-1461. [DOI] [PubMed] [Google Scholar]
- 41.Benchetrit F, Ciree A, Vives V, Warnier G, Gey A, Sautes-Fridman C, Fossiez F, Haicheur N, Fridman WH, Tartour E. Interleukin-17 inhibits tumor cell growth by means of a T-cell-dependent mechanism. Blood. 2002;99:2114–21. doi: 10.1182/blood.v99.6.2114. [DOI] [PubMed] [Google Scholar]
- 42.Ford JW, McVicar DW. TREM and TREM-like receptors in inflammation and disease. Curr Opin Immunol. 2009;21:38–46. doi: 10.1016/j.coi.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouchon A, Dietrich J, Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 2000;164:4991–5. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- 44.Schenk M, Bouchon A, Seibold F, Mueller C. TREM-1--expressing intestinal macrophages crucially amplify chronic inflammation in experimental colitis and inflammatory bowel diseases. J Clin Invest. 2007;117:3097–106. doi: 10.1172/JCI30602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal micr, oflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Niedzielska I, Niedzielski Z, Tkacz M, Orawczyk T, Ziaja K, Starzewski J, Mazurek U, Markowski J. Toll-like receptors and the tendency of normal mucous membrane to transform to polyp or colorectal cancer. J Physiol Pharmacol. 2009;60(Suppl 1):65–71. [PubMed] [Google Scholar]
- 47.Boraska Jelavic T, Barisic M, Drmic Hofman I, Boraska V, Vrdoljak E, Peruzovic M, Hozo I, Puljiz Z, Terzic J. Microsatelite GT polymorphism in the toll-like receptor 2 is associated with colorectal cancer. Clin Genet. 2006;70:156–60. doi: 10.1111/j.1399-0004.2006.00651.x. [DOI] [PubMed] [Google Scholar]
- 48.Hamoudi RA, Appert A, Ye H, Ruskone-Fourmestraux A, Streubel B, Chott A, Raderer M, Gong L, Wlodarska I, De Wolf-Peeters C, MacLennan KA, de Leval L, Isaacson PG, Du MQ. Differential expression of NF-kappaB target genes in MALT lymphoma with and without chromosome translocation: insights into molecular mechanism. Leukemia. 24:1487–97. doi: 10.1038/leu.2010.118. [DOI] [PubMed] [Google Scholar]
- 49.Ye H, Liu H, Attygalle A, Wotherspoon AC, Nicholson AG, Charlotte F, Leblond V, Speight P, Goodlad J, Lavergne-Slove A, Martin-Subero JI, Siebert R, Dogan A, Isaacson PG, Du MQ. Variable frequencies of t(11;18)(q21;q21) in MALT lymphomas of different sites: significant association with CagA strains of H pylori in gastric MALT lymphoma. Blood. 2003;102:1012–8. doi: 10.1182/blood-2002-11-3502. [DOI] [PubMed] [Google Scholar]
- 50.von Haussen J, Koczulla R, Shaykhiev R, Herr C, Pinkenburg O, Reimer D, Wiewrodt R, Biesterfeld S, Aigner A, Czubayko F, Bals R. The host defence peptide LL-37/hCAP-18 is a growth factor for lung cancer cells. Lung Cancer. 2008;59:12–23. doi: 10.1016/j.lungcan.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 51.Weber G, Chamorro CI, Granath F, Liljegren A, Zreika S, Saidak Z, Sandstedt B, Rotstein S, Mentaverri R, Sanchez F, Pivarcsi A, Stahle M. Human antimicrobial protein hCAP18/LL-37 promotes a metastatic phenotype in breast cancer. Breast Cancer Res. 2009;11:R6. doi: 10.1186/bcr2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hensel JA, Chanda D, Kumar S, Sawant A, Grizzle WE, Siegal GP, Ponnazhagan S. LL-37 as a therapeutic target for late stage prostate cancer. Prostate. doi: 10.1002/pros.21282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coffelt SB, Waterman RS, Florez L, Honer zu Bentrup K, Zwezdaryk KJ, Tomchuck SL, LaMarca HL, Danka ES, Morris CA, Scandurro AB. Ovarian cancers overexpress the antimicrobial protein hCAP-18 and its derivative LL-37 increases ovarian cancer cell proliferation and invasion. Int J Cancer. 2008;122:1030–9. doi: 10.1002/ijc.23186. [DOI] [PubMed] [Google Scholar]
- 54.Coffelt SB, Tomchuck SL, Zwezdaryk KJ, Danka ES, Scandurro AB. Leucine leucine-37 uses formyl peptide receptor-like 1 to activate signal transduction pathways, stimulate oncogenic gene expression, and enhance the invasiveness of ovarian cancer cells. Mol Cancer Res. 2009;7:907–15. doi: 10.1158/1541-7786.MCR-08-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chamorro CI, Weber G, Gronberg A, Pivarcsi A, Stahle M. The human antimicrobial peptide LL-37 suppresses apoptosis in keratinocytes. J Invest Dermatol. 2009;129:937–44. doi: 10.1038/jid.2008.321. [DOI] [PubMed] [Google Scholar]
- 56.Hase K, Eckmann L, Leopard JD, Varki N, Kagnoff MF. Cell differentiation is a key determinant of cathelicidin LL-37/human cationic antimicrobial protein 18 expression by human colon epithelium. Infect Immun. 2002;70:953–63. doi: 10.1128/iai.70.2.953-963.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nibbe RK, Koyuturk M, Chance MR. An integrative -omics approach to identify functional sub-networks in human colorectal cancer. PLoS Comput Biol. 6:e1000639. doi: 10.1371/journal.pcbi.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.