Summary
We sought to identify biomarker responses to tuberculosis specific antigens which could 1) improve the diagnosis of tuberculosis infection and 2) allow the differentiation of active and latent infections. Seventy subjects with active tuberculosis (N=12), latent tuberculosis (N=32), or no evidence of tuberculosis infection (N=26) were evaluated. We used the Luminex Multiplexed Bead Array platform to simultaneously evaluate 25 biomarkers in the supernatant of whole blood samples following overnight stimulation using the Quantiferon® Gold In-Tube kit. We defined the response to stimulation as the difference (within an individual patient) between the response to the pooled tuberculosis antigens and the negative control. IP-10 response was significantly higher in tuberculosis-infected (active or latent) subjects compared to the uninfected group (p <0.0001). Among the 25 parameters, expression levels of IL-15 and MCP-1 were found to be significantly higher in the active tuberculosis group compared to the latent tuberculosis group (p = 0.0006 and 0.0030, respectively). When combined, IL-15 and MCP-1 accurately identified 83% of active and 88% of latent infections. The combination of IL-15 and MCP-1 responses was accurate in distinguishing persons with active tuberculosis from persons with latent tuberculosis in this study.
Keywords: tuberculosis, immune response, diagnosis, biomarker
Introduction
Tuberculosis (TB) is one of the oldest and most successful human pathogens and has infected approximately one third of the world’s population 1. After an individual is infected with TB, the infection may have no clinical manifestation (latent TB infection), but 5–10 percent of infected individuals will progress to active TB disease months to years later 2–4. Early detection and treatment of active TB as well as preventive treatment of individuals with latent TB infection are therefore considered two of the cornerstones of TB control.
Current TB diagnostics, however, are in need of improvement. TB infection has been traditionally diagnosed using the Mantoux tuberculin skin test (TST). TST effectiveness is limited by the need for a return visit, inter-reader variability, cross-reactivity with non-tuberculosis mycobacteria as well as the Bacille Calmette-Guerin (BCG) vaccine, and poor sensitivity in immunocompromised patients 5–6. Importantly, the TST only identifies TB infection, but does not provide any information that can distinguish latent TB infection from active TB disease. Microbiologic methods (acid-fast smear and mycobacterial culture) must be used to discriminate latent from active TB. Unfortunately, acid-fast smears are relatively insensitive for TB diagnosis, and mycobacterial culture requires clinicians to wait up to several weeks to obtain a result as well as significant investments in equipment. Recently, Interferon-gamma Release Assays (IGRAs) have been developed which deal with some of the TST’s limitations 7. IGRAs are performed in vitro with a single blood draw (obviating the need for return diagnostic visits), include standards to reduce inter-reader variability, and utilize TB specific antigens to reduce cross-reactivity with other mycobacteria, including BCG. The increased specificity of IGRAs has significant potential to reduce false-positive tests, particularly among BCG-vaccinated, otherwise low-risk populations, permitting public health programs to focus on high-risk persons 8. Much like TST’s, IGRAs suffer from suboptimal sensitivity in immunocompromised individuals and young children. IGRAs are also unable to differentiate latent from active TB 9.
We sought to determine whether other biomarker responses or combinations thereof, after whole-blood stimulation by TB-specific antigens, could improve upon the performance of Interferon-gamma (IFN-γ) by IGRAs. We hypothesized that combinations of multiple biomarkers could a) be more sensitive for TB infection than a single immune marker, and b) potentially distinguish individuals with active TB disease from persons with latent TB infection.
Materials and Methods
Subjects
Our cohort included persons participating in two different ongoing studies: 1) the “TB Epitopes Study” (Immunogenicity of Mycobacterium tuberculosis T Cell Epitopes), which collected a single blood specimen from persons with either active or latent TB and examined immune responses to a number of TB-specific epitopes, or 2) the “GIS-THIS Study” (Geographic Information Systems-based screening for TB, HIV, and syphilis), which screened participants for TB, human immunodeficiency virus, and syphilis in high-incidence neighborhoods. Both studies involved drawing blood for a Quantiferon® Gold In-Tube test, and the leftover supernatant from the Quantiferon tubes was frozen at −80°C and subsequently used in this study. Patients involved in this study were recruited between October 1, 2008 and May 1, 2009. The present study as well as the parent protocols required written informed consent and were approved by the Duke University Institutional Review Board.
Consenting subjects were divided into three experimental groups according to TB status: active TB, latent TB and TB negative. The active TB group included subjects who either grew Mycobacterium tuberculosis from a clinical specimen or were diagnosed clinically per Centers for Disease Control and Prevention criteria 10. Individuals in the active TB group were at variable stages of treatment, including some who had completed treatment. The latent TB group included subjects with either a positive TST, positive Quantiferon® Gold In-Tube assay, or both, but with no signs or symptoms of active TB disease and no positive cultures for M. tuberculosis. Finally, the TB negative group included subjects who tested negative with the TST and/or Quantiferon® Gold In-Tube assays and had no clinical evidence of active TB disease.
Quantiferon Tests
Quantiferon® Gold In-Tube (QFT-GIT) (Cellestis Inc, Valencia CA) tests were performed according to the manufacturer’s instructions by a single ASCP certified medical technologist trained to perform the QFT-GIT assay by Cellestis. Briefly, whole blood was collected by venipuncture from each subject and incubated for 16–24 hours in three separate conditions: 1) a mixture of 3 TB antigens from RD1 and RD11 (ESAT-6, CFP-10 and TB7.7); 2) a mitogen for a positive control; and 3) a mock stimulation for a negative control (Nil). Following the stimulations, 150μl of supernatant was harvested from each tube. 50μL of each supernatant was used to determine its IFN-γ concentration by ELISA (Cellestis Inc, Valencia, CA). In addition, 100μL of each supernatant was frozen at −80°C and analysis was performed within 16 weeks from collection.
Multiplexed Bead Arrays
Supernatants from each stimulation were thawed and analyzed undiluted and at a 1:10 dilution to determine the concentration of 25 potential biomarkers (IL-1β, IL-1RA, IL-2, IL-2R, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12 p40/70, IL-13, IL-15, IL-17, TNF-α, IFN-α, IFN-γ, GM-CSF, MIP-1α, MIP-1β, IP-10, MIG, Eotaxin, RANTES, MCP-1) by using a Human Cytokine 25-plex (Biosource, Camarillo, CA). The assay procedure was performed by a single blinded researcher for all assays and followed manufacturer’s instructions. In summary, sample and standard dilutions were performed with the included assay diluent. Beads coated with antibodies against the 25 potential biomarkers were added to each well of a 96-well filter plate and washed with wash buffer. Beads were resuspended in 100μL of incubation buffer. 100μL of standards that contained a mixture of each analyte with known concentrations were added to the appropriate wells and run in duplicate. The remaining wells received 50μL assay diluents and 50 μL of either Quantiferon® supernatant or 10-fold diluted Quantiferon® supernatant. Plates were at room temperature while shaking for 2 hours to allow sample binding to the appropriate beads. Following two recommended washes with wash buffer, biotinylated antibodies against each biomarker were added to the bead: sample conjugates and incubated at room temperature while shaking for 1 hour. At the end of the incubation time, the plate was washed twice with wash buffer. Streptavidin-RPE was added to each well and the plate was incubated at room temperature while shaking for 30 minutes. After the plate was washed 3 times with wash buffer, the beads were resuspended in wash buffer. The plate was then analyzed on a Luminex 100™ instrument (Luminex, Austin, TX) using Bio-Plex Manager Software (Bio-Rad, Hercules, CA). Analyte concentrations obtained from the undiluted Quantiferon® supernatant were used unless they were above the linear range of the assay, in which case the corresponding 10-fold diluted Quantiferon® sample supernatant was used to determine the analyte concentration. In one case the analyte (MCP-1) concentration in the 10-fold diluted specimen was above the linear range; this response was deleted for analytic purposes. Analyte concentrations reported as below the limit of detection were assigned values of 0 pg/mL for analytic purposes.
Statistical Analysis
The biomarker response was defined as the concentration of the biomarker in the Nil tube supernatant subtracted from the biomarker concentration in the TB antigen tube supernatant (TB-Nil) as detected by the multiplexed bead array. The Wilcoxon rank-sum test was used to test the marginal association between each biomarker response and experimental group 11. Based on the biomarker profiles, we built classification models for the three infection outcomes: negative (uninfected), latent TB, and active TB. To this end conditional inference trees were used 12. For each model, the family-wise error rate was controlled at the two-sided 0.05 level. Receiver operating characteristic curves were generated based on combinations of cytokines. The Spearman rank coefficient was used to assess the relationship between interferon gamma responses in the Quantiferon® vs. the Luminex systems. The R statistical environment along with the party extension package were used to conduct the statistical analyses 12–13 as well as SASv9.3 (SAS Systems, Cary, NC).
Results
Study Subjects
Specimens from 70 subjects were examined; demographic and clinical data are summarized in Table 1. Among the 32 persons with latent TB infection, one was currently taking isoniazid for latent TB, and 8 had received at least some prior treatment for latent TB (range 8–52 weeks of isoniazid, median 38.5 weeks), of whom 5 had completed at least 6 months of isoniazid, and one additional person was currently receiving latent TB treatment (7 weeks received). Three of the persons with latent TB had HIV; two were taking antiretrovirals at the time of blood sampling (CD4+ T-cell count 157/mm3 and 352/mm3), and one was not (CD4+ T-cell count 449 cells/mm3). Among the 12 persons with current or prior active TB disease, 10 were culture-proven and 2 were clinical cases. Nine of the active TB patients had pulmonary disease and 3 had extrapulmonary disease. Four of the active TB patients were currently on TB treatment (3–25 weeks into treatment), and 8 had completed treatment a median 8.5 months previously (range 0.5–17 months). Two of the patients with active TB had HIV, and both were on antiretroviral therapy at the time of blood sampling for the study, with CD4+ T-cell counts of 153 and 358 cells/mm3.
Table 1.
Subject Demographics
| Characteristic | TB Negative | Latent TB | Active TB |
|---|---|---|---|
| N | 26 | 32 | 12 |
| Median Age (range) | 46.5 (26–62) | 50 (2–66) | 43.5 (4–93) |
| Female Gender | 7 (27%) | 15 (47%) | 4 (33%) |
| Race/Ethnicity | |||
| Non-Hispanic Asian | 0 (0%) | 3 (9%) | 1 (8%) |
| Non-Hispanic Black | 22 (85%) | 16 (50%) | 8 (67%) |
| Non-Hispanic White | 2 (8%) | 9 (28%) | 1 (8%) |
| Hispanic | 2 (8%) | 4 (13%) | 2 (17%) |
| US Born | 24 (92%) | 21 (64%) | 7 (58%) |
| HIV Status | |||
| Negative | 25 (96%) | 27 (84%) | 10 (83%) |
| Positive | 1 (4%) | 3 (9%) | 2 (17%) |
| Unknown | 0 (0%) | 2 (6%) | 0 (0%) |
| Diabetes | 3 (12%) | 2 (6%) | 3 (25%) |
| BCG Status | |||
| Unvaccinated | 25 (96%) | 21 (66%) | 7 (58%) |
| Vaccinated | 0 (0%) | 10 (31%) | 4 (33%) |
| Unknown | 1 (4%) | 1 (3%) | 1 (8%) |
| Quantiferon result | |||
| Negative | 26 (100%) | 13 (41%) | 0 (0%) |
| Positive | 0 (0%) | 19 (59%) 5 not done 4 history of prior positive, unknown size |
12 (100%) |
| Tuberculin skin testing | Not done | 23 positive (median 18 mm, range 10–32 mm) | All positive (median 15.5 mm, range 11–25 mm) |
Biomarker Analysis After Antigen Stimulation
In order to verify the performance of our Luminex assay as a diagnostic we initially compared Luminex obtained IFN-γ production values across patient groups. As expected, IFN-γ expression, as detected by Luminex, was significantly higher in the infected subjects (latent TB and active TB combined) than the TB negative group as shown in Figure 1A (p<0.001). Because most subjects’ results were at the upper or lower end of the linear range of the Quantiferon® assay, it was difficult to assess the linearity of the relationship between IFN-γ responses as measured by the Quantiferon® kit vs. Luminex. The Spearman rank correlation between responses in the two kits was 0.47 (p<0.0001).
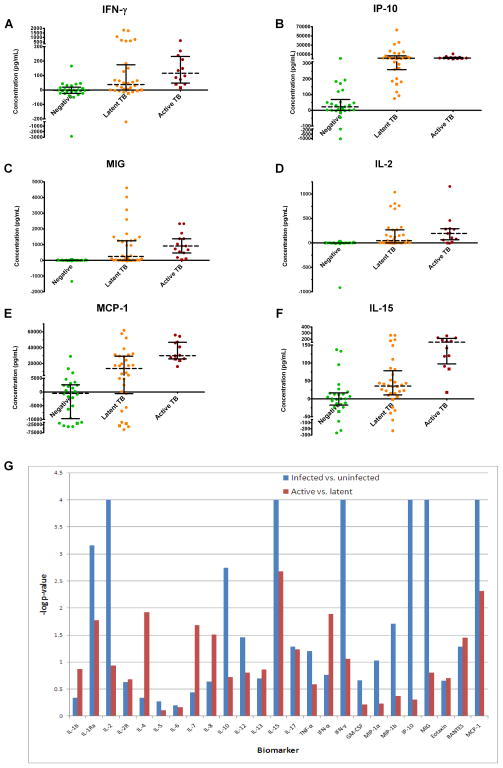
Figure 1. Comparison of biomarker expression between TB negative, latent TB, and active TB subjects.
Secretion of IFN-γ (A), IP-10 (B), MIG (C), and IL-2 (D) are significantly higher in TB infected subjects ( ) than non-infected controls (
) than non-infected controls ( ) but do not significantly differ when active TB subjects are compared to latent TB subjects (
) but do not significantly differ when active TB subjects are compared to latent TB subjects ( ) following TB antigen stimulation of whole blood. MCP-1 (E) and IL-15 (F) secretion are significantly increased following TB antigen stimulation of whole blood in TB infected subjects compared with non-infected controls. These biomarkers are also more highly secreted in active TB subjects than latent TB subjects. Each data point represents the concentration observed following stimulation minus the concentration observed in the negative control condition. HIV positive subjects are denoted by squares. Log p-values from Wilcoxon-rank sum tests comparing biomarker secretion in TB infected subjects to non-infected controls (blue bars) as well as active TB subjects to latent TB subjects (red bars) are also plotted (G). The line (log P=3) represents the Bonferroni-corrected significance threshold (50 comparisons).
) following TB antigen stimulation of whole blood. MCP-1 (E) and IL-15 (F) secretion are significantly increased following TB antigen stimulation of whole blood in TB infected subjects compared with non-infected controls. These biomarkers are also more highly secreted in active TB subjects than latent TB subjects. Each data point represents the concentration observed following stimulation minus the concentration observed in the negative control condition. HIV positive subjects are denoted by squares. Log p-values from Wilcoxon-rank sum tests comparing biomarker secretion in TB infected subjects to non-infected controls (blue bars) as well as active TB subjects to latent TB subjects (red bars) are also plotted (G). The line (log P=3) represents the Bonferroni-corrected significance threshold (50 comparisons).
We next analyzed each of the remaining 24 biomarkers’ ability to differentiate between uninfected and infected subjects (latent TB and active TB combined). The median levels of seven biomarkers (IP-10, MIG, IL-2, MCP-1, IL-15, and IL-1 receptor antagonist) were significantly (p<0.001) higher in the TB-infected group compared to the uninfected group (Figure 1B–F and Supplementary Figure 1A). Of those, IP-10, MCP-1, and IL-15 displayed the least amount of overlap between the uninfected subjects and the TB-infected groups. IL-10 was also identified as being differentially secreted between TB infected and uninfected subjects, but did not meet Bonferroni-corrected significance (corrected for 50 comparisons, so significant p<0.001) (Supplementary Figure 1B). Log-transformed p-values obtained as a result of this analysis are displayed in Figure 1G (blue bars). The complete list of biomarkers and comparison of median values between TB-infected and uninfected persons is in Supplementary Table 1.
Active Versus Latent TB
Next we sought to determine if any biomarkers were differentially expressed between active and latent TB patients with a Wilcoxon rank-sum test. As expected, IFN-γ was not significantly differentially expressed between infection statuses (Figure 1A). Similarly, IP-10, MIG, IL-2 and IL-10 were also not differentially expressed between the active and latent TB groups (Figure 1B–D and Supplementary Figure 1B). Using a Bonferroni-corrected significance threshold for significance (p<0.001), we observed that IL-15 was the only biomarker able to segregate subjects with latent and active TB (Figure 1F). Using a less strict significance cutoff (p<0.01) four biomarkers were identified as having differential secretion in response to TB antigen stimulation in subjects with latent TB compared to those with active TB: MCP-1, IL-1Ra, IFN-α, and IL-4 (Figure 1E and Supplementary Figures 1A, C, and D). We report the p-values for comparisons of all 25 biomarkers’ median production in latently infected patients to that in actively infected patients in Figure 1G (red bars) and Supplementary Table 1.
Although IL-1Ra, IL-4, IL-15, IFN-α, and MCP-1 responses were quantitatively different between the latent and active TB groups, there was still considerable overlap in biomarker responses, as shown in Figures 1A through F. Conditional inference trees were therefore used to explore whether a combination of responses could improve discrimination between the uninfected, latent TB, and active TB groups. A model utilizing MCP-1 followed by IP-10 (Supplementary Figure 2) successfully identified all of the active TB subjects, but also misidentified 23 latent TB subjects and 2 uninfected subjects as having an active infection. In addition, this model misclassified 2 TB uninfected subjects as having a latent TB infection and 3 latent TB subjects as being uninfected.
Since current diagnostic methods perform well at identifying the TB negative individuals we hypothesized that removing this group from consideration for the inference trees would generate a clinically more relevant model. When only the responses of the latent and active TB groups were analyzed, IL-15 response alone was the best discriminator between latent and active TB (Supplementary Figure 3). This model outperformed the previous inference model, reducing the misclassified latent TB subjects from 23 to 7 but it also raised the misclassification of active TB subjects from 0 to 1.
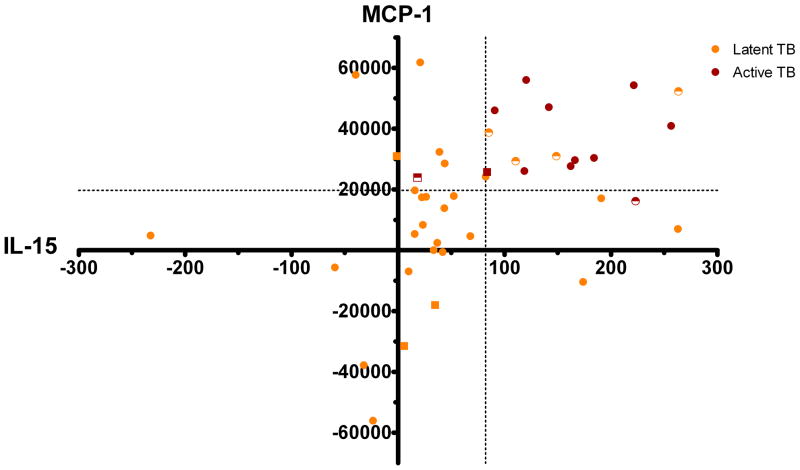
To verify our original hypothesis that combinations of multiple biomarkers could correctly segregate individuals with active TB disease from persons with latent TB infection, we examined the performance of all pairs of biomarker responses that significantly differed between the active and latent TB groups (IL-1Ra, IL-4, IL-15, IFN-α, and MCP-1). This method determined that the combination of an MCP-1 response greater than or equal to 19,696 pg/mL and an IL-15 response greater than or equal to 82 pg/mL achieved the greatest overall accuracy in identifying patients with latent versus active TB (Figure 2). In fact, by using this combination we correctly assigned 38/44 (86.4%) of subjects with either latent or active TB to the correct disease states. The sensitivity and specificity of this two-biomarker combination for active TB (vs. latent TB) were 83% and 88%, respectively. Receiver operating characteristic curves for this combination are shown in Supplementary Figure 4.
Figure 2. Differentiation of active and latent TB subjects.
Simultaneously analyzing IL-15 and MCP-1 concentrations allows the effective separation of active and latent TB disease states. Subjects with active TB are represented by red shapes, and those with latent TB as orange shapes. Using cutoffs of 82pg/mL and 19696pg/mL for IL-15 and MCP-1 respectively, 10/12 (83%) active TB subjects ( ) and 28/32 (88%) latent TB subjects (
) and 28/32 (88%) latent TB subjects ( ) were correctly identified. Incorrectly identified subjects are shown as half-shaded shapes. HIV positive subjects are denoted by squares.
) were correctly identified. Incorrectly identified subjects are shown as half-shaded shapes. HIV positive subjects are denoted by squares.
Of note, there was no significant correlation in the active tuberculosis group between time elapsed since the start of antituberculous treatment and biomarker response for interferon gamma or any of the biomarkers in the models above. The Spearman correlations and p-values between time since start of treatment and biomarker response were r=0.44 (p=0.15) for interferon gamma, r=0.13 (p=0.69) for IP-10, r=−0.02 (p=0.95) for IL-15, and r=−0.36 (p=0.25) for MCP-1.
Discussion
This study suggests that several biomarker responses to TB antigens may be useful for the detection of TB infection as well as for differentiating active and latent TB disease. Of particular promise in detecting TB infection was IP-10. This is consistent with previous studies which found locally high levels of IP-10 in pleural effusions of TB patients as well as within indurations resulting from a positive TST reaction 14–15. In addition, our results confirm recent observations by Ruhwald and collaborators of IP-10 as a marker for TB infection 16–17. Of note, the optimal cutoff for IP-10 response found by Ruhwald et al. (455 pg/ml) did not perform quite as well in our subject population (sensitivity 70.5%, specificity 96.2% in discriminating infected vs. uninfected subjects) as a lower cutoff (200 pg/ml) (sensitivity 84.1%, specificity 96.2%), illustrating that optimal cutoffs will need to be determined from more large, prospective studies. Our results are also in agreement with a smaller study that compared biomarker responses after stimulation in 8 TB patients to 7 healthy controls. The Quantiferon® Gold In-Tube system was also used for that study, which found increased MCP-1, MCP-3, IL-1Ra, and IP-10 responses to TB antigen stimulation among TB patients compared with controls 18. While immunosuppressed individuals often do not mount a detectable IFN-γ response to TB antigen stimulation, the biomarkers identified in our study may be produced at detectable levels in such patients 6, 19. In fact, the four HIV+ TB+ patients in our study mounted IP-10 responses greater than all but one TB-uninfected patient. These limited data support further study of IP-10 response to TB antigens as a potential diagnostic in immunosuppressed populations.
More interestingly, we also identified a biomarker combination which shows promise in distinguishing latent from active TB. Specifically, the combined analysis of IL-15 and MCP-1 responses accurately identified 86% of active and latent TB patients. This combined IL-15 and MCP-1 response pattern could either be a marker for the presence of active TB or instead could represent a response associated with greater susceptibility to development of active TB after infection. Most of our subjects with active TB were sampled after treatment completion, so it is impossible to determine from our data whether these responses might be dynamically influenced by the treatment.
Our study suffers from a number of limitations, including relatively small sample size and a hetereogeneous subject population. In particular, subjects in both the latent and the active tuberculosis group were at different timepoints after the start of treatment. Biomarker responses to the antigens used could potentially vary during and after treatment, but this effect has not been consistent in the literature.20–22 Additionally, there is no gold standard test for latent tuberculosis; all of the available tests have limitations, and discordant results from the same patient are the norm.9, 23 We used standard definitions of latent tuberculosis, but there was heterogeneity in this group (some had positive tuberculin skin tests with negative Quantiferon® tests, others did not have skin testing performed and only had positive Quantiferon® tests) as well. Such heterogeneity is most likely to bias our results toward the null due to increased measurement variability, but we cannot dismiss the potential for spurious associations between biomarker responses and disease group based on misclassification or heterogeneity. Furthermore, the receiver operating characteristic curves are based on a training dataset alone, which likely overestimates model performance. We did not examine a number of potential biomarkers such as EGF or soluble CD40 ligand that have demonstrated potential to discriminate latent from active tuberculosis in other studies; at least one biomarker that seemed promising for this purpose in another study (MIP-1β) did not perform well in our study.24
The particular biomarker responses associated with active TB disease in this study were not surprising given previous investigators’ findings. In vitro studies have shown that IL-15 upregulates the antimicrobial protein cathelicidin, leading to decreased M. tuberculosis survival. 25 Furthermore, Rausch and colleagues have shown that IL-15 is required for proper CD8+ T-cell accumulation in the lungs and therefore increased mortality in knockout mice following M. tuberculosis infection 26. In addition to IL-15, our study identified increased MCP-1 production as an indicator of active TB. Multiple studies indicate high MCP-1 production may be detrimental to the host immune response 27. Included in these findings was discovery of a single nucleotide polymorphism (SNP) in the MCP-1 promoter which correlated with increased MCP-1 expression and increased susceptibility to active TB disease 28. While this finding held true in Mexican, Korean and Peruvian patient populations the SNP had no effect in a Russian population and even correlated with protection in Ghanaian patients 29–30. Additional genetic and/or environmental factors are probably important for determining the role of MCP-1 in fighting TB disease. In light of these contradictory findings it is also important that the present study be extended to diverse TB populations around the world. One possible interpretation is that MCP-1 production in response to M. tuberculosis predisposes certain patient populations to develop active TB, and subsequent to the onset of active TB a strong IL-15 response is mounted. A proper longitudinal study would be needed to determine if IL-15 and MCP-1 production appears concurrently, one precedes the other, or if the “active” response pattern heralds active disease or follows its onset. Additional information might be gained by examining biomarker profiles after prolonged incubation, as opposed to the 16–24 hour incubation period used in our study. A recently published study used the same stimulation platform but a longer incubation (72 hours) time, and found that IL-2 supernatant concentrations at 72 hours (but not 18 hours) discriminated latent from active TB patients 31. This suggests that the simultaneous monitoring of these significant biomarkers may also provide insights into the spectrum of immune responses across the population. Distinct portions of this spectrum (e.g. strong IP-10 response combined with a weak IFN-γ response) may have prognostic significance for pathogenic outcomes such as progression from latent to active disease, extrapulmonary spread of TB, probability of reinfection and/or treatment efficacy. Therefore, it is important that our findings form the basis for a larger study featuring diverse well-defined patient populations who are monitored longitudinally.
Supplementary Material
Acknowledgments
We thank Jeff Hale and Dr. Greg Sempowski for their support of the Luminex assays through the Duke Human Vaccine Institute’s Immune Reconstitution and Biomarker Analysis core facility. This work was funded by a developmental grant from the Duke Center for AIDS Research (CFAR), an NIH funded program P30AI64518. Guido Ferrari, Marc Frahm and Kouros Owzar acknowledge salary support from NIH grant AI050483-05A1. Emily Hecker acknowledges salary support from NIH grant N0140082. Payam Nahid acknowledges salary support from NIH grant K23HL092629. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 1999;282(7):677–86. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 2.World Health O. Global tuberculosis control 2009 - epidemiology, strategy, financing. 2009 Available from: www.who.int/tb/publications/global_report/2009/pdf/full_report.pdf.
- 3.Harries AD, Dye C. Tuberculosis. Annals of Tropical Medicine & Parasitology. 2006;100(5/6):415–31. doi: 10.1179/136485906X91477. [DOI] [PubMed] [Google Scholar]
- 4.Sutherland I, Svandová E, Radhakrishna S. The development of clinical tuberculosis following infection with tubercle bacilli: 1. A theoretical model for the development of clinical tuberculosis following infection, linking from data on the risk of tuberculous infection and the incidence of clinical tuberculosis in the Netherlands. Tubercle. 1982;63(4):255–68. doi: 10.1016/s0041-3879(82)80013-5. [DOI] [PubMed] [Google Scholar]
- 5.Nahid P, Pai M, Hopewell PC. Advances in the Diagnosis and Treatment of Tuberculosis. Proc Am Thorac Soc. 2006;3(1):103–10. doi: 10.1513/pats.200511-119JH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pesanti EL. The negative tuberculin test. Tuberculin, HIV, and anergy panels. American Journal of Respiratory and Critical Care Medicine. 1994;149:1699–709. doi: 10.1164/ajrccm.149.6.7710481. [DOI] [PubMed] [Google Scholar]
- 7.Mori T. Usefulness of interferon-gamma release assays for diagnosing TB infection and problems with these assays. Journal of Infection and Chemotherapy. 2009;15(3):143–55. doi: 10.1007/s10156-009-0686-8. [DOI] [PubMed] [Google Scholar]
- 8.Diel R, Loddenkemper R, Meywald-Walter K, Niemann S, Nienhaus A. Predictive value of a whole blood IFN-gamma assay for the development of active tuberculosis disease after recent infection with Mycobacterium tuberculosis. Am J Respir Crit Care Med. 2008;177(10):1164–70. doi: 10.1164/rccm.200711-1613OC. [DOI] [PubMed] [Google Scholar]
- 9.Mandalakas AM, Hesseling AC, Chegou NN, Kirchner HL, Zhu X, Marais BJ, et al. High level of discordant IGRA results in HIV-infected adults and children. The International Journal of Tuberculosis and Lung Disease. 2008;12:417–23. [PubMed] [Google Scholar]
- 10.Anonymous. Diagnostic Standards and Classification of Tuberculosis in Adults and Children. American Journal of Respiratory & Critical Care Medicine. 2000;161(4 Pt 1):1376–95. doi: 10.1164/ajrccm.161.4.16141. [DOI] [PubMed] [Google Scholar]
- 11.Hájek J, Šidák Ze Sen PK. Theory of rank tests. 2. San Diego, Calif: Academic Press; 1999. [Google Scholar]
- 12.Hothorn T, Hornik K, Zeileis A. Unbiased Recursive Partitioning. Journal of Computational and Graphical Statistics. 2006;15(3):651–74. [Google Scholar]
- 13.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2010. [Google Scholar]
- 14.Okamoto M, Kawabe T, Iwasaki Y, Hara T, Hashimoto N, Imaizumi K, et al. Evaluation of interferon-gamma, interferon-gamma-inducing cytokines, and interferon-gamma-inducible chemokines in tuberculous pleural effusions. J Lab Clin Med. 2005;145(2):88–93. doi: 10.1016/j.lab.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan GLA, Hancock G, Cohn ZA. The expression of a gamma interferon-induced protein (IP-10) in delayed immune responses in human skin. J Exp Med. 1987;166(4):1098–108. doi: 10.1084/jem.166.4.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruhwald M, Bjerregaard-Andersen M, Rabna P, Kofoed K, Eugen-Olsen J, Ravn P. CXCL10/IP-10 release is induced by incubation of whole blood from tuberculosis patients with ESAT-6, CFP10 and TB7.7. Microbes and Infection. 2007;9(7):806–12. doi: 10.1016/j.micinf.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 17.Ruhwald M, Bodmer T, Maier C, Jepsen M, Haaland MB, Eugen-Olsen J, et al. Evaluating the potential of IP-10 and MCP-2 as biomarkers for the diagnosis of tuberculosis. Eur Respir J. 2008;32(6):1607–15. doi: 10.1183/09031936.00055508. [DOI] [PubMed] [Google Scholar]
- 18.Ruhwald M, Bjerregaard-Andersen M, Rabna P, Eugen-Olsen J, Ravn P. IP-10, MCP-1, MCP-2, MCP-3, and IL-1RA hold promise as biomarkers for infection with M. tuberculosis in a whole blood based T-cell assay. BMC Research Notes. 2009;2(1):19. doi: 10.1186/1756-0500-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goletti D, Raja A, Syed Ahamed Kabeer B, Rodrigues C, Sodha A, Carrara S, et al. Is IP-10 an accurate marker for detecting M. tuberculosis-specific response in HIV-infected persons? PLoS ONE. 2010;5(9):e12577. doi: 10.1371/journal.pone.0012577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adetifa IM, Ota MO, Walther B, Hammond AS, Lugos MD, Jeffries DJ, et al. Decay kinetics of an interferon gamma release assay with anti-tuberculosis therapy in newly diagnosed tuberculosis cases. PLoS ONE. 2010;5:9. doi: 10.1371/journal.pone.0012502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pai M, Joshi R, Bandyopadhyay M, Narang P, Dogra S, Taksande B, et al. Sensitivity of a whole-blood interferon-gamma assay among patients with pulmonary tuberculosis and variations in T-cell responses during anti-tuberculosis treatment. Infection. 2007;35(2):98–103. doi: 10.1007/s15010-007-6114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chee CB, KhinMar KW, Gan SH, Barkham TM, Koh CK, Shen L, et al. Tuberculosis treatment effect on T-cell interferon-gamma responses to Mycobacterium tuberculosis-specific antigens. Eur Respir J. 2010;36(2):355–61. doi: 10.1183/09031936.00151309. [DOI] [PubMed] [Google Scholar]
- 23.Hesseling AC, Mandalakas AM, Kirchner HL, Chegou NN, Marais BJ, Stanley K, et al. Highly discordant T cell responses in individuals with recent exposure to household tuberculosis. Thorax. 2009;64(10):840–6. doi: 10.1136/thx.2007.085340. [DOI] [PubMed] [Google Scholar]
- 24.Chegou NN, Black GF, Kidd M, van Helden PD, Walzl G. Host markers in QuantiFERON supernatants differentiate active TB from latent TB infection: preliminary report. BMC Pulm Med. 2009;9:21. doi: 10.1186/1471-2466-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krutzik SR, Hewison M, Liu PT, Robles JA, Stenger S, Adams JS, et al. IL-15 Links TLR2/1-Induced Macrophage Differentiation to the Vitamin D-Dependent Antimicrobial Pathway. J Immunol. 2008;181(10):7115–20. doi: 10.4049/jimmunol.181.10.7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rausch A, Heßmann M, Hölscher A, Schreiber T, Bulfone-Paus S, Ehlers S, et al. Interleukin-15 mediates protection against experimental tuberculosis: A role for NKG2D-dependent effector mechanisms of CD8+ T cells. European Journal of Immunology. 2006;36(5):1156–67. doi: 10.1002/eji.200535290. [DOI] [PubMed] [Google Scholar]
- 27.Lee JS, Song CH, Lim JH, Lee KS, Kim HJ, Park JK, et al. Monocyte chemotactic protein-1 production in patients with active pulmonary tuberculosis and tuberculous pleurisy. Inflammation Research. 2003;52(7):297–304. doi: 10.1007/s00011-003-1176-6. [DOI] [PubMed] [Google Scholar]
- 28.Flores-Villanueva PO, Ruiz-Morales JA, Song C-H, Flores LM, Jo E-K, Montano M, et al. A functional promoter polymorphism in monocyte chemoattractant protein-1 is associated with increased susceptibility to pulmonary tuberculosis. J Exp Med. 2005;202(12):1649–58. doi: 10.1084/jem.20050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thye T, Nejentsev S, Intemann CD, Browne EN, Chinbuah MA, Gyapong J, et al. MCP-1 promoter variant -362C associated with protection from pulmonary tuberculosis in Ghana, West Africa. Hum Mol Genet. 2009;18(2):381–8. doi: 10.1093/hmg/ddn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganachari M, Ruiz-Morales JA, Gomez de la Torre Pretell JC, Dinh J, Granados J, Flores-Villanueva PO. Joint Effect of MCP-1 Genotype GG and MMP-1 Genotype 2G/2G Increases the Likelihood of Developing Pulmonary Tuberculosis in BCG-Vaccinated Individuals. PLoS ONE. 2010;5(1):e8881. doi: 10.1371/journal.pone.0008881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biselli R, Mariotti S, Sargentini V, Sauzullo I, Lastilla M, Mengoni F, et al. Detection of interleukin-2 in addition to interferon-gamma discriminates active tuberculosis patients, latently infected individuals, and controls. Clin Microbiol Infect. 2010;16(8):1282–4. doi: 10.1111/j.1469-0691.2009.03104.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.