Abstract
Activation-induced cytidine deaminase (AID) instigates mutations and DNA breaks in Ig genes undergoing somatic hypermutation (SHM) and class switch recombination (CSR) during B cell activation in response to immunization and infection. This review discusses how AID expression and activity are regulated, including recent discoveries of AID interacting proteins that might recruit AID to immunoglobulin (Ig) genes and also allow it to target both DNA strands. Also discussed is the accumulating evidence that AID binds to, mutates, and creates breaks at numerous non-Ig sites in the genome, initiating cell transformation and malignancies.
AID initiates class switch recombination (CSR) and somatic hypermutation (SHM) of immunoglobulin (Ig) variable region genes
AID was first identified by T. Honjo's group, using subtraction cDNA cloning, as a gene specifically expressed in a B cell line upon induction of CSR (1). Subsequent studies demonstrated that AID is essential for CSR and for SHM of antibody variable (V) region genes (2, 3). Both of these processes occur in B cells during an immune response, resulting in increased diversity of the antibody response, and greatly increasing the efficacy of the antibody response. Due to its similarity to Apobec-1 (see Glossary), AID was first hypothesized to be an RNA cytidine deaminase that edited a mRNA to allow it to encode proteins that would initiate either SHM or CSR. However, AID was subsequently demonstrated to be a DNA cytidine deaminase (4), and results by several other labs have supported this conclusion. AID initiates both SHM and CSR by converting deoxycytidines (dC) to deoxyuracils (dU), which are then processed by one of several mechanisms. DNA replication can cause a C:G transition mutation to T:A bp (4, 5). dU bases can also be excised by uracil DNA glycosylase (UNG), leaving abasic sites, which can be replicated over by error-prone translesion polymerases. AID thereby induces both transition and transversion mutations (see Glossary) into antibody V region genes during an immune response. B cells with V region mutations that result in increased binding affinity to antigen are selectively expanded during an immune response. Abasic sites can also be incised by the AP endonucleases (APE1 and APE2), resulting in single-strand DNA breaks (SSBs) or double stranded breaks (DSBs) if the abasic sites are sufficiently close on opposite strands (6, 7). The U:G mismatches produced by AID activity are also recognized and removed by the mismatch repair (MMR) pathway, which results in conversion of SSBs to DSBs during CSR, and introduction of mutations at A:T bp during SHM and CSR.
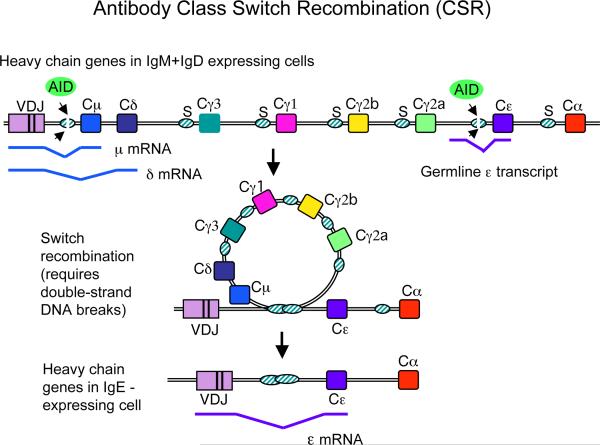
During CSR, DSBs are initiated by AID activity within special tandem repeat sequences termed switch (S) regions, located upstream of Ig heavy chain constant (C) region genes. CSR occurs by recombination of DSBs introduced into the donor Sμ region and acceptor Sx region by non-homologous (both classical and alternative) end joining (C-NHEJ and A-EJ) (8). Fig 1 illustrates CSR, and Fig 2 the role of MMR during DSB formation.
Fig 1. Diagram of Ig class switch recombination (CSR) to IgE.
Top, the mouse Ig H locus in B cells expressing IgM and IgD (by alternative RNA transcription/processing). During CSR, activation-induced cytidine deaminase (AID) deaminates dC residues in the top and bottom strands of transcriptionally active S regions (Sμ and Sε in the diagram shown), initiating a process that results in DSBs in both S regions, and leading to CSR by intrachromosomal deletion (middle). Bottom, the IgH locus after CSR to IgE. Splicing diagrams for the μ, δ, and ε mRNAs and for the ε germline transcript are indicated below the diagrams of the locus. Similar germline transcripts are induced from unrearranged Cγ and Cα genes, depending on the cytokine stimulation received by the B cell.
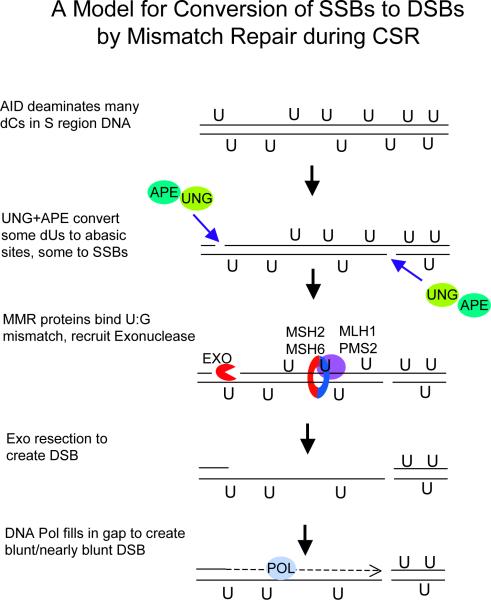
Figure 2. Model for conversion of SSBs to DSBs by mismatch repair during CSR.
AID is hypothesized to introduce several dU residues in S regions during one cell cycle. Some of the dU residues are excised by UNG, and some of the abasic sites are nicked by APE. The U:G mismatches that remain would be substrates for Msh2-Msh6 (90). Msh2-Msh6, along with Mlh1-Pms2, recruit Exo1 (and accessory proteins) to a nearby 5' nick, from where Exo1 begins to excise toward the mismatch (91, 92), creating a DSB with a 5' ss overhang, which can be filled in by DNA polymerase. Fill-in synthesis is probably performed by replication and low fidelity translesion polymerases, when the template strand has an abasic site. Alternatively, the 3' overhang is removed by a 5' flap endonuclease (Fen1) or by Exo1. If the nearest SSB is located 3' to the U:G mismatch bound by Msh2-Msh6, the endonuclease activity of Pms2 creates SSBs on the 5' side of the mismatch, allowing Exo1 to resect 5' to 3' (92) (not illustrated).
As AID is essential for an effective immune response, it has been extensively studied in the 12 years since its discovery, although there are still more questions than answers about its functions, mechanism of action, and regulation. Its expression and activity are highly regulated, and it interacts with numerous other proteins, some of which appear to target and regulate its activity. Nonetheless, in B cells AID has been shown to bind to, to mutate, and to induce DSBs in numerous other genes besides Ig genes, albeit less frequently. Consistent with this, AID promotes B cell leukemia, lymphoma, and myeloma (9, 10). Also, AID is expressed and active in several other types of cells, in which it sometimes promotes cancer (11–13). This review will discuss these issues, focusing on recent findings. Due to space constraints, some important current issues are not discussed here, for example the role of chromatin structure and AID structure and enzymology; see refs (5, 14, 15).
The C terminus of AID is required for CSR but not for SHM
One of the most puzzling aspects of AID function is that the two roles of AID, to induce SHM and CSR, do not appear to usually occur simultaneously. For example, high levels of AID are readily induced in culture by treatment of mouse splenic B cells with cytokines and either lipopolysaccharide (LPS), which activates cells through TLR4, or a ligand for CD40, the major receptor for T cell helper signals. AID is also induced in human peripheral blood or tonsillar B cells by CD40 ligand and cytokines. B cells activated in culture then undergo CSR, but do not mutate their V region genes. There are several reports that human B cell populations, as well as a few human B cell lines, undergo low levels of SHM in response to cytokines, activated T cells and/or ligands that signal through the B cell receptor, and this was not reported to be accompanied by CSR (16–18). In vivo, CSR is initiated earlier than SHM, beginning prior to generation of germinal centers (7). SHM, however, occurs at a high frequency in vivo in germinal center B cells, where it is accompanied by CSR. It is possible that the inability to induce SHM in mouse B cells in culture is simply due to the fact that there are more AID target hotspots, WGCW (where W=A or T), in S regions than in V regions, and perhaps AID levels are lower in cultured B cells than in germinal center B cells. Several features of transcribed S regions might also specifically recruit AID, as will be discussed later. It also seems likely that differential signals induce these processes, and this might result in differences in AID targeting, perhaps due to differential expression of targeting proteins or altered chromatin accessibility.
It is highly likely that there are different interacting partners for AID during SHM and CSR, because the C terminal 10 amino acids of AID are required for CSR but not for SHM (19, 20), and mutation of Gly23 to Ser greatly reduces SHM but has very little effect on CSR (21, 22). The C terminus is not required for targeting AID to the Sμ region, as the C terminally deleted AID can deaminate dC in the Sμ region (19), and instigate DSBs within it (23), although CSR is ablated. Thus, it appears that the role of the C terminus is subsequent to DSB formation in Sμ. Consistent with this, it has recently been reported that the Sμ-Sα switch recombination junctions cloned from human patients expressing AID lacking the C terminus have longer stretches of nucleotide microhomology (see Glossary) than do junctions from normal individuals (24). Increased microhomology is associated with the use of A-EJ rather than C-NHEJ (8), suggesting that the AID C terminus might direct CSR towards C-NHEJ. However, because A-EJ is capable of supporting nearly normal levels of CSR (25), this does not explain the great reduction in CSR in cells expressing C terminally deleted AID.
AID deaminates dC in transcribed Ig S regions and V regions on both strands
In Ig loci, AID only attacks transcribed regions. Only rearranged expressed V genes undergo SHM, and during CSR, cytokines induce transcription from specific promoters located upstream of each S-CH gene segment to synthesize germline (GL) transcripts which are required for CSR (7) (Fig 1). The substrate for AID is ssDNA, which is generated during transcription (5). In general, transcription preferentially induces the non-transcribed, i.e. top strand, to form a very small (~11 nts) stretch of ssDNA at the site where RNA polymerase is transcribing the bottom DNA strand, i.e., the transcription bubble. During transcription of S regions, which consist of G-rich tandem repeats in mouse and humans, the transcribed strand (bottom strand) hybridizes with the S region transcript, forming an R-loop, and leaving the top strand single-stranded over long stretches (26). However, it is clear that AID deaminates dCs on both strands. Uracils have been identified within expressed endogenous V genes and S regions in ung−/− mouse B cells undergoing SHM and CSR, and shown to occur only about 1.4 fold more frequently (per dC) on the top strand (27). Analysis of AID-induced mutations in S regions in ung−/−msh2−/− B cells suggested that AID deaminates dC's on both strands roughly equally (28). In this double-knock out, AID deamination events are not repaired, and they can be observed as C>T transition mutations. Although it has also been shown that transcribed supercoiled, but not linear, plasmids can be attacked by AID on both strands (29), and it is known that supercoiling occurs in vivo just upstream and downstream of the transcription complex (30), the small amount of ss DNA that might be created by supercoiling does not seem adequate to explain the fact that AID deaminates both strands nearly equally in vivo.
In B cell extracts, AID co-purifies with the RNA exosome during deamination of a transcribed DNA substrate (31). The RNA exosome complex is involved in quality control of RNA, and is capable of degrading RNA lacking a poly A tail or a cap (32). This complex can associate with RNA Pol II and remove nascent transcripts from transcribed DNA, thus exposing the transcribed DNA strand (33). ssDNA exposed in this way might then become a substrate for AID. Knock-down of one component of the RNA exosome (Rrp40) reduces CSR in CH12F3 B lymphoma cells, but has no effect on AID levels or GL transcripts. Most interestingly, addition of the 9 core components of the RNA exosome to an in vitro reaction allowed AID to deaminate dC's on both strands of transcribed linear ds templates, which otherwise did not occur (31). Thus, the long-standing mystery of how both strands are targeted by AID might be beginning to be solved.
AID expression is regulated at several levels
Regulation by transcription
The transcriptional regulatory elements for AID have been localized to 4 regions, which extend at least 9 kb upstream of the first exon and 18 kb downstream of the most C terminal exon. Within these 4 regions, there are conserved binding sites for at least 19 transcription factors, both activating and repressive factors, and several sites have been confirmed functionally (34–36). When B cells are activated during an immune response, transcription of AID is induced within one day, but in differentiated antibody secreting cells AID mRNA and protein are no longer expressed. This is likely due to induction of factors that inhibit transcription (36), and as will be discussed, might also due to induction of micro-RNAs that result in degradation of both AID mRNA and reduced AID synthesis. Furthermore, DSBs instigated by AID during CSR in germinal center B cells have been shown to repress the transcription factor CRTC2, which results in differentiation of B cells toward antibody secretion and reduction in AID mRNA levels (37).
Regulation by micro-RNA (miRNA)
AID mRNA levels have been shown to be regulated by two miRNAs, miR-155 and miR-181b. miRNAs are a class of 20–23 nt non-coding RNAs that bind to complementary sequences in mRNAs, causing their degradation or inhibiting their translation. Both miR-155 and miR-181b bind to conserved sites in the 3' untranslated region of AID mRNA, and both of these miRs have numerous additional targets besides AID. miR-155 is transcribed as part of a non-coding RNA precursor, termed Bic, which is induced along with AID, in mouse splenic B cells treated with Ig switch inducers. It is also expressed in germinal center B cells. Mutation of the miR-155 binding site in AID mRNA results in a 2–3 fold increase in AID mRNA and protein levels (38, 39). This causes ~1.5–3 fold increase in CSR in cultured splenic B cells. Interestingly, SHM in VJH4 gene segments and in the Sμ region are not increased, suggesting that these processes are regulated by additional mechanisms besides the amount of AID expressed. However, IgH-c-myc translocations are increased ~5-fold, consistent with many results indicating that higher amounts of AID result in aberrant targeting of AID. Most interestingly, Burkitt lymphoma B cells are deficient in miR-155 (40).
miR-181b was shown to inhibit AID mRNA and protein levels by ~30% when over-expressed in splenic B cells (41). Interestingly, miRNA-181b has a different pattern of expression from miR-155. It is maximal in unstimulated mouse splenic B cells, decreases 5-fold upon activation to switch with LPS+IL4, and then gradually reappears and by day 3 of culture the amount almost returns to the levels in unstimulated cells. Thus, it might be involved in reducing AID levels in resting B cells, and perhaps both of these miRs are involved in down-regulation of AID after activation.
Regulation of AID protein levels
AID is also regulated by nuclear-cytoplasm transport, and is mainly found in the cytoplasm. AID has a strong nuclear export signal (NES) that binds CRM1, which exports AID to the cytoplasm (42, 43). In addition, there is evidence for a cytoplasmic retention mechanism (44). A non-classical nuclear localization signal (NLS) that binds CTNNBL1, a protein involved in RNA splicing, has also been defined (21, 44), and it is possible that CTNNBL1 transports AID to the nucleus. Although CTNNBL1 is not required for CSR (45), this might be due to redundancy. For example, GANP, a protein induced in germinal center cells during an immune response, also appears to interact with AID and to be involved in transporting it into the nucleus (46).
Nuclear AID is less stable than cytoplasmic AID, having an half-life of 2.5 hrs, whereas cytoplasmic AID has a half-life of ~8 hrs. Polyubiquitination of AID is much more active in the nucleus, and results in its degradation in proteasomes, thus explaining the shorter half-life of nuclear AID (47). Hsp90 has been shown to inhibit polyubiquination of AID in the cytoplasm (48). Degradation of nuclear AID is likely to reduce the amount of off-target activity by nuclear AID, and contribute to lowering AID levels later in the immune response.
Phosphorylation of AID regulates its activity and association with chromatin
AID is phosphorylated at several sites, including Ser3, Ser38, Thr140, and Tyr184. Phosphorylation of these Ser and Thr's affect the activity of AID in vivo, although no role for phosphorylation of Tyr184 has been discovered.. The Ser-Thr phosphorylations do not appear to affect the catalytic activity of purified recombinant AID in vitro or when highly expressed in E. coli (5, 49). In vivo, only a small fraction of AID is phosphorylated at S38, and this fraction is found preferentially associated with chromatin (50, 51). S38 is located within a consensus site for protein kinase A (PKA), which has been shown to phosphorylate this site in vitro, and to be associated with Ig S regions in B cells undergoing CSR (52). As PKA is associated with S regions independently of AID, it is hypothesized that AID is recruited to S regions by proteins that will be discussed later, and then becomes phosphorylated. Phoshorylation at S38 is required for association of AID with RPA, a trimolecular ring complex that binds ssDNA (53, 54). RPA increases the binding of AID and its activity on transcribed duplex DNA in vitro when AID levels are limiting, and S38 phosphorylation appears to be important for AID activity in B cells. Mutation of S38 to Ala reduces CSR and SHM by 80–90% relative to that induced by wild type AID in retrovirally transduced splenic B cells, and also in mice with a S38A knock-in mutation (49, 54). T140 is phosphorylated by protein kinase C (49); a T140A mutation has a smaller effect than the S38A mutation, preferentially reducing SHM (49).
By contrast, phosphorylation of S3 inhibits AID activity, as the S3A mutation increases CSR and c-myc-IgH translocations by 1.5 and 2-fold, respectively, in retrovirally transduced splenic B cells, and increases SHM within a GFP transgene in fibroblasts by 2-fold (55). Phosphorylation at S3 does not reduce stability of AID, and preliminary evidence suggests that this phosphorylation reduces association of AID with the IgH Sμ region. The phosphatase PP2A appears to reverse this phosphorylation event in vivo, which is interesting because PP2A inactivation is linked to several types of B cell neoplasias. Taken together, it is clear that several signaling pathways influence AID activity by regulating its phosphorylation.
AID binds to and deaminates numerous non-Ig genes in the mouse genome
One of the most interesting questions regarding AID is how does it preferentially target the Ig loci? Results discussed above suggest that AID might be specifically phosphorylated at Ig S regions. However, the specificity of AID for targeting Ig genes is not absolute. It has been clear for awhile that AID also mutates other genes expressed in germinal center B cells, including bcl6, cd79, and cd95 (56–59). Recently, by using a candidate gene approach, it was estimated that AID mutates ~25% of all genes transcribed in germinal center B cells, although the mutation frequency is much lower than at the Ig loci, and many of the mutations are repaired in an error-free manner in cells with intact base excision and mismatch repair systems (60). Surprisingly, the ability to repair AID-induced lesions in an error-free manner at different genomic sites appears to differ, but the mechanism for this is unknown.
By using the genome-wide approach of chromatin immunoprecipitation (ChIP) followed by massively parallel sequencing of the ChIP'ed DNA (ChIP-seq), it was found that AID, expressed at endogenous levels, binds to 5,910 genes (at 12,200 sites) in B cells induced to undergo CSR in culture (61). The binding is highest at Sμ, but is almost as high at several other genes, including genes previously shown to be mutated by AID. Most of the binding sites are in transcribed genes, and they correlate well with RNA Pol II binding sites. In this study, the investigators also performed ChIP-seq to identify genome-wide RPA binding sites in these same cells. Most interestingly, RPA binding was restricted to IgH genes, plus a few other non-Ig genes. RPA binding was dependent upon AID, as it was not detected in aid−/− cells, and was reduced about 3-fold in cells expressing AID with S38A or T140A mutations. To reconcile these findings with previous evidence suggesting that RPA is required for AID binding to DNA (53), it is possible that RPA binds co-operatively with AID and stabilizes both AID- and RPA-binding. In the ChIP-seq experiments (61), RPA binding might also be stimulated by resection of the DNA from a SSB instigated by AID, which creates ss DNA. AID might be more active at S regions due to the presence of high levels of WGCW AID targeting hotspots and due to the presence of PKA, thus increasing RPA binding at S regions. These results clearly show that in B cells induced to undergo CSR in culture, AID binds to and mutates numerous sites in the genome, but preferentially binds and mutates IgH S regions (61).
Another recent genome-wide study used ChIP followed by hybridization to tiling arrays containing the entire mouse genome (ChIP-chip) to demonstrate that physiological levels of AID induce hundreds of reproducible DSBs throughout the genome in B cells induced to undergo CSR in culture (62). In this case, the DSBs were detected by ChIP for Nbs1, a protein component of the Mre11-Rad50-Nbs1 complex (MRN), which binds within 1 kb of DSBs, including DSBs induced by AID activity (63, 64). As in the AID-binding study, the greatest amount of Nbs1-binding was detected in the Ig Sμ region. Some of the additional sites identified occur in or near genes that are amplified or translocated in B cell lymphomas.
The results suggest that AID deaminates dCs at many of the sites it binds to (60, 61). However, even if many of these lesions lead to SSBs, most are unlikely to spontaneously form DSBs, as this requires the breaks to be on opposite DNA strands and quite near each other. Also, because most sites do not have a high concentration of hotspot targets for AID, unlike S regions, it is unlikely that there will be sufficient dUs and SSBs to create substrates for mismatch repair. Thus, at most sites, SSBs will not cause DSBs until the cell attempts to replicate through them during S phase. This differs from AID-induced DSBs in S regions, which are introduced and repaired during G1 phase (64, 65). DSBs that form during S phase will be repaired mostly by NHEJ, but also by homologous recombination (HR) (66, 67). DSBs present in S phase can lead to chromosome breaks and translocations. The hypothesis that HR is involved in repair of some of the AID-induced DSBs is consistent with the recent report that xrcc2−/− mouse B cells induced to switch sustain numerous γH2AX foci, chromosome breaks, and arrest in late S/G2 phase (68). XRCC2 is required for HR. XRCC2-deficient aid−/− cells do not have these chromosome breaks. Taken together, these studies demonstrate that AID is indeed a dangerous enzyme, as it frequently induces mutations and DNA breaks at sites other than the Ig loci, consistent with a great deal of evidence indicating that it is involved in initiating and promoting chromosomal translocations leading to B cell lymphomas and leukemias (69, 70).
AID might be recruited to stalled RNA Pol II by Spt5
By screening an shRNA library for ability to inhibit CSR in CH12F3 B cells, Spt5 was recently identified as a protein that is important for CSR (71). Spt5 is a component of DSIF, a heterodimer that is associated with stalled RNA Pol II. Spt5 was found to directly bind AID in vitro, and by the use of ChIP-seq to identify genome-wide binding sites, Spt5, RNA Pol II, and AID were found to mostly co-localize throughout the genome. A previous finding that AID co-IPs with RNA Pol II in extracts from activated B cells (72) might be due to interaction of AID and RNA Pol II with Spt5. Not all transcribed genes bind Spt5, but the genes that bind Spt5 also generally have AID binding. Knock-down of Spt5 decreased AID binding to the Ig Sμ regions, further suggesting that Spt5 recruits AID to DNA. Similarly to RNA Pol II, both Spt5 and AID binding are highest near transcription start sites of most genes, although they are also spread throughout the gene bodies. The high concentration of AID, Spt5, and RNA Pol II near transcription initiation sites might explain why most AID-induced mutations occur within <1 kb of the initiation site in V genes. These results fits with a model proposed several years ago, which posited that a mutator factor (which we now know is AID) associates with RNA Pol II, introducing mutations when RNA Pol II stalls and delivers the factor to DNA (73).
Unlike its behavior at other genes, RNA Pol II appears to stall and accumulate across the entire Ig Sμ region, and this has been proposed to be due to R-loop formation (74, 75). Spt5 and AID are also bound in high amounts across Sμ in activated B cells undergoing CSR, suggesting that stalled RNA Pol II recruits Spt5, which in turn recruits AID (61, 71). Challenging this model is the finding that high concentrations of RNA Pol II, Spt5 and AID extend throughout the Cμ region in these B cells (71), as the C region does not form R-loops (26). Taken together, Spt5 appears to be important, and perhaps essential, for recruiting AID to S regions, and perhaps to all its targets, and perhaps for increasing ssDNA formation due to stalled RNA Pol II. Also very interesting is the fact that Spt5 binds the RNA exosome in Drosophila (76), and thus it is possible that Spt5 recruits the RNA exosome, which then would allow AID to target both DNA strands.
PTBP2 binds S region RNA and recruits AID to S regions
Another layer of specificity for recruitment of AID to S regions appears to be provided by PTBP2, a protein reported to inhibit RNA splicing and to bind to polypyrimidine RNA tracts (77). This protein was identified due to its ability to bind AID in the CH12F3 B cell line induced to undergo CSR. Also, PTBP2 binds to both sense and anti-sense Sμ transcripts. Knockdown of PTBP2 reduces CSR and association of AID with Sμ DNA in cells, but has no effect on cell proliferation, AID levels, GL transcript levels, or AID activity in vitro. Thus, the fact that this protein has specificity for RNA transcribed from the Sμ region and also increases AID binding to Sμ and Sγ1 DNA suggests that it might recruit AID specifically to S regions. An appealing hypothesis is that PTBP2 binds to S region RNA in R-loops, and thus increases the specificity of recruitment of AID to S regions.
14-3-3 scaffold proteins bind S regions and AID, dependent on the AID C terminus
Another candidate for proteins that recruit AID to S regions is 14-3-3, a family of proteins with numerous functions, including an involvement in DNA replication, and an ability to bind cruciform DNA structures (78). In vitro experiments demonstrated that 14-3-3 proteins can bind a segment of dsDNA with repeating AGCT or AGCA motifs alternating with 4 T residues. These motifs are AID target hotspots and common in S regions. 14-3-3 proteins also bind transcribed S regions in B cells induced to undergo CSR, and can also directly bind AID in vitro (79). These data suggest that 14-3-3 helps recruit AID to S regions. Splenic B cells deficient for just one of the isoforms (γ) switch about 50% as well as wild-type B cells. Most interestingly, the interaction between AID and 14-3-3 requires the C terminal 9 amino acids of AID. Note, however, as already mentioned AID lacking the C terminus still targets the Sμ region at least as well as full-length AID. Thus it seems likely that 14-3-3 contributes to CSR and might help to recruit AID to S regions, but this does not explain the role of the AID C terminus in CSR.
How is AID is recruited to Ig V regions?
Although the data discussed above suggest how AID might be recruited to S regions, much less is known about V region targeting. This process is harder to study due to a lack of a robust cell culture model, and unlike S regions, V region gene segments do not have obvious unique characteristics that distinguish them from non-Ig genes. In early studies using a transgene substrate, the sequence motif CAGGTG, which resembles a binding site for E box proteins, e.g. E47, was found to be essential for AID targeting for SHM, and this was not due to an effect on transcription of the transgene (80). However, an E box motif does not appear likely to provide enough specificity to explain why SHM is restricted to V genes, and many other studies performed using Ig light chain genes in the chicken DT40 B cell line suggest that the requirements for AID-dependent V region SHM are much more complex than simply an E box (81–83). Thus, how AID is specifically targeted to Ig V regions remains a major unanswered question.
AID functions in other cell types and might be involved in DNA demethylation
Although AID is expressed at the highest levels in activated B cells, it is also expressed in other cells types, including oocytes, ES cells (84), breast tissue (85), and prostate epithelial cells (86). The available evidence suggests that AID induces mutations, DNA breaks, and translocations at non-Ig genes in H. pylori-infected gastric epithelial cells, in prostate cells, and in breast tissue, and that AID expression can lead to tumorigenesis in these tissues (11, 13, 86, 87). It is not understood why AID would be expressed in these other tissues. Is it simply due to a lack of negative selection or does it have a beneficial role? One hypothesis is that AID might be involved in deamination of 5-meC in DNA, a process that would result in demethylation, and which occurs primarily, but not exclusively, during development (84, 87). AID is also expressed at very low levels in developing B cells in the bone marrow and there is evidence that it induces a low level of CSR and SHM in IgH V genes in these cells (88, 89). A most interesting suggestion is that AID mutates autoreactive Ig V genes in developing B cells to prevent autoimmunity, but more work needs to be done to establish this.
Concluding remarks
In conclusion, AID is essential for producing antibodies with exquisite specificity for any infectious agent, but at the same time it contributes to genome instability, apparently due to imprecise targeting. As befitting an enzyme whose activity appears to verge toward creating a disaster, its regulation is extremely complex. It is clear that AID activity is regulated by numerous mechanisms including: (1) regulation of aid transcription by activators and repressors, as well as histone methylation and acetylation at the locus (14), (2) regulation of mRNA stability and translation by miRNAs, (3) regulation of protein stability by nuclear/cytoplasmic transport and polyubiquitination, (4) regulation of protein activity by phosphorylation and dephosphorylation by at least two different pathways, and (5) regulation of protein recruitment to Ig genes and to numerous other transcribed genes by several proteins. Although most of the proteins that recruit AID to DNA are not specific for Ig genes, it is possible that AID is preferentially recruited to Ig S regions through a combination of non-specific and somewhat specific interactions. For example, RNA Pol II stalls at S regions, perhaps due to formation of R-loops, and this appears to recruit Spt5, which in turn helps to recruit AID. Although these proteins also bind at numerous other sites in the genome, RNA Pol II stalling might be more severe at S regions compared to other genes. AID might also be recruited by 14-3-3, which appears to bind preferentially to S regions. Likewise, AID binds PTBP2, an RNA-binding protein that might preferentially bind GL transcripts at S regions. Also, if AID is preferentially phosphorylated when associated with S regions, this might also increase its association and activity at S regions. Thus, we might have an outline of how AID preferentially instigates DSBs in S region. However, most of these mechanisms do not appear to operate for V regions, which are also specific targets of AID. How AID specifically targets V genes is just one of the many important discoveries still to be made about AID.
Acknowledgements
I thank Dr Jayanta Chaudhuri for several helpful comments, and Drs Karuna Ganesh and Michael Neuberger for sharing unpublished information about CTNNBL1. JS is supported by grants from NIH RO1 AI023283 and R21 AI088578.
Glossary
- Apobec1
(Apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1) is a member of a family of cytidine deaminases, which includes AID, some of which edit mRNA while others edit DNA. Apobec1, partnered with a targeting protein, edits the mRNA for apolipoprotein B-100, introducing a stop codon, which converts it to the mRNA for ApoB-48. Both Apo-B proteins are important in lipid metabolism. Little is known about the functions of the other Apobecs, although some are important for resisting viral infections.
- Microhomology
As S-S junctions are formed by an end-joining type of recombination, they often appear to occur by ligation of two blunt DSBs, resulting in 0 bp of microhomology at the junction. Alternatively, sometimes they occur at sites of short identical sequences in the donor and acceptor S regions, so one cannot determine exactly where the junction occurs. These short bits of homology, often just 1 or 2 nucleotides in length, are termed microhomology. Junctions formed by classical (C)-NHEJ mostly have 0–3 bp or microhomology, whereas junctions formed by alternative (A)-EJ or also called A-NHEJ mostly have greater than 5 bp of microhomology. This can extend to 10–15 nucleotides, depending on the similarity between the donor and acceptor S regions.
- Transition and transversion mutations
Transitions are mutation from one pyrimidine to another, i.e. dC>dT or dT>dC, or from one purine to the other purine, i.e. dG>dA or dA>dG. Transversions are mutations from a pyrimidine to either purine or a purine to either pyrimidine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 3.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 4.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–104. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 5.Peled JU, Kuang FL, Ussel M. D. Iglesias, Roa S, Kalis SL, Goodman MF, Scharff MD. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 6.Guikema JE, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, Schrader CE. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med. 2007;204:3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Ann Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stavnezer J, Bjorkman A, Du L, Cagigi A, Pan-Hammarstrom Q. Mapping of switch recombination junctions, a tool for studying DNA repair pathways during immunoglobulin class switching. Adv. Immunol. 2010;108:45–109. doi: 10.1016/B978-0-12-380995-7.00003-3. [DOI] [PubMed] [Google Scholar]
- 9.Chesi M, Robbiani DF, Sebag M, Chng WJ, Affer M, Tiedemann R, Valdez R, Palmer SE, Haas SS, Stewart AK, Fonseca R, Kremer R, Cattoretti G, Bergsagel PL. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer cell. 2008;13:167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klemm L, Duy C, Iacobucci I, Kuchen S, vonLevetzow G, Feldhahn N, Henke N, Li Z, Hoffmann TK, Kim YM, Hofmann WK, Jumaa H, Groffen J, Heisterkamp N, Martinelli G, Lieber MR, Casellas R, Muschen M. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer cell. 2009;16:232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okazaki IM, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morisawa T, Marusawa H, Ueda Y, Iwai A, Okazaki IM, Honjo T, Chiba T. Organ-specific profiles of genetic changes in cancers caused by activation-induced cytidine deaminase expression. International journal of cancer. 2008;123:2735–2740. doi: 10.1002/ijc.23853. [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto Y, Marusawa H, Kinoshita K, Endo Y, Kou T, Morisawa T, Azuma T, Okazaki IM, Honjo T, Chiba T. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nature medicine. 2007;13:470–476. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- 14.Storck S, Aoufouchi S, Weill JC, Reynaud CA. AID and partners: for better and (not) for worse. Curr Opin Immunol. 2011;23 doi: 10.1016/j.coi.2011.02.002. in press. [DOI] [PubMed] [Google Scholar]
- 15.Chelico L, Pham P, Goodman MF. Stochastic properties of processive cytidine DNA deaminases AID and APOBEC3G. Philos Trans R Soc Lond B Biol Sci. 2009;364:583–593. doi: 10.1098/rstb.2008.0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurrieri C, McGuire P, Zan H, Yan XJ, Cerutti A, Albesiano E, Allen SL, Vinciguerra V, Rai KR, Ferrarini M, Casali P, Chiorazzi N. Chronic lymphocytic leukemia B cells can undergo somatic hypermutation and intraclonal immunoglobulin V(H)DJ(H) gene diversification. J Exp Med. 2002;196:629–639. doi: 10.1084/jem.20011693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faili A, Aoufouchi S, Gueranger Q, Zober C, Leon A, Bertocci B, Weill JC, Reynaud CA. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat Immunol. 2002;3:815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- 18.Woo CJ, Martin A, Scharff MD. Induction of somatic hypermutation is associated with modifications in immunoglobulin variable region chromatin. Immunity. 2003;19:479–489. doi: 10.1016/s1074-7613(03)00261-9. [DOI] [PubMed] [Google Scholar]
- 19.Barreto V, Reina-San-Martin B, Ramiro AR, McBride KM, Nussenzweig MC. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol Cell. 2003;12:501–508. doi: 10.1016/s1097-2765(03)00309-5. [DOI] [PubMed] [Google Scholar]
- 20.Ta VT, Nagaoka H, Catalan N, Durandy A, Fischer A, Imai K, Nonoyama S, Tashiro J, Ikegawa M, Ito S, Kinoshita K, Muramatsu M, Honjo T. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 21.Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, Sakakibara Y, Hijikata H, Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- 22.Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol. 2011;12:264–270. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- 23.Doi T, Kato L, Ito S, Shinkura R, Wei M, Nagaoka H, Wang J, Honjo T. The C-terminal region of activation-induced cytidine deaminase is responsible for a recombination function other than DNA cleavage in class switch recombination. Proc Natl Acad Sci U S A. 2009;106:2758–2763. doi: 10.1073/pnas.0813253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kracker S, Imai K, Gardes P, Ochs HD, Fischer A, Durandy A. Impaired induction of DNA lesions during immunoglobulin class switch recombination in humans influences end-joining repair. Proc. Nat. Sci. USA. 2010 doi: 10.1073/pnas.1012591108. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boboila C, Yan C, Wesemann DR, Jankovic M, Wang JH, Manis J, Nussenzweig A, Nussenzweig M, Alt FW. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J Exp Med. 2010;207:417–427. doi: 10.1084/jem.20092449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 27.Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, Gramlich HS, Schatz DG, Woodgate R, Wilson DM, 3rd, Gearhart PJ. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat Immunol. 2011;12:70–76. doi: 10.1038/ni.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 29.Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci U S A. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins I, Weber A, Levens D. Transcriptional consequences of topoisomerase inhibition. Mol Cell Biol. 2001;21:8437–8451. doi: 10.1128/MCB.21.24.8437-8451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, Januszyk K, Gregory RI, Deng H, Lima CD, Alt FW. The RNA Exosome Targets the AID Cytidine Deaminase to Both Strands of Transcribed Duplex DNA Substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends in biochemical sciences. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 33.El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crouch EE, Li Z, Takizawa M, Fichtner-Feigl S, Gourzi P, Montano C, Feigenbaum L, Wilson P, Janz S, Papavasiliou FN, Casellas R. Regulation of AID expression in the immune response. J Exp Med. 2007;204:1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SR, Zan H, Pal Z, Zhang J, Al-Qahtani A, Pone EJ, Xu Z, Mai T, Casali P. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat Immunol. 2009;10:540–550. doi: 10.1038/ni.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran TH, Nakata M, Suzuki K, Begum NA, Shinkura R, Fagarasan S, Honjo T, Nagaoka H. B cell-specific and stimulation-responsive enhancers derepress Aicda by overcoming the effects of silencers. Nat Immunol. 2010;11:148–154. doi: 10.1038/ni.1829. [DOI] [PubMed] [Google Scholar]
- 37.Sherman MH, Kuraishy AI, Deshpande C, Hong JS, Cacalano NA, Gatti RA, Manis JP, Damore MA, Pellegrini M, Teitell MA. AID-induced genotoxic stress promotes B cell differentiation in the germinal center via ATM and LKB1 signaling. Mol Cell. 2010;39:873–885. doi: 10.1016/j.molcel.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kluiver J, van den Berg A, de Jong D, Blokzijl T, Harms G, Bouwman E, Jacobs S, Poppema S, Kroesen BJ. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene. 2007;26:3769–3776. doi: 10.1038/sj.onc.1210147. [DOI] [PubMed] [Google Scholar]
- 41.de Yebenes VG, Belver L, Pisano DG, Gonzalez S, Villasante A, Croce C, He L, Ramiro AR. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito S, Nagaoka H, Shinkura R, Begum N, Muramatsu M, Nakata M, Honjo T. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci U S A. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geisberger R, Rada C, Neuberger MS. The stability of AID and its function in class-switching are critically sensitive to the identity of its nuclear-export sequence. Proc Natl Acad Sci U S A. 2009;106:6736–6741. doi: 10.1073/pnas.0810808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patenaude AM, Orthwein A, Hu Y, Campo VA, Kavli B, Buschiazzo A, Di Noia JM. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nature structural & molecular biology. 2009;16:517–527. doi: 10.1038/nsmb.1598. [DOI] [PubMed] [Google Scholar]
- 45.Han L, Masani S, Yu K. Cutting edge: CTNNBL1 is dispensable for Ig class switch recombination. J Immunol. 2010;185:1379–1381. doi: 10.4049/jimmunol.1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maeda K, Singh SK, Eda K, Kitabatake M, Pham P, Goodman MF, Sakaguchi N. GANP-mediated recruitment of activation-induced cytidine deaminase to cell nuclei and to immunoglobulin variable region DNA. J Biol Chem. 2010;285:23945–23953. doi: 10.1074/jbc.M110.131441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aoufouchi S, Faili A, Zober C, Orlando OD, Weller S, Weill JC, Reynaud CA. Proteasomal degradation restricts the nuclear lifespan of AID. J Exp Med. 2008;205:1357–1368. doi: 10.1084/jem.20070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orthwein A, Patenaude AM, Affar el B, Lamarre A, Young JC, Di Noia JM. Regulation of activation-induced deaminase stability and antibody gene diversification by Hsp90. J Exp Med. 2010;207:2751–2765. doi: 10.1084/jem.20101321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McBride KM, Gazumyan A, Woo EM, Schwickert TA, Chait BT, Nussenzweig MC. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J Exp Med. 2008;205:2585–2594. doi: 10.1084/jem.20081319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McBride KM, Gazumyan A, Woo EM, Barreto VM, Robbiani DF, Chait BT, Nussenzweig MC. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc Natl Acad Sci U S A. 2006;103:8798–8803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasqualucci L, Kitaura Y, Gu H, Dalla-Favera R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc Natl Acad Sci U S A. 2006;103:395–400. doi: 10.1073/pnas.0509969103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vuong BQ, Lee M, Kabir S, Irimia C, Macchiarulo S, McKnight GS, Chaudhuri J. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat Immunol. 2009;10:420–426. doi: 10.1038/ni.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 54.Cheng HL, Vuong BQ, Basu U, Franklin A, Schwer B, Astarita J, Phan RT, Datta A, Manis J, Alt FW, Chaudhuri J. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc Natl Acad Sci U S A. 2009;106:2717–2722. doi: 10.1073/pnas.0812304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gazumyan A, Timachova K, Yuen G, Siden E, Di Virgilio M, Woo EM, Chait BT, Reina San-Martin B, Nussenzweig MC, McBride KM. Amino-Terminal Phosphorylation of Activation-Induced Cytidine Deaminase Suppresses c-myc/IgH Translocation. Mol Cell Biol. 2011;31:442–449. doi: 10.1128/MCB.00349-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 57.Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti RS, Klein U, Kuppers R, Rajewsky K, Dalla-Favera R. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci U S A. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muschen M, Re D, Jungnickel B, Diehl V, Rajewsky K, Kuppers R. Somatic mutation of the CD95 gene in human B cells as a side-effect of the germinal center reaction. J Exp Med. 2000;192:1833–1840. doi: 10.1084/jem.192.12.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gordon MS, Kanegai CM, Doerr JR, Wall R. Somatic hypermutation of the B cell receptor genes B29 (Igbeta, CD79b) and mb1 (Igalpha, CD79a) Proc Natl Acad Sci U S A. 2003;100:4126–4131. doi: 10.1073/pnas.0735266100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 61.Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Staszewski O, Baker RE, Ucher AJ, Martier R, Stavnezer J, Guikema JEJ. Activation-induced cytidine deaminase induces reproducible DNA breaks at many non-Ig loci in activated B cells. Mol Cell. 2011;41:232–242. doi: 10.1016/j.molcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berkovich E, Monnat RJ, Jr., Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 64.Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, Redon C, Ried T, Bonner WM, Honjo T, Nussenzweig MC, Nussenzweig A. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 66.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell cycle (Georgetown, Tex. 2008;7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hasham MG, Donghia NM, Coffey E, Maynard J, Snow KJ, Ames J, Wilpan RY, He Y, King BL, Mills KD. Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat Immunol. 2010;11:820–826. doi: 10.1038/ni.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, 3rd, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 70.Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, Ried T, Nussenzweig A, Nussenzweig MC. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, San-Martin BR, Barreto V, Nieland TJ, Root DE, Casellas R, Nussenzweig MC. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 73.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 74.Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, Gearhart PJ. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med. 2009;206:1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 77.Nowak U, Matthews AJ, Zheng S, Chaudhuri J. The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nat Immunol. 2011;12:160–166. doi: 10.1038/ni.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zannis-Hadjopoulos M, Yahyaoui W, Callejo M. 14-3-3 cruciform-binding proteins as regulators of eukaryotic DNA replication. Trends in biochemical sciences. 2008;33:44–50. doi: 10.1016/j.tibs.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 79.Xu Z, Fulop Z, Wu G, Pone EJ, Zhang J, Mai T, Thomas LM, Al-Qahtani A, White CA, Park SR, Steinacker P, Li Z, Yates J, 3rd, Herron B, Otto M, Zan H, Fu H, Casali P. 14-3-3 adaptor proteins recruit AID to 5'-AGCT-3'-rich switch regions for class switch recombination. Nature structural & molecular biology. 2010;17:1124–1135. doi: 10.1038/nsmb.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Michael N, Shen HM, Longerich S, Kim N, Longacre A, Storb U. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 2003;19:235–242. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 81.Kothapalli N, Norton DD, Fugmann SD. Cutting edge: a cis-acting DNA element targets AID-mediated sequence diversification to the chicken Ig light chain gene locus. J Immunol. 2008;180:2019–2023. doi: 10.4049/jimmunol.180.4.2019. [DOI] [PubMed] [Google Scholar]
- 82.Blagodatski A, Batrak V, Schmidl S, Schoetz U, Caldwell RB, Arakawa H, Buerstedde JM. A cis-acting diversification activator both necessary and sufficient for AID-mediated hypermutation. PLoS genetics. 2009;5:e1000332. doi: 10.1371/journal.pgen.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kothapalli NR, Fugmann SD. Targeting of AID-mediated sequence diversification to immunoglobulin genes. Curr Opin Immunol. 2011;23 doi: 10.1016/j.coi.2010.12.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 85.Pauklin S, Sernandez IV, Bachmann G, Ramiro AR, Petersen-Mahrt SK. Estrogen directly activates AID transcription and function. J Exp Med. 2009;206:99–111. doi: 10.1084/jem.20080521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, Rosenfeld MG. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fritz EL, Papavasiliou FN. Cytidine deaminases: AIDing DNA demethylation? Genes Dev. 2010;24:2107–2114. doi: 10.1101/gad.1963010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han JH, Akira S, Calame K, Beutler B, Selsing E, Imanishi-Kari T. Class switch recombination and somatic hypermutation in early mouse B Cells are mediated by B Cell and toll-like receptors. Immunity. 2007;27:64–75. doi: 10.1016/j.immuni.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuraoka M, McWilliams L, Kelsoe G. AID expression during B-cell development: searching for answers. Immunologic research. 2011;49 doi: 10.1007/s12026-010-8185-7. in press. [DOI] [PubMed] [Google Scholar]
- 90.Wilson TM, Vaisman A, Martomo SA, Sullivan P, Lan L, Hanaoka F, Yasui A, Woodgate R, Gearhart PJ. MSH2-MSH6 stimulates DNA polymerase {eta}, suggesting a role for A:T mutations in antibody genes. J Exp Med. 2005;201:637–645. doi: 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Genschel J, Modrich P. Mechanism of 5'-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 92.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]