Abstract
Yeast lacking copper-zinc superoxide dismutase (sod1Δ ) have a number of oxygen-dependent defects, including auxotrophies for lysine and methionine and sensitivity to oxygen. Here we report additional defects in metabolic regulation. Under standard growth conditions with glucose as the carbon source, yeast undergo glucose repression where mitochondrial respiration is de-emphasized, energy is mainly derived from glycolysis, and ethanol is produced. When glucose is depleted, the diauxic shift is activated, in which mitochondrial respiration is re-emphasized and stress resistance increases. We find that both of these programs are adversely affected by the lack of Sod1p. Key events in the diauxic shift do not occur and sod1Δ cells do not utilize ethanol and stop growing. The ability to shift to growth on ethanol is gradually lost as time in culture increases. In early stages of culture, sod1Δ cells consume more oxygen and have more mitochondrial mass than wild-type cells, indicating that glucose repression is not fully activated. These changes are at least partially dependent on the activity of the Hap2,3,4,5 complex, as indicated by CYC1-lacZ reporter assays. These changes may indicate a role for superoxide in metabolic signaling and regulation and/or a role for glucose derepression in defense against oxidative stress.
Keywords: Oxidative stress, superoxide, ROS, yeast, glucose repression, diauxic shift, Sod1p, copper-zinc superoxide dismutase
INTRODUCTION
Although the metabolic pathways used by eukaryotic organisms are generally very similar, some are quite specific to the type of organism. For example, wild-type (WT) yeast (Saccharomyces cerevisiae) grown in batch cultures under standard conditions (in 2% glucose) typically go through three phases—log phase, diauxic shift, and stationary phase—and the expression of the respiratory machinery is initially repressed in a glucose-dependent fashion (glucose repression), favoring energy generation via fermentation as long as glucose is abundant. This log phase growth rate is rapid, and most of the glucose is converted, via glycolysis, to ethanol, which is secreted rather than being fully metabolized via mitochondrial respiration. As the level of glucose drops, the diauxic shift occurs: Glucose-mediated repression of respiratory components is lifted (derepression), synthesis of hundreds of proteins involved in mitochondrial respiration machinery is upregulated, respiratory metabolism of the previously produced ethanol becomes the primary mode of ATP generation, and the overall growth rate becomes much slower. Finally, when nutrients are fully exhausted, the yeast culture enters a quiescent stationary phase with very low energy generation (or utilization) and no growth [1,2]. The metabolism of growing yeast can be further manipulated and/or tested by providing alternate carbon sources instead of glucose. Raffinose, a trisaccharide of galactose, fructose, and glucose, is nonrepressing but fermentable. Thus, the respiratory apparatus is induced in the culture even at very low cell density, but fermentation is also active. Cells with defective respiration can still utilize raffinose to grow, although they grow more slowly than they do on glucose. Pyruvate, lactate and ethanol cannot be metabolized through glycolysis and thus require functional respiration to support growth. Again, growth is slower than on glucose.
These changes, that begin with sensing of glucose levels via the SNF3/RGT2 complex in the plasma membrane [reviewed in [3,4], are regulated through a number of pathways that direct yeast responses to carbon source shifts. Perhaps the earliest such pathways to be discovered were those regulated by Hap1 and the Hap2,3,4,5 complex. These transcription factors are part of the cascade of responses that results in glucose de-repression in response to oxygen and carbon source changes, respectively, and exert their effects by binding to specific promoter sequences (UASes) and stimulating transcription of regulated genes. These genes include many components of the respiratory machinery, but the response has been particularly well characterized for the cytochrome c promoter, which is often used as an indicator of changes in these pathways. Reviewed in [5,6].
Copper-zinc superoxide dismutase (Sod1p) is one of the principal defenses against superoxide stress in eukaryotic cells, particularly in the cytoplasm and the mitochondrial intermembrane space. When Sod1p is inactivated in S. cerevisiae, detrimental phenotypes emerge across many key cellular processes, including amino acid biosynthesis, cation storage and metabolism, and general cellular fitness[7]. 4Fe-4S clusters are particularly susceptible to damage by the superoxide anion, and the activity of enzymes with exposed 4Fe-4S clusters (e.g., aconitase and homoaconitase) is inhibited by elevated levels of superoxide, although expression of these enzymes apparently remains unaltered [8]. Most obvious among the metabolic defects exhibited by sod1Δ yeast is a growth defect, which is observed to different degrees in different media. sod1Δ yeast exhibit impaired aerobic growth under fermentative conditions, and, although they can grow via respiration under some conditions, this growth is often not nearly as robust as in the WT strain and varies depending on the carbon source. sod1Δ yeast grow better on some nonfermentable carbon sources (lactate, pyruvate) than on others (ethanol, glycerol) for reasons that are not clear. sod1Δ yeast also show increased oxygen consumption in log phase and shorter survival times in stationary phase [9].
We began our study with a closer examination of the nature of some of the defects observed in sod1Δ yeast, focusing on the stages of a typical batch culture in glucose. We find defects in some basic metabolic pathways in sod1Δ yeast, especially as they pertain to normal fermentative and respiratory function and the switch between them at the diauxic shift. Additionally, we find an increase in oxygen consumption and in log phase mitochondrial mass in sod1Δ yeast.
MATERIALS AND METHODS
Yeast Strains, Plasmids, Media and Growth Conditions
The yeast strains used in this study: EG103 (Matα, leu2-3, 112, his3Δ 1, trp1-289a, ura3-52), referred to as WT, and EG118 (EG103 with sod1Δ::URA3), referred to as sod1Δ. In the last figure only, EG314 (EG103 with sod1Δ::KanR) was used instead of EG118 as the sod1Δ strain to enable selection for URA3 gene-containing plasmids. Unless otherwise indicated, cells were grown in synthetic complete (SC) medium with 2% glucose (SDC), raffinose (SRC) or ethanol (SEC) as described [10], except that the supplements Leu, His, Trp, Met, Ura and Ade were increased 4-fold. Before each experiment, frozen stocks were streaked on rich (YPD) plates and grown in an atmosphere of 95% nitrogen, 5 % oxygen for 3 days. Overnight cultures were grown from a single colony in SDC for approximately 20 hours. For all experimental samples, unless otherwise noted, the initial pH of the medium was 6.0, the growth temperature was 30 degrees C, and cultures were well aerated by shaking at 220 rpm in flasks in which the ratio of flask volume to medium volume was 5:1. Growth was followed by measuring the optical density at 600 nm (OD600) and each flask was inoculated at a starting OD600 of 0.1 (1 × 106 cells/ml). For oxygen consumption experiments, cells were inoculated as above, grown until they reached an OD600 of 1, then diluted 1:10 in fresh medium for further growth.
The reporter constructs pLG669Z, carrying the full CYC1 promoter fused to the lacZ gene, and pLG669ZΔAX, carrying the same gene with the binding site for Hap2,3,4,5 mutated, were kindly provided by L. Guarente [11,12]. For lacZ assays, plasmid-containing cells were inoculated at OD600 of 0.05 in SC-uracil with the indicated carbon source and cultured to an OD600 to 0.5–1.0 (early exponential phase) unless otherwise stated.
Measurement of ethanol and glucose in medium
Cells were grown in SDC medium (2% glucose), as described above. Every two to four hours, a small aliquot of spent medium was analyzed. Ethanol concentration was measured according to Beutler [14]. Briefly, NAD+, alcohol dehydrogenase, and aldehyde dehydrogenase were added, and formation of NADH, which is proportional to the amount of ethanol present, was followed at 340 nm. Glucose was measured by the end-point method as described [15]. Briefly, ATP, magnesium phosphate, hexokinase, glucose-6-phosphate dehydrogenase, and NADP+ were added to a small volume of the spent medium. The final absorbance at 339 nm is proportional to the amount of glucose present in the medium.
Measurement of glycogen
Glycogen contents were measured as described by Sillje et al [16]. Briefly, cells were grown in SDC medium as described above. Every two to four hours a sample containing 5 × 107 cells was harvested and washed in ice-cold water. The pellet was suspended in 1 ml of 0.25 M Na2CO3 at 60°C and placed in boiling water. After 2 hours, 450 μl was removed for glycogen determination. The sample was acidified, digested with amyloglucosidase, neutralized, and centrifuged. The liberated glucose was determined as described above.
Oxygen Consumption
Molecular oxygen consumption was measured with a model 5300 Biological Oxygen Monitor and Clark-type oxygen electrode (Model YSI 5300, Yellow Springs, OH), following the manufacturer’s protocols. Aliquots of cells were transferred into a sealed chamber containing 5.0 ml of air-saturated spent medium, magnetically stirred, and maintained at 30 °C. The oxygen electrode response was recorded every ten seconds, and cellular oxygen consumption was calculated as the rate of oxygen loss from the sample per 107 cells.
Flow cytometry
Cell cultures were grown to an OD600 of 0.5 (log phase) in SDC medium and diluted in spent medium to an OD600 of 0.1 (about 1×106 cells/ml). Flow cytometry was carried out as described [17]. Briefly, 50 nM Rhodamine 123 (Rh123), a fluorescent mitochondrial probe, was added and cells were incubated for 10 minutes at room temperature with occasional shaking to allow cellular dye content to reach steady state. Samples were analyzed on a FACSCalibur flow cytometer (BDIS) equipped with a 15 mW air-cooled 488 nm argon-ion laser for excitation of Rh123. Green Rh123 fluorescence data was collected after a 530/30 nm band pass filter. Forward scatter, side scatter and fluorescence data were all displayed on four-decade log scales. A minimum of 20,000 events was collected for each sample. Analysis of the multivariate data was performed with CELLQuest™ software (BDIS). Values represent the average of 3 separate colonies with error bars indicating the standard deviation. This experiment was repeated at least five different times.
Western blotting
Protein extracts were prepared from whole cells according to Yaffe and Schatz [18]. Briefly, cell pellets were lysed in 1.85 M NaOH, 7.4% β-mercaptoethanol. Proteins were precipitated with trichloroacetic acid (TCA), washed in ethanol, and resuspended in 4% sodium dodecylsulfate (SDS), 0.16 M Tris-HCl, ph 6.8. Samples were boiled for 5 minutes in loading buffer containing β-mercaptoethanol, resolved by 12% SDS-PAGE under reducing conditions and transferred to nitrocellulose membranes. The membranes were blocked in 10% dry milk, 0.1% Tween-20, PBS (PBST), incubated with primary antibody for 2 hours at room temperature with rocking in blocking buffer, washed three times, incubated with horseradish peroxidase-conjugated secondary antibodies for 1 hour in blocking buffer, and washed three times with PBST and twice with PBS. The ECL kit (Amersham Biosciences) was used for detection. Polyclonal antibodies against porin and cytochrome c oxidase were kindly provided by C. Koehler (UCLA). Antibody against actin was purchased from Santa Cruz Biotechnology, Inc.
Quantitative real-time PCR
Cells lysates were prepared by glass bead lysis, after which total cellular DNA was prepared using the DNeasy kit (Qiagen). Primers for PCR were designed with Primer3 software (http://frodo.wi.mit.edu), and checked by nucleotide BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/) to avoid cross-reactivity. The primer sequences were as follows: ATP9, 5′ CCATA CTCAT TTGCA TTATC AGCTC 3′ (forward) and 5′ CAGCA GGTAC GAATA ATGAG AAGAA 3′ (reverse); COX3, 5′ TGTTG ACCAC CCGTA GGTAT TG 3′ (forward) and 5′ ACTGA ACCAT AAACA CCATC AGAGA 3′ (reverse); FRE1, 5′ ATTTG AGAGC ACCTG AGAAA AGTGA 3′ (forward) and 5′ AGTAAGCACAGCCACCCAGAAG 3′ (reverse); VMA1, 5′ GCTAC CTACC AGACT TACGC TCCA 3′ (forward) and 5′ CAGAC AATCC ATCAC CAATC CA 3′ (reverse). The PCR mixture contained 0.25 pmol of 5′ and 3′ primers, an empirically optimized amount of total DNA, and was carried out using taq DNA polymerase with QuantiTect SYBR Green PCR kit (Qiagen) under the following conditions: 95°C for 2 min, 96°C for 20 s, 55°C for 45 s, and 72°C for 45 s, for a total of 41 cycles in an ABIPrism 7000 sequence detector system (Applied Biosystems, Foster City, CA). Data are reported as the relative ratio of each mitochondrial gene to each nuclear gene for WT and sod1Δ as determined by fluorescence measurements.
Cyc1 promoter activity
β-Galactosidase reporter assays were performed using a plate reader, with reagents as described [19]. Kinetic readings were taken over a period of 1–2 hours, and the initial slope (representing the initial velocity of the reaction and therefore enzyme activity) was calculated from the linear portion of the graph. Protein concentration was determined by the Bradford assay as described in [8] and β-galactosidase activity was calculated using the formula: β-galactosidase activity = (1000 x initial slope)/[(ml extract assayed) x (mg/ml protein conc.)]. All β-galactosidase reporter assay experiments include three independent samples each for WT and sod1Δ yeast strain.
RESULTS
sod1Δ yeast exhibit several features indicative of a disruption in normal metabolism
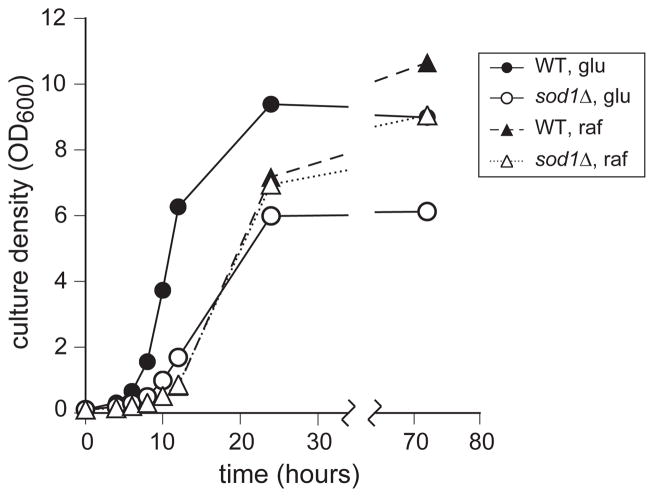
Two of the predominant phenotypes of sod1Δ yeast are observed when they are grown on standard glucose-containing medium. In the typical growth curve shown in Figure 1, it can be seen that sod1Δ yeast (1) grow more slowly than WT in the early phase of the culture and (2) stop growing at a significantly lower cell density. By contrast, their growth on medium containing the nonrepressing sugar, raffinose, as the carbon source was much more similar to growth of WT on the same medium (Figure 1).
Figure 1.
Growth of sod1Δ and wild yeast in glucose or raffinose. WT (EG103) (filled symbols) and sod1Δ (EG118) (empty symbols) were grown in SC medium with 2% glucose (circles and solid lines) or raffinose (triangles and broken lines) as the carbon source. At the indicated times the turbidity of the culture was measured as OD600 as an indication of growth. Data represent an average of 3 independent colonies, errors were 5% or less, and, for clarity, are not shown. This experiment was repeated at least five times, with similar results.
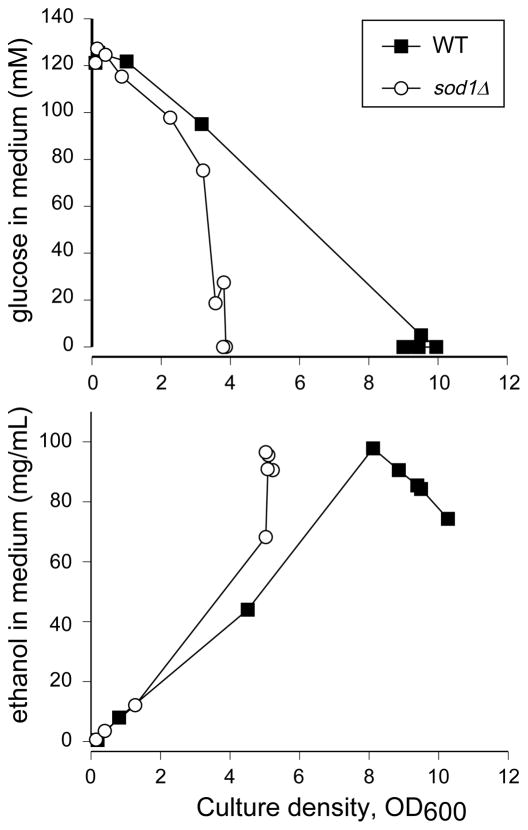
To examine the reasons for the poor growth in glucose, we began with an examination of the glucose content of the spent medium at various time points, as a measure of the ability of WT and sod1Δ yeast to take up glucose (Figure 2A). In these figures, glucose concentration is plotted versus cell density, rather than time, to correct for the slower growth rate of sod1Δ cells. As the data in Figure 2A demonstrate, the concentration of glucose in the medium is lower for sod1Δ cultures than for WT at each given optical density reading, possibly indicating less efficient use of glucose in the mutant strain.
Figure 2.
Glucose consumption and ethanol production by WT and sod1Δ yeast. WT (EG103) and sod1Δ (EG118) cells were grown in SC media at pH 6.0 containing 2% glucose. Every 2 to 4 hours, a small aliquot of the culture was removed and the A. glucose content or B. ethanol content of the medium was measured as described in Materials and Methods. Because the sod1Δ cells grow more slowly than WT, the data are plotted versus culture density rather than time. Experiments were repeated at least twice with similar results. A representative one is shown. Closed squares, WT; open circles, sod1Δ.
Under fermentative growth, yeast produce ethanol from glucose, so we next tested the ability of the strains to metabolize glucose by measuring the ethanol content of the medium at different time points. Ethanol content of the medium from both WT and sod1Δ cultures was observed to be low initially and to increase with cell density (Figure 2B), due to the fermentative conversion of glucose to ethanol. This observed ethanol buildup was similar in WT and sod1Δ yeast at early stages, indicating a normal ability to produce ethanol. However, at later stages, the ethanol content of the medium from WT cultures peaked and then began to decrease as the cell density continued to increase. By contrast, the ethanol content of spent medium from sod1Δ cultures reached a maximum but then remained unchanged and the culture failed to increase in density. Thus, it appears that the sod1Δ yeast do not utilize the ethanol they have produced and do not grow further.
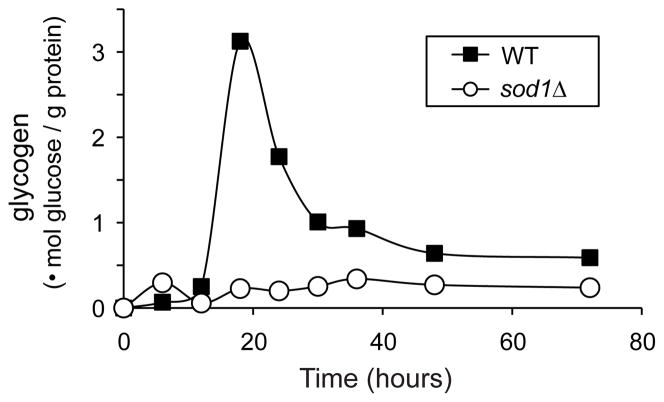
At the end of log phase, yeast store glucose in the form of glycogen, a process that is considered a hallmark of the diauxic shift, in order to provide energy during the diauxic phase. We therefore measured the glycogen content of WT and sod1Δ cells at various time points during growth on glucose. WT yeast accumulated (and then used) glycogen as expected, while sod1Δ cells did not accumulate glycogen at all (Figure 3). These data prompted us to ask whether sod1Δ cells have difficulty undergoing the diauxic shift.
Figure 3.
sod1Δ cells do not accumulate glycogen. WT and sod1Δ cells (EG103 and EG118) were grown in SDC medium. At the indicated times, cells were harvested, digested, and their glycogen content was determined as described in Materials and Methods. Data represent an average of 3 independent colonies and the experiment was repeated at least 3 times.
The diauxic shift is in essence a carbon source change, from glucose to ethanol, i.e., from a fermentable, repressing carbon source to a nonfermentable non-repressing one (ethanol). To determine if this difficulty was specific to glucose and ethanol, or if it applied to other carbon source changes as well, we carried out media-change experiments. Cultures were grown with either glucose (fermentable, repressing) or raffinose (fermentable, non-repressing) as their carbon source for 24 hours and then diluted into medium containing glucose, raffinose, or ethanol (nonfermentable, nonrepressing). sod1Δ cells grown in glucose for 24 hours did not grow after transfer to ethanol, but did grow when transferred to glucose or raffinose (Figure 4). sod1Δ cells initially grown in raffinose for 24 hours were capable of growth when transferred to any of the three tested carbon sources. We next mimicked that change at different stages of culture in order to test the timing of onset of the above phenomenon. WT and sod1Δ cells were grown in glucose and then transferred, at different time points, to medium containing either glucose or ethanol (Figure 4B). sod1Δ cells grew when transferred to ethanol at earlier time points (8 or 18 hours) before or at the diauxic shift, but not at later time points (24 or 48 hours) after diauxic shift or in stationary phase. The same cultures inoculated into fresh medium containing glucose had no difficulty growing. Thus, the sod1Δ mutant strain is capable of growing in non-fermentable carbon sources, but has a defect in its ability to change from repressing to non-fermentable carbon sources at later stages of growth.
Figure 4.
sod1Δ cells are defective in executing the switch from glucose-based to ethanol-based growth. A. sod1Δ cells grown in glucose are unable to switch to growth in ethanol after 24 hours, although they can switch if grown in raffinose. WT and sod1Δ cells were grown in either SDC (bars labeled “from glucose”) or SRC (bars labeled “from raffinose”) for 24 h and were then inoculated into either SDC, SRC, or SEC medium and grown for an additional 72 h before their optical density was measured. B. sod1Δ cultures or WT cultures treated with paraquat are able to switch from growth in glucose to growth in ethanol at earlier stages of growth but not at later stages. WT with or without 1 mM paraquat and sod1Δ cells were grown in SDC medium for the indicated amount of time, and then inoculated into SC medium containing either 2 % glucose or 2% ethanol. After 72 h the OD600 was measured. Data represent the average of 3 independent colonies and each experiment was repeated at least twice.
Intracellular superoxide can be increased chemically by addition of the redox-cycling herbicide, paraquat. Treatment with this compound has been shown to induce all known sod1Δ phenotypes in WT yeast cultures [20], and it is thus a useful control. Carbon source-switching experiments as described above were conducted on WT cells treated with 1 mM paraquat. As with the sod1Δ cultures, paraquat-treated cultures did not grow when transferred at the later time points (Figure 4B), providing evidence that the observed effect is directly related to elevated levels of superoxide [20].
sod1Δ yeast respire at an elevated rate when growing on glucose
In the current work, we found sod1Δ yeast to consume nearly twice as much oxygen as WT starting early in log phase growth. (Figure 5A). Since glucose normally represses the expression of respiratory enzymes, the observed increase in oxygen consumption might indicate that sod1Δ yeast do not undergo proper or full repression, which could explain their slower log phase growth. To test this hypothesis, WT and sod1Δ cells were grown on the nonrepressing sugar raffinose. Both WT and sod1Δ cells grown in raffinose exhibited increased oxygen consumption compared to those grown in glucose, as would be expected (Figure 5B). However, yeast lacking Sod1p still consumed more oxygen than WT. This result suggests that the increase in oxygen consumption seen in sod1Δ cells is not due simply to a defect in glucose repression of the mutant strain.
Figure 5.
Oxygen consumption is elevated in sod1Δ as compared to WT cultures grown in glucose or raffinose as the carbon source. A. WT (EG103, closed triangles) and isogenic sod1Δ yeast (EG118, open squares) were grown in SDC medium until they reached an OD600 of 1.0 and diluted 10-fold into fresh SDC medium (time zero). At the indicated times, oxygen consumption was measured as described in Materials and Methods. Values represent the average of 3 separate colonies with error bars indicating the standard deviation. B. WT (EG103, black bars) and isogenic sod1Δ yeast (EG118, gray bars) were seeded at OD600 of 0.1 in SC medium containing 2% raffinose as the carbon source and grown under standard (high aeration) conditions. When cultures reached an OD600 of 0.5, oxygen consumption was measured. Values represent the average of 3 separate colonies with error bars indicating the standard deviation. Experiments were repeated at least five different times with similar results.
sod1Δ yeast maintain about twice as much mitochondrial mass as compared to WT
The observed increase in oxygen consumption during glucose growth might indicate an overall increase in metabolic rate or simply uncoupling of the mitochondria, leading to more oxygen consumption without more energy generation. We reasoned that increased energy generation would require higher mitochondrial content or an upregulation of the respiratory apparatus, while oxygen consumption due to uncoupling would not. Therefore, we next addressed the question of relative mitochondrial content.
We measured mitochondrial content using a variety of different methods, beginning with the commercially available, mitochondrion-specific fluorescent dye, Rhodamine 123. Rhodamine 123 is imported into the mitochondrial matrix in a membrane potential-dependent fashion; the total amount of dye taken up is proportional to the overall potential across the inner mitochondrial membrane as well as the total volume of mitochondria. Use of this dye allowed us to measure functional mitochondria in WT and sod1Δ strains, quantified in arbitrary units. WT and sod1Δ yeast were cultured under standard conditions and subjected to a 10-minute incubation in the presence of Rhodamine 123. Flow cytometry was performed to measure dye uptake. sod1Δ cells showed about 60% more Rhodamine 123 fluorescence than WT cells (geometric mean of rhodamine fluorescence: 5.7 ± 0.5 for WT compared to 9.1± 0.7 for sod1Δ ). This result could be interpreted as indicating either that the membrane potential of sod1Δ mitochondria is higher than that of WT (which we consider unlikely) or that sod1Δ yeast maintain proportionally more mitochondrial mass.
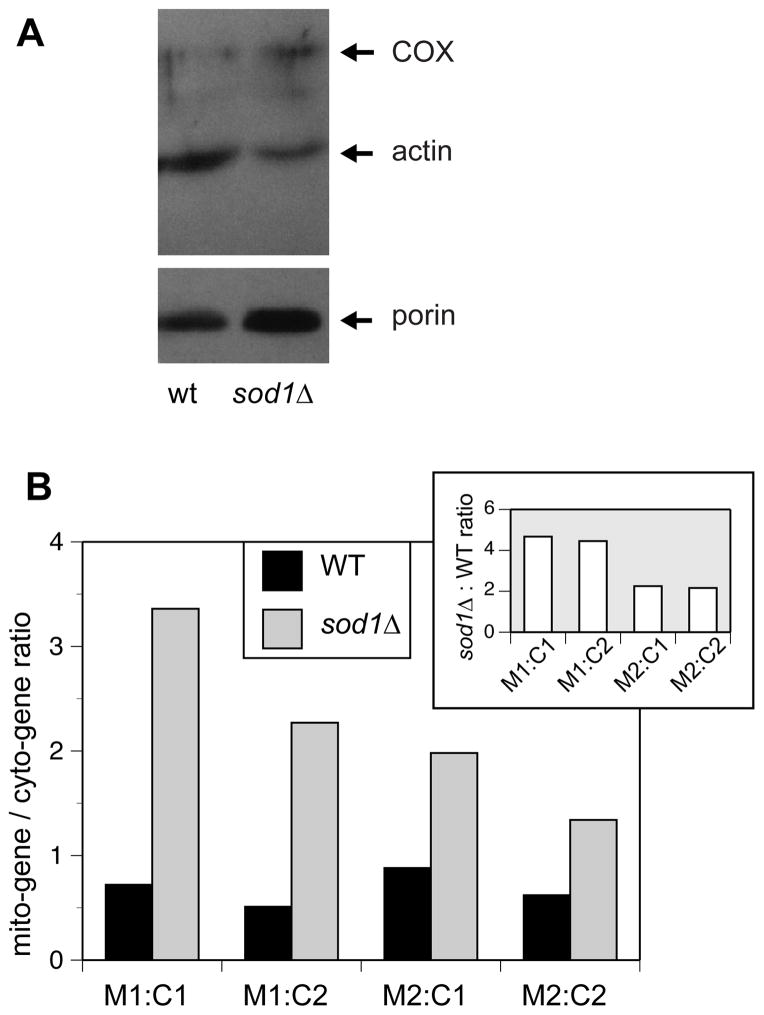
Next, we quantified specific cellular proteins in order to test the hypothesis that sod1Δ yeast do in fact contain more mitochondria. By Western blot analysis, we examined the levels of three proteins: 1) The outer mitochondrial membrane protein, porin, which is commonly used as a mitochondrial marker and whose levels are correlated with mitochondrial content; 2) The inner mitochondrial membrane protein, Cox1, a component of the respiratory complex cytochrome oxidase, also expected to vary according to mitochondrial content; and 3) The ubiquitous cytosolic protein, actin, a cytoskeletal protein whose levels are expected to remain constant under the experimental conditions presented here. Therefore, elevated porin and Cox1 content relative to actin indicates an increase in cellular mitochondrial mass.
Western blot analysis, using antibodies raised against actin, porin and Cox1, was carried out on cells grown in glucose and harvested in early log phase (5 × 106 cells per ml). In all cases, the porin and cytochrome oxidase signals in sod1Δ were higher than those in WT, while the actin signal was similar or reduced in sod1Δ. Thus the ratios of either porin or cytochrome c oxidase to actin were higher in sod1Δ yeast compared to WT, consistent with increased mitochondrial mass in the mutant strain (Figure 6A).
Figure 6.
Relative levels of mitochondrial proteins and DNA indicate a higher mitochondrial mass in sod1Δ yeast. A. Expression of mitochondrial proteins relative to cytosolic ones is increased in sod1Δ. WT (EG103) and sod1Δ (EG118) cells were grown to an OD600 of 0.5 in SDC medium and lysates were prepared as described in “Materials and Methods”. Immunoblotting was performed on equal amounts of lysate protein using antibodies that recognize porin, actin, and cytochrome oxidase (COX1). This experiment was repeated at least three different times with similar results. A representative experiment is shown in which it can be seen that there are increased amounts of mitochondrial proteins porin and COX1 in the sod1Δ yeast relative to WT, while the amount of the cytosolic protein, actin, is slightly decreased. The break indicates different exposures of the same gel. B. The ratio of mitochondrial DNA to nuclear DNA is increased in sod1Δ compared to WT yeast. WT EG103 and isogenic sod1Δ yeast were seeded at OD600 of 0.1 in SDC medium and grown to an OD600 of 0.5 (early exponential phase). Cells were harvested and whole cell DNA was purified as described in Materials and Methods. Real time - PCR on segments of the mitochondrial genes COX3 (M1) and ATP9 (M2) and nuclear genes FRE1 (C1) and VMA1 (C2) was used to quantitate each gene by fluorescence measurements using SYBR Green as the fluorescent reporter. The data are reported as the ratio of each mitochondrial DNA segment to each nuclear DNA segment. This experiment was repeated three times and a representative trial is shown. The inset represents the sod1Δ:WT ratio, showing that sod1Δ cells have a 2 to 4 fold higher ratio of mitochondrial to nuclear DNA.
As a further test, we used real-time quantitative PCR to measure the ratio of nuclear to cytoplasmic DNA in the two strains. Since the ratio of mitochondrial DNA to mitochondrial volume is believed to stay relatively constant, an increase in gene copy number in the mitochondrial genome relative to gene copy number in the nuclear genome would indicate an increase in mitochondrial mass. We chose regions within representative genes, two mitochondrial, ATP9 and COX3, and two nuclear, FRE1 and VMA1, synthesized primers for each gene, and looked at the ratios of pair-wise combinations of the genes by qPCR. Results are reported as ratios of each mitochondrial gene to each nuclear gene for WT and sod1Δ --COX3:FRE1, COX3:VMA1, ATP9:FRE1, and ATP9:VMA1. Our results indicate that the relative ratio of mitochondrial gene to nuclear gene DNA in sod1Δ cells is an average of 3.1±1.1 fold higher than that of WT for all four gene combinations (Figure 6B), again supporting the idea that there are more mitochondria per cell in sod1Δ as compared to WT cells.
sod1Δ yeast show evidence of a defect in glucose repression
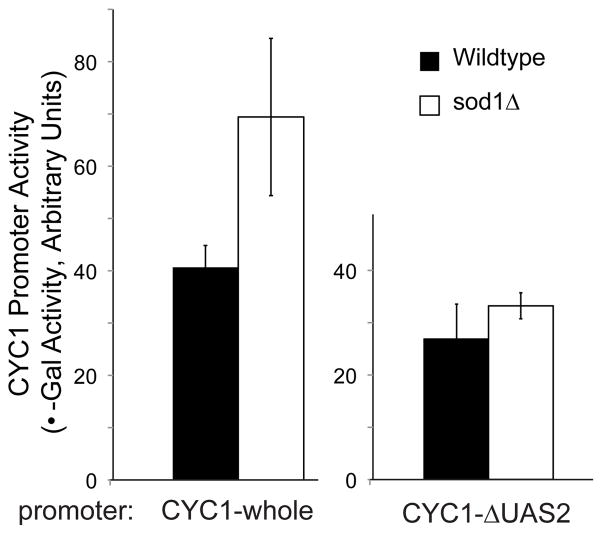
Higher oxygen consumption coupled with increased mitochondrial mass may indicate a defect in glucose repression. If this is the case, we would expect to see increased expression of specific respiratory genes. To test this possibility, we used lacZ assays to examine gene expression driven by the cytochrome c (CYC1) promoter. Expression from this promoter in response to carbon source change is driven by the Hap2,3,4,5 transcription factor complex, which is a major player in the alleviation of glucose repression. sod1Δ cells showed increased CYC1-lacZ expression relative to WT when grown on glucose (Figure 7). Mimicking the sod1Δ phenotype by growing WT cells in the redox-cycling drug paraquat showed a similar result—paraquat-treated cells had more expression from this promoter (data not shown). When the binding site for Hap 2,3,4,5 was deleted from the promoter, the difference between the strains was not observed, indicating the involvement of this complex (Figure 7).
Figure 7.
sod1Δ yeast show decreased glucose repression. A. sod1Δ yeast show increased expression from the CYC1 promoter when grown on glucose and this expression is Hap2,3,4,5 driven. WT (EG103) and sod1Δ (EG314) strains containing the reporter construct pLG669Z, which has the full CYC1 promoter fused to the lacZ gene (CYC1-whole), or the construct pLG669Z-ΔAX, containing the CYC1 promoter with a mutated Hap2,3,4,5 binding site (CYC1-ΔUAS2) were grown to OD600 of 0.5 to 1.0 (exponential phase), and the mean beta-galactosidase activities were determined. It is apparent that there is higher CYC1 induction in sod1Δ yeast compared to WT, and that this effect is greatly reduced if the Hap2,3,4,5 binding site is absent. Graphs represent the results of 3 colonies per strain, done in duplicate assays.
DISCUSSION
The absence of Sod1p leads to some unexpected metabolic consequences in S. cerevisiae, and this work was undertaken to increase our understanding of the reasons for these changes. Our original observation, made many years ago, was that sod1Δ strains of yeast grow more slowly and to a lower final cell density in liquid cultures, particularly in defined medium. In the current study, by measuring glucose and ethanol levels in the medium over time, we find that, compared to WT, the sod1Δ cells consume similar (or slightly higher) amounts of glucose and accumulate similar (or slightly higher) amounts of ethanol per cell in the medium at early stages. However, once glucose levels fall, the sod1Δ cultures fail to switch to utilization of ethanol and abruptly stop growing, or, in other words, the diauxic shift does not take place.
A normal diauxic shift is accompanied by accumulation and then utilization of the storage carbohydrate glycogen. In the sod1Δ strain, there was no accumulation of glycogen (Figure 3). Our results do not fully address the underlying reasons for the failure of glycogen to accumulation. One possibility is that glucose-6-phosphate, the allosteric activator of glycogen synthase as well as the starting material for glycogen synthesis, is chronically low in the sod1Δ yeast due to competition from an overactive pentose phosphate pathway. (The pentose phosphate pathway is upregulated in sod1Δ strains, presumably to cope with the excess demand for NADPH [21]. If this pathway takes priority and uses most of the G6P synthesized, it could result in a lack of substrate for glycogen synthesis.) Alternatively, some regulatory process(es) involved in the diauxic shift or in glycogen synthesis itself may be affected by the presence of excess superoxide.
Because the diauxic shift is, in effect, a response to a change in the carbon source (from glucose to ethanol), we tested the effect of changing the timing of the carbon source shift. Interestingly, we found this timing to be critical—sod1Δ or paraquat-treated WT yeast moved from glucose medium to fresh medium with ethanol as the carbon source during log phase (after 8 hours of growth) can adapt to the respiratory carbon source and grow. However, if they have reached the diauxic shift or stationary phases (24 or 48 hr in culture), they do not grow after transfer (Figure 4B). The carbon source present in the original culture is also important, as raffinose-grown sod1Δ cells grew when transferred to ethanol, while glucose-grown cells did not (Figure 4A). It is important to note this defect is not due to a global inability to respire (petite phenotype) since (1) sod1Δ cells grow on the respiratory carbon source, lactate (data not shown) and (2) raffinose-grown sod1Δ cells and glucose-grown sod1Δ cells transferred early continue growing when moved to ethanol (Figure 4A). This behavior points to a difficulty in executing the diauxic shift under certain conditions rather than an absolute deficiency.
We conclude that sod1Δ has a defect in its ability to respond to changing carbon sources at later stages of glucose-fueled fermentative growth. One explanation for this behavior could be that insufficient energy is available to fuel the execution of the switch in the sod1Δ mutant strains. Lack of glycogen accumulation could be responsible, as it is normally used during the shift. Since this strain diverts more glucose to the pentose phosphate pathway to provide extra NADPH reducing power [21], it may be that it uses up glucose prematurely, leaving no material to fuel the diauxic shift. The fact that the switch is more easily made from raffinose to ethanol than from glucose to ethanol supports this idea: Since glucose represses many respiratory genes and raffinose does not, the changes in cellular machinery required to go from glucose- to ethanol-fueled growth are much larger than those required to go from raffinose to ethanol. It is possible that the sod1Δ strain is unable to muster the resources to make the more demanding switch from glucose.
An alternative explanation involves signaling—excess O2- and/or H2O2 may interfere with some signaling mechanism or deliver a false signal. In mammalian cells, H2O2 is used as a second messenger to regulate growth [22], and something similar may happen in yeast, although it has not been definitively shown [23,24]. By this analogy, it is possible that excess superoxide-derived H2O2 might continue to drive the growth program inappropriately even as glucose levels drop.
sod1Δ yeast growing on glucose consumed approximately twice as much oxygen as WT at early and late stages of culture (Figure 5A). Reasoning that this might be due to increased mitochondrial content on the sod1Δ strain, we used several different assays, including flow cytometry using a dye sensitive to mitochondrial membrane potential, mitochondrial DNA copy number relative to that of nuclear DNA (Figure 6B), and levels of common mitochondrial and cytosolic protein markers (Figure 6A), to compare the relative mitochondrial mass in mutant and WT strains. In each of these determinations of mitochondrial mass, the sod1Δ yeast show an elevated signal—approximately twice that of the WT—which is in agreement with the twofold increase in oxygen consumption we observe in the mutant strain. It should be noted that the increase is seen in the flow cytometry experiment, in which the uptake of dye is dependent on membrane potential, that is, on mitochondrial function. If the mitochondrial mass was the same in the two strains, it would have meant that the sod1Δ strain had a higher membrane potential than the WT, which seems unlikely. If respiration was less efficient in the sod1Δ strain, then the dye uptake should have been lower and the approximately two fold difference would not have been observed. Therefore, taken together, these data convinced us that the increased oxygen consumption can be attributed to increased respiration due to an increase in functional mitochondria, i.e., that the effect is not due to inefficient use of oxygen or uncoupling, which would not have engendered extra mitochondrial volume or altered gene expression.
A logical extension of this reasoning says that inappropriate glucose derepression could be occurring in the sod1Δ yeast. We explored this possibility by measuring lacZ expression driven by the CYC1 promoter. This promoter is turned on by the transcription factor Hap2,3,4,5 when glucose becomes limiting and is thus a good indicator of glucose repression status. We found that this promoter is more active in the sod1Δ strain, indicating some level of glucose derepression.
Why glucose repression is affected is not clear from our work to date. One possible explanation is that some step in the glucose sensing and signaling pathway, from glucose sensing by the Snf3/Rtg2 complex, to the Ras/cAMP cascade, to transcriptional activation of Hap4 and other genes is impaired by excess superoxide. Another possibility is that it could be a natural regulatory response to an elevated energy requirement in the mutant strain relative to WT yeast. sod1Δ yeast are subject to greater oxidative stress than WT, leading to higher turnover of damaged cellular components and, perhaps more importantly, an increased difficulty in maintaining redox balance. To regenerate cellular reducing equivalents in the mutant strain (i.e., NADH and NADPH, which are required to maintain an optimal GSH:GSSG ratio), increased flux through the pentose phosphate pathway is required [21], diverting glycolytic intermediates that would otherwise be put toward ATP generation. Increased mitochondrial ATP production (via increased mitochondrial mass) seems a reasonable response to the altered energy demands of the mutant cell. A third possibility is that an increase in respiration can function as a physiological response to create a “relief valve” when oxidative stress is high. Bonawitz et al. [25] proposed this idea to explain the increased life span observed in tor1Δ yeast strains, which also show glucose derepression. Increased oxygen consumption could certainly lower ROS generation, and a role for TOR signaling in the phenotypes observed in sod1Δ yeast is possible, but these possibilities will have to be addressed through further research.
Although the phenomena we observe in sod1Δ yeast could arise from some strategic or “rational” response by the cell to its situation of elevated superoxide stress, it is equally possible that all of these defects arise simply because excess superoxide interferes with the ability of the cell to signal the necessity for and/or properly execute its metabolic changes. Particularly for the diauxic shift defect, everything we have seen is consistent with the notion that the sod1Δ yeast cultures are stuck in a state prior to transition into stationary phase, incapable of completing some of the required steps (e.g., glycogen accumulation, optimal ethanol consumption level), while “spinning their wheels” by continuously carrying out the steps they can complete until nutrients run out and catastrophic failure occurs.
Interestingly, the growth of sod1Δ in raffinose is more WT-like than its growth in glucose, supporting the idea that a derepressed condition (or an inability to repress) is good for these mutants. The difference in final cell density between WT and sod1Δ that is observed in glucose medium virtually disappears for growth in raffinose, indicating a rescue of the growth phenotype of sod1Δ cells (Figure 1). Thus, the sod1Δ cells seem to do best on a “dual-purpose” carbon source, which both supports glycolysis and allows higher levels of respiration. This compromise might allow more glucose to be diverted to the pentose phosphate pathway while extracting higher energy yield from the remaining glucose via mitochondrial respiration. Indeed, it has been noted that glucose derepression turns on the expression of both Sod1p and Sod2p prior to the presumed oxidative stress generated by respiration, and thus can be considered a preventative measure against oxidative stress [26].
Our studies demonstrating effects on the diauxic shift and glucose repression provide a more complete picture of the metabolic implications of oxidative stress in yeast. These phenomena may be particular to this model organism and closely related species—certainly cells in higher multicellular organisms are exposed to far less variation in oxygen levels and nutritional supply and thus may have lost those response pathways over the course of evolution. On the other hand, cancer cells are known to undergo a shift to aerobic glycolytic metabolism, which may be related at some level. It is also interesting to compare the effects of Sod1p deletion in S. cerevisiae with those observed in genetically manipulated mice. As described above, the absence of Sod1p in yeast has a significant detrimental effect on the health of the cell and particularly on its rate of growth, but our studies imply that many of these effects can be attributed to defects in metabolic pathways that are specific to yeast. By contrast, sod1−/−mice appear healthy at birth, suggesting that there are no major defects in the metabolic pathways most critical to growth of mammalian cells. Nevertheless, as they grow, elevated levels of oxidative stress are clearly present in the sod1−/− mice as evidenced by significantly increased levels of lipid peroxidation, protein carbonyls, oxidative damage to DNA, and DNA mutation rates. Unlike the intact sod1−/− mice, cultured fibroblasts from these mice do not grow well, possibly because the cultured cells experience elevated levels of oxygen relative to those in the intact animal [27]. Thus, sod1Δ yeast may be a better model for individual cultured cells than for whole animals. Overall, the simplicity and flexibility of the budding yeast, Saccharomyces cerevisiae as a model organism has served scientific researchers well, and will likely continue to do so, provided special care is taken in applying results to higher organisms.
CONCLUSIONS
The loss of cytoplasmic SOD activity causes metabolic changes in yeast grown in batch cultures. Glucose repression is not fully “on” in sod1Δ cultures in the early phases of growth, and the diauxic shift is not properly executed in the late stages. Glucose derepression may be adaptive in that it causes increased resistance to stress and/or depletes oxygen, but it is difficult to imagine that the difficulties in executing the diauxic shift are anything but pathological—it seems likely that excess superoxide either interferes with signaling by redox-active molecules or damages key cellular components.
Acknowledgments
This work was supported by grant DK46828 to JSV from the National Institute of Diabetes and Digestive and Kidney diseases. MHSC acknowledges support from the Chemistry-Biology Interface NIH Predoctoral Training Grant; JKM received support from a Ruth L. Kirschstein National Research Service Award. We are grateful to Mike Fenton for experimental help with RT-PCR, Steven Claypool for help with western blot analysis, and Prof. Carla Koehler for providing antibodies.
List of Abbreviations
- Sod1p
copper zinc superoxide dismutase
- ROS
reactive oxygen species
- WT
wild type
- sod1Δ
yeast lacking Sod1p
- GSH
GSSG, reduced and oxidized glutathione, respectively
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. “Sleeping beauty”: Quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Werner-Washburne M, Braun EL, Crawford ME, Peck VM. Stationary phase in Saccharomyces cerevisiae. Mol Microbiol. 1996;19:1159–1166. doi: 10.1111/j.1365-2958.1996.tb02461.x. [DOI] [PubMed] [Google Scholar]
- 3.Gancedo JM. The early steps of glucose signaling in yeast. FEMS Microbiol Rev. 2008;32:673–704. doi: 10.1111/j.1574-6976.2008.00117.x. [DOI] [PubMed] [Google Scholar]
- 4.Schneper L, Düvel K, Broach J. Sense and sensibility: nutritional response and signal integration in yeast. Current Opinion in Microbiology. 2004;7:624–630. doi: 10.1016/j.mib.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Schuller HJ. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet. 2003;43:139–160. doi: 10.1007/s00294-003-0381-8. [DOI] [PubMed] [Google Scholar]
- 6.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 7.Gralla EB. In: Superoxide dismutase: Studies in the yeast Saccharomyces cerevisiae in Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Scandalios J, editor. Cold Spring Harbor Laboratory Press; Plainview: 1997. pp. 495–525. [Google Scholar]
- 8.Wallace MA, Liou LL, Martins J, Clement MH, Bailey S, Longo VD, Valentine JS, Gralla EB. Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis. Cross-compartment protection by CuZn-superoxide dismutase. J Biol Chem. 2004;279:32055–32062. doi: 10.1074/jbc.M403590200. [DOI] [PubMed] [Google Scholar]
- 9.Longo VD, Gralla EB, Valentine JS. Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae. Mitochondrial production of toxic oxygen species in vivo. J Biol Chem. 1996;271:12275–12280. doi: 10.1074/jbc.271.21.12275. [DOI] [PubMed] [Google Scholar]
- 10.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. [Google Scholar]
- 11.Guarente L, Lalonde B, Gifford P, Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984;36:503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- 12.Haldi ML, Guarente L. Multiple domains mediate heme control of the yeast activator HAP1. Mol Gen Genet. 1995;248:229–235. doi: 10.1007/BF02190805. [DOI] [PubMed] [Google Scholar]
- 13.Guarente L, Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an Upstream Activation Site. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- 14.Beutler H. Ethanol. In: Bergmeyer H, editor. Methods of Enzymatic Analysis. 3. Weinheim; Deerfield Beach, Fl: 1984. pp. 598–606. [Google Scholar]
- 15.Kunst A, Draeger B, Ziegenhorn J. D-Glucose: UV-methods with hexokinase and glucose-6-phosphate dehydrogenase. In: Burgmeyer H, editor. Methods of Enzymatic Analysis. 3. Weinheim; Deerfield Beach, Fl: 1984. pp. 163–172. [Google Scholar]
- 16.Sillje HH, ter Schure EG, Rommens AJ, Huls PG, Woldringh CL, Verkleij AJ, Boonstra J, Verrips CT. Effects of different carbon fluxes on G1 phase duration, cyclin expression, and reserve carbohydrate metabolism in Saccharomyces cerevisiae. J Bacteriol. 1997;179:6560–6565. doi: 10.1128/jb.179.21.6560-6565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludovico P, Sansonetty F, Corte-Real M. Assessment of mitochondrial membrane potential in yeast cell populations by flow cytometry. Microbiology. 2001;147:3335–3343. doi: 10.1099/00221287-147-12-3335. [DOI] [PubMed] [Google Scholar]
- 18.Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose M, Botstein D. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- 20.Wallace M, Bailey S, JM F, JS V, EB G. Induction of phenotypes resembling CuZn-superoxide dismutase deletion in wild-type yeast cells: an in vivo assay for the role of superoxide in the toxicity of redox-cycling compounds. Chem Res Toxicol. 2005;18:1279–1286. doi: 10.1021/tx050050n. [DOI] [PubMed] [Google Scholar]
- 21.Slekar KH, Kosman DJ, Culotta VC. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J Biol Chem. 1996;271:28831–28836. doi: 10.1074/jbc.271.46.28831. [DOI] [PubMed] [Google Scholar]
- 22.Veal E, Day AM, Morgan BA. Hydrogen Peroxide sensing and signaling. Molecular Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 23.D’Autréaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nature Reviews, Molec Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 24.Aguirre J, Rios-Momberg M, Hewitt D, Hansberg W. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 2005;13:111–118. doi: 10.1016/j.tim.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metabolism. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maris AF, Assumpcao AL, Bonatto D, Brendel M, Henriques JA. Diauxic shift-induced stress resistance against hydroperoxides in Saccharomyces cerevisiae is not an adaptive stress response and does not depend on functional mitochondria. Curr Genet. 2001;39:137–149. doi: 10.1007/s002940100194. [DOI] [PubMed] [Google Scholar]
- 27.Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H. Trends in oxidative aging theories. Free Radic Biol Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]