Abstract
Proteomics has been applied to study intracellular bacteria and phagocytic vacuoles in different host cell lines, especially macrophages (Mφs). For mycobacterial phagosomes, few studies have identified over several hundred proteins for systems assessment of the phagosome maturation and antigen presentation pathways. More importantly, there has been a scarcity in publication on proteomic characterization of mycobacterial phagosomes in dendritic cells (DCs). In this work, we report a global proteomic analysis of Mφ and DC phagosomes infected with a virulent, an attenuated, and a vaccine strain of mycobacteria. We used label-free quantitative proteomics and bioinformatics tools to decipher the regulation of phagosome maturation and antigen presentation pathways in Mφs and DCs. We found that the phagosomal antigen presentation pathways are repressed more in DCs than in Mφs. The results suggest that virulent mycobacteria might co-opt the host immune system to stimulate granuloma formation for persistence while minimizing the antimicrobial immune response to enhance mycobacterial survival. The studies on phagosomal proteomes have also shown promise in discovering new antigen presentation mechanisms that a professional antigen presentation cell might use to overcome the mycobacterial blockade of conventional antigen presentation pathways.
Keywords: proteomics, phagosome, macrophage, dendritic cell, antigen presentation, systems biology, Mycobacterium tuberculosis
Introduction
Phagocytosis is a form of receptor-mediated endocytosis carried out by specialized cells, particularly professional antigen presentation cells (APCs) such as macrophages (Mφs) and dendritic cells (DCs)1. The process plays a central role in defense against bacteria by enabling mechanisms including antigen presentation2. Many intracellular bacterial pathogens have evolved strategies to survive in the host by replicating within the host-cell cytoplasm, which is exploited as protected niches. To escape immune defense, these bacteria trigger their internalization into host cells by subverting the cellular actin cytoskeleton. This actin-based phagocytosis process is mediated by a multitude of receptors such as Fc3, Complement4, PAMP5, and others6.
Phagosomes are fully competent antigen-processing organelles7 capable of presenting exogenous antigens such as bacterial antigens via class I and II pathways8. While MHC class I molecules generally do not present exogenous antigens, antigens from some intracellular pathogens have been shown to elicit an MHC class-I-dependent CD8+ T-cell response by the process referred to as cross-presentation9.
Proteomic studies on phagosomes have contributed significantly to the understanding of phagosome biogenesis and immunity-related functions10–12. Proteomic analyses of phagosomes have demonstrated that phagosomes’ self-sufficiency for antigen presentation arises from the assembly of proteins needed for cross-presentation on them13. In addition, Jutras et al. found that γ-secretase is a functional component of phagosomes that remains associated with newly formed phagosomes through their maturation into phagolysosomes14. One of the γ-secretase complex members, Nicastrin, was found to be present on BCG- and latex bead-containing phagosomes in human THP-1 Mφs15. We further found with proteomics that Nicastrin was enriched on the phagosome of the wild-type Mycobacterium tuberculosis (Mtb) and to a lesser extent on the BCG vaccine phagosome. Because γ-secretase has been implicated in proteolysis14, the Nicastrin enrichment on Mtb phagosomes suggests its potential role in phagosome functions16.
Proteomics has been applied to study intracellular bacteria in different host cells, especially Mφs. Pathogenic mycobacteria infect Mφs where they modulate the intersection of phagosomes with the intracellular trafficking pathways of antigen presentation molecules17. However, few analyses have quantified several hundred phagosome proteins from mycobacteria-infected Mφs to allow a systems assessment of the modulation of phagosomes by mycobacteria15, 16. More importantly, there has been a dearth of proteomic characterization of mycobacterial phagosomes from DCs, another type of potent professional APC.
In this work, we compare the global Mφ and DC phagosomal proteomes infected with three different mycobacterial strains. We used label-free comparative proteomics and a systems data analysis approach to decipher the regulation of phagosome maturation and antigen presentation pathways in Mφs and DCs.
Materials and Methods
Preparation of mycobacterial phagosomes
Mφs and DCs were derived from the C57BL/6 mouse bone marrow and cultured as previously described16, 18. Mφs and DCs are both professional APCs.
C57BL/6 bone marrow-derived Mφ-like cell line BMA.A3 was kindly provided by Dr. Kenneth L. Rock (University of Massachusetts Medical School). This cell line was maintained in the same medium and has been well characterized to study phagocytosis and antigen presentation for Mtb19. The C57BL/6-derived Mφs were cultured in McCoy’s medium with 10% FBS, penicillin and gentamicin supplemented with 10 ng/ml recombinant mouse GM-CSF (Cell Sciences, Canton, MA). Mφs were rested in GM-CSF-free medium for 2 days and used for mycobacterial infections. DCs were cultured from mouse bone marrow cells grown in GM-CSF-containing medium for 7 days and CD11c+ cells purified using anti-CD11c-coated magnetic beads as described by the manufacturer (Miltenyi, Auburn, CA). The cells were more than 97% pure20. Uninfected APCs were also pelleted to prepare whole cell lysate samples.
The APCs were infected by the wild type reference Mtb H37Rv lab strain (H37Rv), the fbpA-knockout mutant strain derived from Mtb H37Rv (KO)21, and the BCG Pasteur vaccine strain (BCG), respectively. We harvested bacillus-containing phagosomes from APCs infected by the three mycobacterial strains respectively based on the method described earlier17 and adapted later18. Uninfected APCs were also pelleted to prepare whole cell lysate samples.
Specifically, the APCs were infected with mycobacteria (MOI 1:5) for 4 hrs with gentle mixing, washed thrice with McCoys medium and incubated for another 18 hrs. The APCs were scraped, washed thrice in a phagosome fractionation buffer (PFB) with 10 mM Hepes, 5 mM EDTA, 5 mM EGTA, pH 7.0 and suspended in PFB with an anti-protease mix consisting of 1 µg/mL leupeptin, 1 µg/mL pepstatin and 1 mM phenylmethyl sulfonylfluoride. Pellets were then homogenized in a glass tissue homogenizer 10 times and passed 10 times through a 28-gauge needle. Lysates were centrifuged at 500 g for 5 min to sediment nuclei and the post nuclear supernatant was layered on a step gradient of 50% and 12% sucrose in the PFB. After centrifugation at 1,000 g for 60 min, the interphase of phagosome fraction was collected and further purified by passing through two successive cushions of 70 kDa and 400 kDa ficoll in the PFB. The final purified phagosome pellet was collected by centrifugation at 10,000 g for 15 min. The purified phagosome pellets or whole cell pellets were lysed in a SDS/PAGE sample buffer with bead-beating as before16 for the following proteomic analysis. We routinely used anti-Calnexin and anti-GS28 antibodies in Western blot to examine ER and Golgi contaminations in phagosome preparations. Western blot of selected markers has been used to examine the purities of mycobacterial phagosome preparations for global proteomic analysis15, 16.
Label-free proteomic analysis
The SDS-solubilized phagosome and whole cell lysate samples were processed for tryptic digestion by following the filter-assisted sample preparation (FASP) protocol22, 23 with some modifications. Briefly, an aliquot of 100 µg proteins from each sample prepared as above was buffer-exchanged into a solution of 8M urea, 10 mM DTT, and 25 mM Tris-HCl and was further diluted 8X with 25 mM Tris-HCl. Trypsin (Promega, Madison, WI) was added at a 1:20 ratio to digest the proteins at 37°C overnight followed by acidification with 5% formic acid. The resulting tryptic peptides were further purified with C18 ZipTip columns (Milipore, Bedford, MA) and eluted in 80% acetonitrile with 0.1% TFA. Three elutions were pooled, dried under a SpeedVac (Eppendorf, Hauppauge, NY), and reconstituted in 20 µl of 0.1% TFA.
The purified peptide samples were analyzed with a 90-min 5–35% acetonitrile gradient on a Thermo Scientific nano-electrospray LC/LTQ-FTMS instrument as described16. To identify proteins, the RAW MS data files were converted to mzXML format and processed with the MassMatrix search engine (http://searcher.rrc.uic.edu/) housed in a server at the Proteomics and Informatics Services Facilities at University of Illinois at Chicago24. The mzXML data files were searched against the mouse IPI database downloaded from the European Bioinformatics Institute website (www.ebi.ac.uk; version 3.39). The peptide mass tolerance was set at ±10 ppm with methionine oxidation as a differential modification. The fragment mass tolerance was set at ±0.6 Da. Methionine oxidation and phosphorylation on serine, threonine, or tyrosine were set as differential modifications. We allowed up to three modifications and two missed cleavages per peptide and an automated isotope check to include a 1.003-amu mass shift. An in-fly randomized decoy database was used for false discovery rate estimation to accept identified proteins and peptides at a false discovery rate <5%. There were 37,890 peptide charge state (PCS) identifications accepted from the 16 LC/MS runs. A PCS is a peptide identified at a specific charge state. Although some peptides were identified at multiple PCSs, a majority of the peptides were identified at a single PCS.
The identified PCSs were quantified with the label-free proteomics method as described25, 26. Using the cross-reference technique27, a PCS identified in one run is also quantified in another run even though it is not identified by an MS2 spectrum in that run25. This procedure increases the number of proteins that can be quantified from each run26. The intensity of a PCS is represented by an extracted ion chromatographic (XIC) intensity. The XIC intensity is the integration of a PCS MS ion current intensity over an LC elution-time window to accommodate a peptide peak28. For each LC/MS run, the abundance of a protein is represented by the summation of the XIC intensities from all PCSs quantified for that protein in the run.
Because the same set of PCSs were used to calculate the XIC intensity of a protein in all of the samples and because the protein XIC intensities were normalized to the summed XIC intensity in each sample respectively, we used the XIC intensities to assess the protein relative abundances in the samples28.
The Mφ and DC whole cell lysates had 900 and 870 proteins quantified with non-zero intensities, respectively. The Mφ phagosome samples had 890, 780, and 680 proteins quantified with non-zero intensities for BCG, KO, and H37Rv, respectively. The DC phagosome samples had 830, 860, and 810 proteins quantified with non-zero intensities for BCG, KO, and H37Rv, respectively. In total, 1001 unique proteins were quantified with non-zero intensities in at least one of the 16 LC/MS runs. Approximately 20% of the 1001 × 8 × 2 protein abundance values were below threshold. These missing values were imputed with 25% of the lower 20-percentile abundance values. Subsequently, the protein abundances were normalized to parts-per-million based on the summed PCS XIC intensities for each LC/MS run (Table S1).
To assess significance of a protein abundance ratio, we used a combination of Power Law Global Error Modeling-Signal to Noise Ratio (PLGEM-STN), fold-change, and the rule of Minimum number of Permuted Significant Pairings (MPSP) as previously described25, 26. PLGEM-STN takes into account the protein abundance but typically over-penalizes low-abundance proteins. Fold-change typically over-penalizes high-abundance proteins. MPSP reduces false discovery rate for a small dataset where neither PLGEM-STN nor fold-change alone can achieve a satisfactorily low false discovery rate25. To perform a PLGEM-STN analysis here, we estimated a PLGEM using LC/MS replicates of a sample and averaged the PLGEM parameters over four selected phagosome samples (Table S2). We applied the fitted PLGEM parameters to establish the STN threshold to determine differentially abundant proteins. We selected a STN threshold such that no positives were detected from all four pairs of LC/MS duplicates in the four selected samples (Table S3). Similarly, a 4-fold change threshold was selected so that no positives were detected from all four pairs of LC/MS duplicates of those four samples. Although the use of LC/MS replicates instead of sample preparation replicates tends to underestimate false discovery rates when applying the MPSP rule26, an MPSP of four still significantly reduces false discovery rates25, and our choice of a zero false discovery rate threshold mitigates that underestimation. After selecting differentially abundant proteins with the PLGEM-STN and fold-change thresholds respectively, we accepted a protein as differentially abundant with a statistical significance if its abundance ratio met at least one of these two thresholds after MPSP filtering25.
Gene set enrichment analysis of selected GO terms and pathways
We performed gene set enrichment analysis (GSEA) (www.broadinstitute.org/gsea)29, a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between biological states. Although the GSEA software was originally designed for analyzing genomic datasets, it can be used to analyze a proteomic dataset when the abundance of a protein has been determined across multiple samples, such as in this study (Table S1).
An a priori set of proteins was selected with the DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/)30 to identify enriched functional categories and examine the mechanisms associated with the regulation of the identified proteins. DAVID allows an assessment of over-represented gene ontology (GO) terms, KEGG pathways, and other gene set definitions simultaneously, and the output is readily compatible with GSEA analysis. For the phagosomal proteome profile data in this study, we focus on gene sets annotated in GO (www.geneontology.org)31 and KEGG Pathway (www.genome.jp)32 at a p-value <.05 with a medium classification stringency. Due to the limited number of sample replicates, we used gene set permutation instead of phenotype permutation in GSEA. We set the number of permutations to 1000, and the weighted enrichment statistic was applied. Signal to noise ratio was used to rank proteins. The inclusive gene set size range was set to 20–100. When applicable, a leading edge analysis was performed following a GSEA analysis to identify the subset of proteins within a gene set that drove the regulation of the gene set.
Clustering and GO analysis
Clustering analysis was carried out using Cluster33. GO analysis was carried out with the BINGO plugin34 in the CytoScape platform (www.cytoscape.org)35 as before36.
Results
We employed label-free quantitative proteomics to analyze the Mφ and DC whole cell lysates (WC) and the phagosomes (Φs) harvested from Mφs and DCs infected by the H37Rv, KO and BCG strains, respectively. We quantified 830 ± 70 proteins in each sample to result in a total of 1001 unique proteins from these eight samples (Table S1).
Regulation of overrepresented GO terms and KEGG pathways
We evaluated the regulation of enriched GO terms and KEGG pathways among the eight samples (Table 1). We compared six groups of sample pairs. Twenty-two clusters of GO terms or KEGG pathways were found with statistical significance in at least one sample pair. The acceptable statistical significance was set at a false discovery rate <25% as recommended in the GSEA user manual for a small dataset. A GSEA normalized enrichment score shown in bold indicates that the GO term or KEGG pathway was regulated in that sample pair. Hereafter, the proteins in a GO term or KEGG pathways are called a protein set.
Table 1. Regulation of the overrepresented GO terms and KEGG pathways.
The values in bold are statistically significant (see Methods).
| Groups of pair-wise comparison among the samples | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | ||||||||
| Description of the overrepresented GO terms and KEGG pathways |
ΦMφ,avg/ΦDC,avg | ΦMφ,KO/ΦDC,KO | ΦMφ,H37Rv/ΦDC,H37Rv | ΦMφ,BCG/ΦDC,BCG | ΦMφ,KO/ΦMφ,H37Rv | ΦMφ,KO/ΦMφ,BCG | ΦMφ,H37Rv/ΦMφ,BCG | ΦDC,KO/ΦDC,H37Rv | ΦDC,KO/ΦDC,BCG | ΦDC,H37Rv/ΦDC,BCG | WCMφ/ΦMφ,avg | WCDC/ΦDC,avg | WCMφ/WCDC |
| Cluster 1 | |||||||||||||
| MITOCHONDRIAL INNER MEMBRANE | −1.20 | 1.09 | 0.77 | −1.21 | 1.20 | 0.93 | −1.14 | 0.94 | −1.38 | −1.60 | −1.16 | 1.29 | 1.90 |
| ORGANELLE INNER MEMBRANE | −1.11 | 1.05 | 0.91 | −1.14 | 1.24 | 0.93 | −1.21 | 1.07 | −1.29 | −1.62 | −1.14 | 1.20 | 1.91 |
| MITOCHONDRIAL MEMBRANE | −1.18 | 0.91 | 0.80 | −1.30 | 1.16 | 0.98 | −1.10 | 0.99 | −1.41 | −1.73 | −1.19 | 1.32 | 1.88 |
| MITOCHONDRIAL ENVELOPE | −1.13 | 0.95 | 0.78 | −1.27 | 1.19 | 1.02 | −1.08 | 0.96 | −1.42 | −1.70 | −1.20 | 1.22 | 1.84 |
| Cluster 2 | |||||||||||||
| ELECTRON TRANSPORT CHAIN | −1.54 | 0.86 | −0.69 | −1.26 | 1.01 | −0.88 | −1.00 | 0.61 | −1.51 | −1.68 | −0.79 | 1.09 | 1.21 |
| GENERATION OF PRECURSOR METABOLITES AND ENERGY | 0.68 | 1.19 | −0.94 | −1.25 | 1.39 | 1.22 | −1.20 | 0.81 | −1.50 | −1.38 | −0.58 | 0.86 | 1.24 |
| OXIDATION REDUCTION | −0.89 | 1.21 | 0.84 | −1.26 | 1.08 | 1.28 | −0.82 | −1.05 | −1.49 | −1.50 | 1.00 | −1.12 | 1.00 |
| Cluster 3 | |||||||||||||
| STRUCTURAL CONSTITUENT OF RIBOSOME | 1.06 | 0.89 | −0.79 | 1.57 | 1.56 | 1.23 | −1.48 | 1.10 | 1.02 | −0.88 | 1.01 | −0.83 | 1.65 |
| RIBOSOME | 1.14 | 1.06 | −0.82 | 1.46 | 1.55 | 1.52 | −1.41 | 0.97 | 0.97 | −0.91 | 0.71 | −0.95 | 1.68 |
| TRANSLATION | 1.08 | 0.92 | −1.34 | 1.51 | 1.51 | 1.12 | −1.44 | 1.13 | 1.20 | 1.10 | 1.01 | 0.83 | 1.48 |
| RIBONUCLEOPROTEIN COMPLEX | 1.34 | 0.98 | −1.07 | 1.77 | 1.58 | 1.13 | −1.52 | 1.16 | 1.47 | 1.04 | 0.79 | −1.07 | 1.32 |
| Cluster 4 | |||||||||||||
| CELLULAR RESPIRATION | 0.65 | 1.37 | 0.76 | −1.29 | 1.42 | 1.33 | −0.96 | 0.73 | −1.58 | −1.66 | −0.47 | 0.98 | 1.78 |
| ENERGY DERIVATION BY OXIDATION OF ORGANIC COMPOUNDS | 0.70 | 1.52 | −0.83 | −1.54 | 1.56 | 1.62 | −0.88 | 0.64 | −1.64 | −1.50 | 0.64 | 0.88 | 1.67 |
| Cluster 5 | |||||||||||||
| CYTOSKELETON | −1.74 | −1.41 | −1.31 | −0.79 | 0.73 | −0.76 | −0.56 | 0.83 | 1.10 | 1.05 | 0.83 | 1.21 | −1.01 |
| Cluster 6 | |||||||||||||
| INTRACELLULAR PROTEIN TRANSPORT | 1.60 | 1.06 | 1.02 | 1.19 | 0.96 | 0.73 | −1.02 | 0.58 | 0.78 | −0.73 | −1.12 | −1.99 | −1.13 |
| CELLULAR PROTEIN LOCALIZATION | 1.55 | 1.00 | 0.97 | 1.17 | 0.93 | −0.79 | −1.00 | 0.55 | 0.76 | −0.73 | −1.17 | −1.93 | −1.11 |
| CELLULAR MACROMOLECULE LOCALIZATION | 1.53 | 1.00 | 0.96 | 1.19 | 0.96 | −0.79 | −1.00 | 0.55 | 0.75 | −0.73 | −1.19 | −1.95 | −1.09 |
| INTRACELLULAR TRANSPORT | 1.55 | 1.12 | 1.23 | 1.00 | 0.83 | 0.79 | −0.72 | −0.97 | −0.80 | −0.72 | −1.36 | −1.78 | −0.86 |
| PROTEIN TRANSPORT | 1.62 | 1.12 | 0.99 | 1.51 | 1.05 | −1.17 | −1.26 | −0.99 | 0.84 | −0.70 | −0.87 | −1.62 | −1.02 |
| ESTABLISHMENT OF PROTEIN LOCALIZATION | 1.56 | 1.08 | 0.99 | 1.50 | 1.02 | −1.25 | −1.25 | −0.95 | 0.81 | −0.69 | −0.91 | −1.65 | −1.02 |
| Cluster 7 | |||||||||||||
| ACTIN CYTOSKELETON ORGANIZATION | −1.58 | −1.91 | −1.64 | 1.02 | 0.81 | −0.94 | −0.96 | 1.03 | 1.78 | 1.05 | −1.07 | −1.01 | −1.60 |
| ACTIN FILAMENT-BASED PROCESS | −1.71 | −1.94 | −1.70 | 0.95 | 0.77 | −0.83 | −0.91 | 1.03 | 1.72 | 1.00 | −1.14 | −1.00 | −1.46 |
| CYTOSKELETON ORGANIZATION | −1.59 | −1.57 | −1.69 | −0.99 | 0.89 | −0.65 | −0.74 | −0.76 | 1.29 | 1.22 | −0.99 | −1.03 | −1.52 |
| Cluster 8 | |||||||||||||
| ACTIN CYTOSKELETON | −2.03 | −1.46 | −1.72 | −0.98 | 1.13 | 0.73 | −0.70 | 0.89 | 1.44 | 1.04 | −0.92 | 1.05 | −1.32 |
| ACTIN BINDING | −2.20 | −1.68 | −1.81 | 0.72 | 1.00 | −0.85 | −0.83 | 0.86 | 1.57 | 1.14 | −0.66 | −1.08 | −1.61 |
| CYTOSKELETAL PROTEIN BINDING | −2.16 | −1.68 | −1.85 | −0.83 | 0.94 | −0.83 | −0.79 | 0.82 | 1.46 | 1.16 | 0.80 | −1.04 | −1.66 |
| CYTOSKELETAL PART | −1.74 | −1.12 | −1.48 | −0.82 | 0.84 | 1.00 | 0.71 | −0.97 | 1.10 | 1.28 | −0.97 | 1.23 | −0.90 |
| Cluster 9 | |||||||||||||
| CELL CORTEX | −1.36 | −1.21 | −1.22 | 0.93 | 0.93 | −1.16 | −0.60 | 1.06 | 1.29 | 1.26 | −1.29 | −1.15 | −1.13 |
| Cluster 10 | |||||||||||||
| HYDROGEN ION TRANSMEMBRANE TRANSPORTER ACTIVITY | 0.74 | 0.66 | 0.71 | 0.71 | 0.76 | −1.05 | −1.22 | −1.09 | −1.44 | −1.10 | −1.37 | 0.92 | 1.56 |
| MONOVALENT INORGANIC CATION TRANSMEMBRANE TRANSPORTER ACTIVITY | 0.65 | −0.72 | 0.85 | −0.90 | 0.61 | −1.00 | −1.10 | −1.08 | −1.54 | −1.06 | −1.56 | 0.79 | 1.56 |
| INORGANIC CATION TRANSMEMBRANE TRANSPORTER ACTIVITY | 0.57 | 0.73 | 0.82 | 0.75 | 0.91 | −0.58 | −1.11 | 0.86 | −1.18 | −1.02 | −1.28 | 0.97 | 1.61 |
| Cluster 11 | |||||||||||||
| MRNA PROCESSING | 1.54 | 0.92 | 0.96 | 1.62 | 1.44 | −1.18 | −1.45 | 1.13 | 1.32 | 1.00 | −1.02 | −1.03 | 0.63 |
| RNA PROCESSING | 1.46 | 0.74 | 1.01 | 1.75 | 1.38 | −1.26 | −1.49 | 1.18 | 1.31 | 1.01 | −0.97 | −1.00 | 0.68 |
| Cluster 12 | |||||||||||||
| RESPIRATORY CHAIN | −1.99 | −1.40 | −1.20 | −1.42 | 0.75 | −1.37 | −1.23 | 0.66 | −1.68 | −1.52 | −0.83 | 0.96 | 1.31 |
| Cluster 13 | |||||||||||||
| ENDOPLASMIC RETICULUM MEMBRANE | 1.36 | 0.92 | −0.89 | 1.06 | 0.80 | −0.84 | −0.88 | −0.82 | 0.86 | 0.86 | −1.08 | 1.23 | 1.81 |
| NUCLEAR ENVELOPE-ENDOPLASMIC RETICULUM NETWORK | 1.39 | 1.05 | −0.64 | 1.24 | 0.94 | −0.78 | −0.94 | 0.63 | 0.88 | 0.81 | −1.05 | 0.99 | 1.70 |
| ENDOPLASMIC RETICULUM PART | 1.46 | 0.93 | 1.25 | 1.20 | 0.49 | −0.96 | −0.75 | 0.83 | 0.71 | −0.80 | 1.58 | 1.36 | 1.07 |
| Cluster 14 | |||||||||||||
| MICROSOME | 1.09 | 1.24 | 0.80 | 0.65 | 1.26 | 0.93 | −0.90 | 1.00 | −0.68 | −0.97 | −1.65 | −1.12 | 1.46 |
| VESICULAR FRACTION | 1.10 | 1.26 | 0.81 | 0.64 | 1.26 | 0.91 | −0.89 | 1.00 | −0.69 | −0.95 | −1.65 | −1.11 | 1.48 |
| MEMBRANE FRACTION | 1.11 | 0.96 | 0.90 | 1.25 | 1.37 | 0.68 | −1.19 | 1.16 | 1.33 | 0.98 | −1.84 | −0.76 | 1.44 |
| INSOLUBLE FRACTION | 1.14 | 1.00 | −0.85 | 1.22 | 1.31 | 0.78 | −1.23 | 1.19 | 1.31 | 0.91 | −1.77 | −0.77 | 1.49 |
| CELL FRACTION | 1.25 | 1.11 | −0.85 | 1.19 | 1.32 | 0.85 | −1.21 | 1.04 | 1.19 | 0.90 | −1.72 | −0.67 | 1.55 |
| Cluster 15 | |||||||||||||
| REGULATION OF CELLULAR COMPONENT SIZE | −1.55 | −1.43 | −1.18 | 0.86 | 1.19 | −1.07 | −0.86 | 1.27 | 1.53 | −0.81 | 0.67 | 1.17 | −0.84 |
| Cluster 16 | |||||||||||||
| CHROMATIN | 0.64 | 0.95 | −1.42 | 1.29 | 1.16 | −0.83 | −1.15 | 0.89 | 0.87 | −0.86 | −1.37 | −1.51 | 1.05 |
| CELLULAR MACROMOLECULAR COMPLEX ASSEMBLY | 1.02 | 1.51 | −1.17 | 1.21 | 1.35 | 0.97 | −1.10 | −1.27 | 0.84 | 1.24 | −0.96 | −1.65 | −0.97 |
| CELLULAR MACROMOLECULAR COMPLEX SUBUNIT ORGANIZATION | 0.93 | 1.42 | −1.15 | 1.20 | 1.32 | 0.88 | −1.08 | −1.17 | 0.81 | 1.13 | −0.92 | −1.60 | −1.02 |
| MACROMOLECULAR COMPLEX ASSEMBLY | 1.09 | 1.69 | −1.16 | 1.15 | 1.35 | 1.09 | −1.07 | −1.33 | 0.73 | 1.27 | −1.00 | −1.62 | −0.96 |
| MACROMOLECULAR COMPLEX SUBUNIT ORGANIZATION | 1.06 | 1.69 | −1.12 | 1.14 | 1.36 | 1.08 | −1.05 | −1.32 | 0.73 | 1.22 | −1.00 | −1.56 | −1.00 |
| Cluster 17 | |||||||||||||
| LYSOSOME | 1.70 | 1.65 | 1.48 | 1.16 | 1.16 | −0.70 | −1.05 | −0.80 | −1.26 | −0.73 | 0.99 | −1.06 | 0.76 |
| LYTIC VACUOLE | 1.72 | 1.66 | 1.50 | 1.15 | 1.19 | −0.69 | −1.04 | −0.84 | −1.23 | −0.74 | 1.00 | −1.05 | 0.75 |
| VACUOLE | 1.81 | 1.76 | 1.40 | 1.11 | 1.20 | −0.66 | −0.99 | −0.78 | −1.35 | −0.84 | 0.91 | −1.04 | 0.87 |
| Cluster 18 | |||||||||||||
| HEMOPOIESIS | −1.26 | −1.06 | −1.04 | 0.91 | −1.30 | −1.40 | −1.04 | 0.84 | −0.95 | −0.83 | −0.78 | 1.05 | 0.78 |
| HEMOPOIETIC OR LYMPHOID ORGAN DEVELOPMENT | −1.37 | −1.28 | −1.28 | 1.03 | −1.24 | −1.46 | −1.19 | −0.97 | 0.93 | −0.69 | −0.74 | 0.95 | 0.76 |
| Cluster 19 | |||||||||||||
| MEMBRANE ORGANIZATION | 1.26 | 0.72 | −0.94 | 1.09 | 0.81 | −0.48 | −1.06 | 0.95 | 0.83 | −0.64 | −1.06 | −1.24 | 1.72 |
| VESICLE-MEDIATED TRANSPORT | 1.38 | 0.86 | 1.00 | 1.25 | 0.95 | −1.01 | −1.02 | 0.77 | 0.98 | 0.64 | −1.20 | −1.33 | 1.64 |
| Cluster 20 | |||||||||||||
| CELLULAR HOMEOSTASIS | 0.59 | 0.76 | 0.92 | −0.79 | −0.87 | 0.89 | −0.75 | 1.36 | 0.95 | −1.07 | 1.72 | −0.66 | −0.74 |
| Cluster 21 | |||||||||||||
| ALZHEIMER'S DISEASE (KEGG) | −1.46 | −1.01 | −0.97 | −1.18 | 0.83 | −0.86 | −1.20 | 0.74 | −1.46 | −1.41 | −1.31 | 0.90 | 1.80 |
| OXIDATIVE PHOSPHORYLATION (KEGG) | −1.57 | −0.71 | −1.05 | −1.33 | 0.93 | −0.78 | −1.22 | 0.81 | −1.80 | −1.61 | −1.13 | 0.99 | 1.56 |
| PARKINSON'S DISEASE (KEGG) | −1.31 | −0.68 | −1.15 | −1.17 | 0.89 | −1.09 | −1.43 | −0.91 | −1.62 | −1.39 | −1.27 | 0.83 | 1.95 |
| HUNTINGTON'S DISEASE (KEGG) | −1.10 | 0.72 | −0.76 | −1.11 | 0.99 | −0.90 | −1.32 | 0.64 | −1.57 | −1.43 | −1.47 | 0.74 | 1.96 |
| Cluster 22 | |||||||||||||
| ANTIGEN PROCESSING AND PRESENTATION (KEGG) | 1.69 | 1.47 | 0.85 | 1.27 | 1.23 | 0.68 | −1.12 | 0.89 | 0.91 | 1.04 | 1.65 | 1.15 | 0.92 |
Group I contains one sample pair to compare the average Mφ and DC phagosomes (ΦMP,avg/ΦDC,avg). This comparison was done in GSEA by assigning the three Mφ phagosomes as one class and the three DC phagosomes as the second class. The protein sets more enriched in DC phagosomes include electron transfer chain (Cluster 2), cytoskeleton (Cluster 5), actin organization and process (Cluster 7), cell cortex (Cluster 9), respiration chain (Cluster 12), regulation of cellular component size (Cluster 15), hemopoiesis (Cluster 18), and several disease related KEGG pathways (Cluster 21).
The protein sets involved in cellular protein localization (Cluster 6) and mRNA processing (Cluster 11) were enriched in Mφ phagosomes. More interestingly, the protein sets in lysosome and lytic vacuole (Cluster 17) and antigen processing and presentation (Cluster 22) are enriched in Mφ phagosomes.
In Group II, the Mφ and DC phagosomes were compared for each mycobacterial strain. The normalized enrichment scores of these three phagosome pairs support the regulation patterns observed between the Mφ and DC phagosomes in Group I. Additionally, 2, 6, and 11 protein sets were regulated between the Mφ and DC phagosomes for the BCG, H37Rv, and KO strains, respectively. No protein sets were regulated between the Mφ phagosomes in Group III.
For the DC phagosomes in Group IV, the KO and H37Rv phagosomes were more similar to each other than to the BCG phagosome. There was no significant difference between the KO and H37Rv phagosomes. On the other hand, 18 protein sets were regulated between the KO and BCG phagosomes. Ten protein sets were regulated between the H37Rv and BCG phagosomes.
We also compared the whole cell lysate to the average phagosome in DCs or Mφs (Group V). For Mφs and DCs respectively, we assigned the KO, H37Rv, and BCG phagosomes as one class and the whole cell lysate as the other class to compare. Seven and 11 protein sets were regulated in Mφs and DCs, respectively. Twenty-five protein sets were regulated between the Mφ and DC whole cell lysates (Group VI).
Clustering analysis of the phagosomal and whole cell proteomes
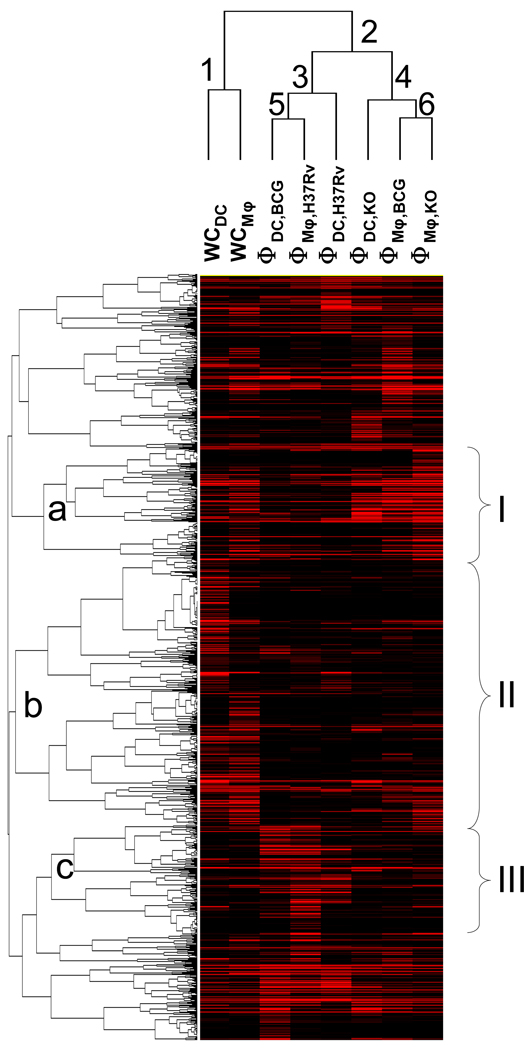
The GSEA analysis focuses on overrepresented protein clusters grouped by their functional similarities. To complement the GSEA analysis, we perform a cluster analysis to compare the eight samples based on the abundance values of the 1001 quantified proteins without relying on a priori definition of their functions (Fig. 1).
Figure 1. Clustering analysis of the six phagosome and two whole cell lysate samples based on the abundance profiles of the 1001 quantified proteins.
Φ – phagosome. WC – whole cell lysate.
The whole cell lysates (under node 1) are clearly distinct from the phagosomes (under node 2), supporting the label-free proteomics methodology used for the analysis.
The KO phagosomes are more similar to each other than to the H37Rv ones (nodes 3 and 4). The H37Rv phagosomes are more similar to each other than to the KO ones. The BCG phagosomes appear to depend more on the host cell type.
A qualitative examination of the protein clusters indicates that nodes a (cluster I), b (cluster II), and c (cluster III) are enriched with proteins from the KO phagosomes, whole cell lysates, and H37Rv phagosomes, respectively. We performed gene ontology analysis to identify the main themes represented by these three different protein groups (Fig. S1).
Not surprisingly, nucleus and mitochondrial proteins are enriched among those more closely associated with the whole cell lysates (cluster II, Fig. 1). Because nuclei proteins are present only in the whole cell lysate preparations, their enrichment in cluster II is expected and confirms that GO analysis indeed reveals major biological components in samples. The enrichment of mitochondrial proteins indicates that higher amounts of mitochondria were present in the whole cell lysates than in the phagosome preparations. Several macromolecule complexes are also enriched, including proteasome, ribosome, microsome, synaptosome, COPI vesicle coat, and chaperon-containing T-complex.
GO terms lysosome, chaperone-containing T-complex, and mitochondria are enriched among the proteins more abundant in the KO phagosomes (cluster I, Fig. 1). The enrichment of lysosomal proteins suggest that KO phagosomes proceeded further in maturation, consistent with the phenotype observed by independent methods18. The lack of enrichment of lysosomal proteins among the proteins more abundant in H37Rv phagosomes suggests that H37Rv blocked phagosome maturation. ER proteins are enriched among the proteins more abundant in H37Rv phagosomes.
An overview of the eight proteome profiles in Fig. 1 reveals that H37Rv blocks phagosome maturation by excluding lysosomal proteins while the KO phagosomes recruit more lysosomal proteins. This result obtained with proteomics is consistent with observations in a prior study using Western blot and fluorescence microscopy18. The KO phagosomes also recruit more chaperon-containing T-complex proteins, consistent with the result in a prior proteomic analysis16. Interestingly, the H37Rv phagosomes appear to contain more ER proteins.
Antigen presenting pathways in mycobacterial phagosomes
During phagocytosis, a phagosome is formed when specific receptors on the APC surface recognize ligands on the bacterium surface. The nascent phagosome progresses to maturation, which involves regulated interaction with other membrane organelles including recycling endosomes, late endosomes, and lysosomes. The maturation of phagosomes results in killing and degradation of bacteria to lead to antigen presentation.
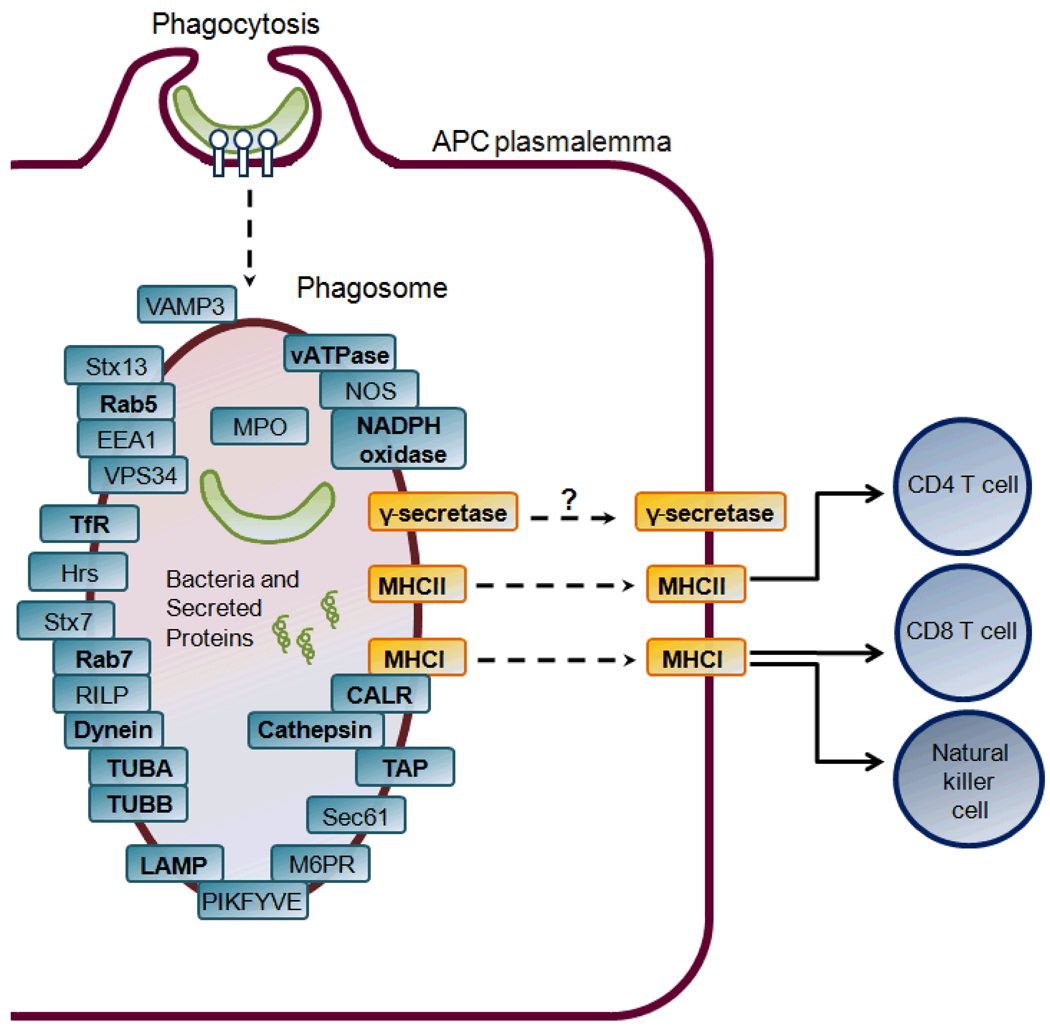
Fig. 2 shows the phagosome components (i.e., proteins or protein complexes) that are known to be involved in the processes of phagosome biogenesis, maturation, and antigen presentation based on the KEGG Phagosome Pathway annotation32.
Figure 2. Phagosome and antigen presentation pathways.
The rounded rectangular text boxes represent the phagosome components that are a protein or protein complex. All the shown phagosome components except γ-secretase are annotated in the KEGG Phagosome Pathway (www.genome.jp). The bold fonts indicate that a protein is detected in this study for the phagosome component.
In this study, a total of 41 proteins were detected for 22 phagosome components (Table 2). Each protein is presented with the total numbers of spectral counts and detected peptides and the log2-transformed abundance ratios between a sample pair. Abundance ratios were calculated for the three permuted pairs of Mφ phagosomes, the three permuted pairs of DC phagosomes, the three pairs of Mφ phagosome versus DC phagosome for the three mycobacterial strains respectively, and the Mφ versus DC whole cell lysate. Together, 10 abundance ratios are presented for each protein.
Table 2. List of quantified proteins involved in the phagosome and antigen presentation pathways.
The values are Log2-transformed abundance ratios between sample pairs. Abundance ratios in bold are statistically significant (see Methods).
| In Mφs | In DCs | In Mφs vs in DCs | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene symbol |
Protein description |
Phagosome pathway components |
Spectral counts | Peptide counts | ΦMφ,KO/ΦMφ,H37Rv | ΦMφ,KO/ΦMφ,BCG | ΦMφ,H37Rv/ΦMP,BCG | ΦDC,KO/ΦDC,H37Rv | ΦDC,KO/ΦDC,BCG | ΦDC,H37Rv/ΦDC,BCG | ΦMφ,KO/ΦDC,KO | ΦMφ,H37Rv/ΦDC,H37Rv | ΦMφ,BCG/ΦDC,BCG | WCMφ/ WCDC |
| Actg1 | actin, gamma, cytoplasmic 1 | F-actin | 7136 | 26 | −0.6 | 0.1 | 0.7 | −0.4 | 0.0 | 0.4 | −0.3 | −0.1 | −0.4 | 0.4 |
| Atp6v0c | ATPase, H+ transporting, lysosomal V0 subunit C | vATPase | 10 | 1 | 12 | 0.8 | −11 | −0.7 | −7.9 | −7.1 | 13 | 1.0 | 4.6 | 11.0 |
| Atp6v1a | ATPase, H+ transporting, lysosomal V1 subunit A | 32 | 6 | −1.3 | 0.8 | 2.1 | −2.9 | −1.7 | 1.1 | 1.0 | −0.5 | −1.6 | 0.7 | |
| Atp6v1b2 | ATPase, H+ transporting, lysosomal V1 subunit B2 | 12 | 5 | 1.4 | 0.6 | −0.8 | 3.6 | 0.5 | −3.2 | 0.4 | 2.7 | 0.3 | 1.4 | |
| Atp6v1e1 | ATPase, H+ transporting, lysosomal V1 subunit E1 | 57 | 7 | 1.7 | −0.2 | −1.9 | −3.5 | 1.2 | 4.7 | 3.1 | −2.0 | 4.5 | −2.1 | |
| Atp6v1f | ATPase, H+ transporting, lysosomal V1 subunit F | 1 | 1 | −8.2 | −7.0 | 1.2 | 1.0 | 1.3 | 0.2 | −7.3 | 1.9 | 0.9 | −8.7 | |
| Calr | calreticulin | CALR | 477 | 14 | −1.2 | −0.6 | 0.6 | 0.8 | −1.0 | −1.7 | 0.1 | 2.1 | −0.3 | −1.7 |
| Cd14 | CD14 antigen | Toll-like receptor | 25 | 4 | −1.4 | 2.3 | 3.7 | 0.0 | −0.2 | −0.2 | 1.0 | 2.4 | −1.5 | 3.7 |
| Coro1a | coronin, actin binding protein 1A | coronin | 44 | 2 | −2.0 | −7.4 | −5.4 | −0.3 | −2.7 | −2.4 | −11 | −9.7 | −6.7 | −4.9 |
| Ctss | cathepsin S | cathepsin | 28 | 2 | 3.7 | 0.0 | −3.7 | 1.4 | 2.9 | 1.4 | 2.2 | −0.1 | 5.1 | −0.6 |
| Dync1h1 | Dynein cytoplasmic 1 heavy chain 1 | Dynein | 204 | 29 | −0.2 | 0.1 | 0.3 | −1.0 | −0.2 | 0.9 | 0.0 | −0.8 | −0.2 | −0.2 |
| Dync1i2 | Dynein cytoplasmic 1 intermediate chain 2 | 2 | 1 | −2.0 | −0.4 | 1.6 | −3.1 | −1.9 | 1.1 | −0.3 | −1.3 | −1.8 | −5.3 | |
| H2-Ab1 | histocompatibility 2, class II antigen A, beta 1 | MHCII | 17 | 2 | 12 | −1.8 | −14 | 9.6 | 8.4 | −1.2 | 3.3 | 1.0 | 14 | 0.0 |
| H2-Eb1 | histocompatibility 2, class II antigen E beta | 7 | 1 | 11 | 0.2 | −11 | −10 | −1.9 | 8.3 | 13 | −8.5 | 11 | −0.3 | |
| H2K1 | histocompatibility 2, K1, K region | MHCI | 9 | 5 | −3.5 | −3.3 | 0.1 | −2.2 | −3.3 | −1.1 | −2.0 | −0.8 | −2.0 | −1.1 |
| H2-Q1 | histocompatibility 2, Q region locus 1 | 19 | 2 | 2.3 | 1.3 | −1.0 | 6.2 | 12 | 5.9 | 0.3 | 4.2 | 11 | 0.2 | |
| H2-T10 | histocompatibility 2, T region locus 10 | 2 | 2 | −1.8 | −0.2 | 1.6 | −0.4 | −0.4 | 0.0 | −0.1 | 1.4 | −0.3 | 3.9 | |
| Itgb1 | integrin beta 1 (fibronectin receptor beta) | integrin | 3 | 3 | −3.0 | −0.6 | 2.4 | −3.4 | −4.5 | −1.1 | 1.0 | 0.6 | −2.9 | 1.8 |
| Itgam | integrin alpha M | Complement | 64 | 8 | 1.1 | 0.2 | −0.9 | −0.5 | 1.3 | 1.8 | 0.4 | −1.2 | 1.5 | 2.0 |
| Itgb2 | integrin beta 2 | receptors | 81 | 5 | 1.5 | 0.6 | −0.9 | 6.0 | 2.3 | −3.7 | −0.3 | 4.2 | 1.5 | −1.1 |
| Lamp2 | lysosomal-associated membrane protein 2 | LAMP | 49 | 6 | 1.9 | −0.4 | −2.3 | 1.9 | 1.7 | −0.2 | 1.0 | 1.0 | 3.1 | 1.4 |
| M6pr | mannose-6-phosphate receptor, cation dependent | M6PR | 20 | 2 | 13 | 5.0 | −7.5 | −0.7 | −1.9 | −1.2 | 14 | 1.0 | 7.3 | 6.5 |
| Msr1 | macrophage scavenger receptor 1 | Scavenger receptors | 2 | 1 | 3.3 | 8.8 | 5.6 | −0.7 | −1.9 | −1.2 | 8.9 | 4.9 | −1.8 | 0.2 |
| Ncstn | Nicastrin | gamma-secretase | 4 | 1 | 3.1 | 5.0 | 1.9 | −2.8 | 0.1 | 2.9 | 5.4 | −0.5 | 0.5 | 4.8 |
| Rab5c | RAB5C, member RAS oncogene family | Rab5 | 33 | 3 | 0.6 | 0.0 | −0.5 | 2.2 | 3.0 | 0.8 | −1.8 | −0.1 | 1.2 | −1.7 |
| Rab7 | RAB7, member RAS oncogene family | Rab7 | 121 | 5 | 0.5 | 0.0 | −0.5 | 0.4 | −0.1 | −0.5 | 1.0 | 0.9 | 0.9 | 1.3 |
| Ncf1 | neutrophil cytosolic factor 1 | NADPH oxidase | 9 | 2 | 0.0 | −0.9 | −1.0 | 2.6 | 3.4 | 0.9 | −1.7 | 0.8 | 2.6 | 3.1 |
| Rac1 | RAS-related C3 botulinum substrate 1 | 76 | 6 | −0.7 | 0.0 | 0.7 | 5.4 | 4.3 | −1.2 | −2.4 | 3.8 | 1.9 | 1.3 | |
| Sec22b | SEC22 vesicle trafficking protein homolog B (S. cerevisiae) | Sec22 | 25 | 3 | 5.1 | 0.1 | −5.0 | 1.6 | 2.8 | 1.3 | 1.8 | −1.7 | 4.5 | 3.0 |
| Tap1 | transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | TAP | 4 | 1 | 1.6 | 1.6 | 0.0 | −2.4 | −0.4 | 2.0 | 4.5 | 0.5 | 2.5 | 2.0 |
| Tap2 | transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) | 7 | 3 | −1.1 | 1.0 | 2.1 | 0.9 | −1.3 | −2.2 | 3.1 | 5.1 | 0.8 | 3.8 | |
| Tfrc | transferrin receptor | TfR | 6 | 1 | −1.5 | 1.9 | 3.4 | −2.4 | 8.1 | 11 | −0.3 | −1.2 | 5.9 | −7.7 |
| Tuba1a | tubulin, alpha 1A | TUBA | 10 | 2 | 0.3 | 3.1 | 2.7 | −2.7 | 1.6 | 4.4 | 5.4 | 2.3 | 3.9 | 1.7 |
| Tuba1b | tubulin, alpha 1B | 560 | 12 | −0.4 | −0.4 | −0.1 | −0.2 | 0.2 | 0.4 | 0.0 | 0.2 | 0.6 | 0.4 | |
| Tubb1 | tubulin, beta 1 | TUBB | 1 | 1 | −11 | −0.4 | 11 | 5.6 | −4.3 | −9.9 | −6.6 | 10 | −11 | −4.0 |
| Tubb2a | tubulin, beta 2A | 4 | 2 | −2.7 | −0.2 | 2.5 | −4.9 | −1.3 | 3.6 | 2.7 | 0.5 | 1.5 | −0.8 | |
| Tubb2c | tubulin, beta 2C | 13 | 4 | −6.0 | 1.0 | 7.0 | 4.7 | 7.1 | 2.4 | −3.1 | 7.5 | 3.0 | −2.0 | |
| Tubb3 | tubulin, beta 3 | 6 | 3 | −7.7 | −5.0 | 2.7 | −1.3 | 7.6 | 8.9 | −2.8 | 3.6 | 9.8 | −1.6 | |
| Tubb4 | tubulin, beta 4 | 91 | 5 | 0.3 | −1.2 | −1.5 | 1.3 | 4.6 | 3.3 | 1.0 | 2.0 | 6.8 | 2.0 | |
| Tubb5 | tubulin, beta 5 | 696 | 20 | −1.0 | −0.5 | 0.5 | −1.3 | 1.2 | 2.4 | −0.5 | −0.7 | 1.2 | 1.0 | |
| Tubb6 | tubulin, beta 6 | 17 | 3 | 0.7 | 1.2 | 0.5 | −6.1 | −3.6 | 2.4 | 5.0 | −1.7 | 0.2 | 0.7 | |
Actin represents the most abundant protein in the sample preparations with total spectral counts >10 times higher than that of other proteins. Its abundance level is consistent across all phagosome samples with <2-fold fluctuation. It thus serves as a quality control for the label-free proteomics method. Except Actg1, Dync1h1, Tubb2a, and Itgam, all other 37 proteins are differentially abundant with statistical significance in at least one sample pair (Table 2).
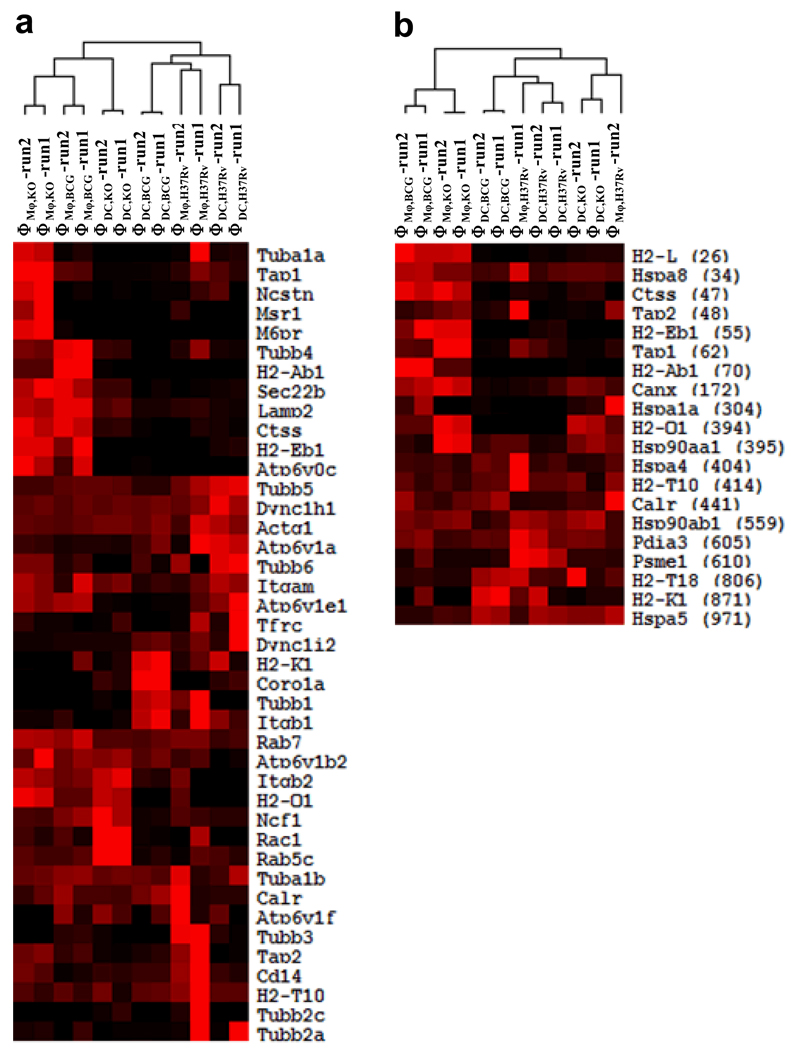
Fig. 3 illustrates the clustering of the six phagosome samples based on the proteins in Table 2 (panel a) or the proteins found by a leading edge analysis to drive the upregulation of the antigen processing and presentation pathway in Mφ phagosomes (panel b). A leading edge analysis in GSEA identifies the proteins that contribute to the core enrichment of a protein set. In panel b for example, the first seven proteins with rankings between 26 and 70 were identified to be responsible for the core enrichment to drive the upregulation of the antigen processing and presentation pathway in Mφ phagosomes (Table 1, cluster 22).
Figure 3. Clustering of the six phagosome samples based on the phagosome and antigen presentation pathway (panel a) or the antigen processing and presentation pathway (panel b).
The numbers in parenthesis in panel b indicate the rankings of the proteins in a GSEA leading edge analysis. In panel a, both the samples and proteins were clustered. In panel b, the samples were clustered while the proteins were ordered by their rankings in the GSEA analysis. The duplicate runs for each sample are shown.
Coro1a was regulated between many samples (Table 2). Specifically, the protein was upregulated in BCG phagosomes, DC phagosomes, and DC whole cell lysate. The upregulation of Coro1a was confirmed with Expander that is a java-based tool to analyze gene expression data37 (Fig. S2).
Discussion
Professional APCs such as Mφs and DCs internalize pathogens via phagocytosis and display foreign antigen complex with MHC on their surface for the recognition by T cells (Fig. 2). While DCs are potent initiators of immune response, DC phagosomes are much less characterized by proteomics than Mφ phagosomes, especially for mycobacteria-infected ones. Previously, we analyzed Mφ phagosomal proteomes modulated by the wild-type Mtb strain H37Rv, the ΔfbpA mutant strain KO, and the BCG vaccine strain BCG with label-free proteomics and systems biology analysis16. With improved proteome coverage in this study, we analyzed the mycobacterial phagosomes from the APCs infected by the H37Rv, KO, and BCG strains respectively to include DCs and their phagosomes.
Although we did not fractionate the samples in this study, we were able to quantify approximately 1000 unique host proteins, which is 3-fold more than those quantified in the previous study. At least two factors contributed to more proteins being quantified here. One is an improved sample preparation based on the FASP method22. The other is an increased number of sample runs for cross reference in label-free quantitative proteomics25, 27. 830 ± 70 proteins were detected in each sample to result in a total of 1001 unique proteins quantified from all eight samples (Table S1).
It remains an open question how many proteins can ultimately be extracted and characterized from a phagosome. It was previously estimated that 1000 proteins could be present on a phagosome11, but a recent in-depth proteomic analysis of latex-bead containing phagosomes identified 2415 phagosome proteins with a high level of sensitivity13. Latex-bead containing phagosomes are exceedingly pure preparations of phagosomes38. Proteomics is the most plausible approach to provide molecular details of a phagosome at a global level. It has been shown that 3000–7000 proteins could be identified from a Hela cell sample including membrane proteins based on the FASP sample preparation and gel-free peptide fractionation23, 39. Such a high-sensitivity proteomic analysis has the potential to identify very low-level proteins in a phagosome preparation. Trace contaminants might also be detected with the high sensitivity.
Biochemical purification of phagosomes is unlikely to be perfect because the complex interaction of phagosomes with other organelles in the cytoplasm makes a complete separation of phagosomes from other organelles difficult40. Thus, we caution that identifying a protein in a phagosome preparation by sensitive proteomic approaches such as the methods presented does not necessarily represent the definitive evidence for the true localization of the proteins on a phagosome. Secondary large-scale methods are still lacking to validate the localization of proteins identified from a phagosome preparation by proteomics.
With proper experimental design, the existence of contaminating proteins from mitochondria and other organelles might not prevent the determination of phagosome protein relative abundances12. In an earlier time-course study of phagosome dynamics, Roger et al. suggested that the organelle preparations of early and late phagosomes were identical thus contaminants should not fluctuate over time40. They discerned contaminating proteins based on this principle. Similarly, if multiple phagosome preparations are performed under identical conditions16, proteins from contaminating organelles should not differ among different phagosome preparations. Thus, the proteins found differentially abundant among different phagosome preparations represent the effect of modulation by different infecting bacterial strains or the effect of phagosome biogenesis in different host cells.
The above assumptions require additional experimental validation because it is also likely that non-identical changes in cellular compartments might be induced at different time points or by different bacterial infections. These potential changes might co-purify with phagosomes to lead to false-positive differentially abundant phagosomal proteins.
Electron microscopy has been used to examine the purity of phagosome preparations at a structural level for entrapment of other organelles or their fragments. With a higher sensitivity in LC/MS-based proteomic analysis, however, trace contaminants that cannot be discerned by electron microscopy might also be detected. Therefore, molecular-level sensitivity will be required to pinpoint which phagosomal proteins contain contaminating fractions from other organelles.
Lee et al. used radioactively labeled THP-1 macrophages that never saw latex bead or bacterial particles as internal standard to estimate the degree of contamination from other organelles during phagosome preparations15. They reported purities of 82.5 to 92% for BCG phagosome preparations using density gradient centrifugation. This radioisotopic mixing approach estimated the percentage of total contaminating proteins but did not provide a quantitation of the contaminating fraction for individual proteins.
Conceivably, the radioisotopic mixing approach could be adapted for using stable-isotope labeling to estimate the contaminating fraction of individual proteins in a phagosome preparation. Stable-isotope labeled host cells that never see bacteria or bead particles can be used as internal standard to mix with unlabeled infected cells that contain phagosomes to purify. The purified phagosomes can be analyzed with LC/MS to quantify the labeled and unlabeled fractions of a protein. The proteins with only unlabeled fraction are not contaminated and those with a labeled fraction can be estimated for the degree of contamination that can be corrected. This approach to quantify unlabeled and labeled fractions of a phagosomal protein was used to determine the dynamics of phagosome proteomes40. In that process, slow-turnover fraction of a protein was corrected from mixed phagosome populations40. Though promising for quantification of contamination, application of the stable isotope-mixing approach requires validation of stable isotope labeling of bone-marrow-derived APCs, which is beyond the scope of current study and will be pursued in the future.
We examined the purities of phagosome preparations in this study with Western blot for Calnexin and GS28 as ER and Golgi markers, respectively. Their levels were low and comparable in the three phagosome preparations for each host cell system. While Calnexin was detected in the six phagosome preparations by proteomics, GS28 was absent from these samples to suggest that the phagosome preparations were pure at least in regard to Golgi. The degree of ER involvement in phagosomes has been under debate but it is generally recognized that ER membrane contributes to the phagosome composition in many cases (see review12). Our proteomic results indicate that Calnexin abundance shows a trend of increase in the order of DC phagsomes, Mφ phagosomes, and whole cell lysates (Table S1).
Regardless of the complicated issue of phagosome preparation purity, studies have examined the proteome profiles of bacteria-containing vacuoles or latex-bead containing phagosomes41–43 to decipher the endosomal-lysosomal pathway44 and to identify the determinants of phagosome-lysosome fusion45. Several more recent studies have been carried out at larger scales with more sensitive proteomic techniques to enable a systems-level investigation of phagosomal proteomes from mycobacteria-infected Mφs15, 16. As proteins regulated among different phagosome preparations or time-point samples are most likely to be authentic members of phagosomes12, we here compare the Mφ and DC phagosomes and their whole cell lysates with emphases on regulated proteins.
Compared to the spectral counting method of proteomics15, the label-free approach with peptide and protein cross-reference provides more quantitative results, especially for lower abundant proteins25. A cluster analysis of the eight proteome profiles reveals three major protein groups of interest (Fig. 2). The overview of these proteome profiles indicates that H37Rv blocks phagosome maturation by excluding lysosome proteins while the KO phagosomes recruit lysosome proteins as well as chaperon-containing T-complex proteins. Interestingly, the H37Rv phagosomes appear to contain more ER proteins that might contribute to cross-presentation12. The BCG phagosomes from Mφs and DCs, however, show mixed results.
BCG is the only approved vaccine for humans. BCG gives variable protection against tuberculosis depending upon geographic regions because of the loss of some immunogenic antigens, strain variation and population factors. In addition, the BCG vaccine sequesters within early endosomes that do not fuse with lysosomes46.
Peptide processing and presentation through MHCII is essential. The blockade of phagosome acidification by BGC suggests that APCs like Mφs and DCs would be deficient in presenting MHCII-dependent peptides from BCG. Indeed, BCG phagosomes are near neutral in pH due to the exclusion of vacuolar-type H+-ATPase enzyme, and as a consequence, intra-phagosomal cathepsin-D is inactive. Using a T-cell hybridoma specific for a major secreted antigen from BCG antigen 85B, we found that Mφs and DCs showed a decrease in antigen processing and presentation.
Tuberculosis is an important disease for which vaccination strategies more efficient than BCG have been sought. For this reason, we analyzed alternative mechanisms of antigen processing that could lead to immune recognition of the pathogen. From the previous proteomics and systems biology analyses of phagosomes containing BCG as well as Rv purified from mouse Mφs16, we found Nicastrin, a component of γ-secretase, was enriched on the phagosome of the wild-type H37Rv and to a lesser extent on the BCG vaccine phagosomes. This trend was repeated in this study (Table 2). Nicastrin was also present on BCG and latex-bead containing phagosomes in human THP-1 Mφs possibly with greater abundances at the earlier time points of phagosome biogenesis15. Because γ-secretase has been implicated in proteolysis14 and a putative function of recycling phagosomal proteins can be envisaged, we analyzed the function of Nicastrin and that of the other two constituents of γ-secretase, i.e. Aph and Presenilin. The analysis suggested that the γ-secretase members might be involved in antigen processing and presentation process (unpublished results). Inadvertently, a recent study suggests a role for the γ-secretase pathway in susceptibility to HIV-1 infection among different ethnic groups47. These results invite further study of the functional roles of γ-secretase in infectious diseases.
GSEA analysis indicates that the antigen processing and presentation pathway is upregulated in Mφ phagosomes compared to DC phagosomes (Table 1, ΦMP,avg/ΦDC,avg, cluster 22). Consistent with upregulation of the antigen processing and presentation pathway, the pathways associated with lysosome, lytic vacuole, and vacuole GO terms are also upregulated in Mφs (Table 1, cluster 17). Because clusters 17 and 22 are not upregulated in Mφ whole cell lysate versus DC whole cell lysate (Table 1, WCMP/ WCDC), the upregulation of antigen presentation pathway in Mφ phagosomes is likely due to differential enrichment of the antigen presentation proteins on the phagosome rather than in the whole cell lysate. This notion is further supported by the observation that pathways with GO terms related to protein localization are significantly enriched on Mφ phagosomes while showing a trend of downregulation in the Mφ whole cell lysate (Table 1, cluster 6).
The pathways with GO terms of intracellular protein transport and localization are upregulated in Mφ phagosomes vs DC phagosomes while they are downregulated in DC phagosomes compared to the DC whole cell lysate (Table 1, cluster 6). This result suggests that Mφ phagosomes are more permissible to content trafficking, leading to improved intraphagosomal survival or proliferation of mycobacteria over DC phagosomes. The downregulation of actin binding and organization proteins (Table 1, clusters 7 and 8) in Mφ phagosomes, especially for the H37Rv and KO infected ones, also suggests that H37Rv and KO strains might interfere with F-actin remodeling process to avoid delivery to phagolysosomes.
While mitochondrial membranes are significantly more abundant in the Mφ whole cell lysate, they are less abundant on Mφ phagosomes (Table 1, cluster 1). A further examination indicates that DC BCG phagosomes are enriched with mitochondrial membranes. In our preliminary studies, we observed that the KO strain colocalized into autophagosomes that are thought to include ER48 or mitochondrial49 membranes for biogenesis. During formation, autophagosomes sequester cytoplasmic contents that might include other organelles such as mitochondria50. It is also likely that autophagosomes fuse with phagosomes or phagolysosomes to lead to the entrapment of other organelles within the phagosome51. The enrichment of mitochondrial membranes on DC BCG phagosomes relative to DC KO phagosomes, DC H37Rv phagosomes, and Mφ BCG phagosomes suggests that mitochondrial membranes might indeed be more involved in phagosome biogenesis in BCG-infected DCs.
An attempt to derive more detailed pictures about the regulation of phagosome proteomes based on the relative abundance ratios of individual proteins has proved to be complicated in some cases (Table 2). For example, while the relative abundance of Atp6v0c in Mφ phagosomes suggests that the H37Rv strain more effectively excludes the vacuolar-type H+-ATPase enzyme from Mφ phagosomes than the KO and BCG strains, the results for Atp6v1f do not agree. Furthermore, although relative abundances of Atp6v1b2 in DC phagosomes indicate that the H37Rv strain more effectively excludes the vacuolar-type H+-ATPase enzyme from DC phagosomes than the KO and BCG strains, those of Atp6v1e1 do not agree. It remains to determine whether members of a common enzyme complex are truly regulated in a discordant fashion or the discordance in regulation for members of a common enzyme complex is due to technical variability. For such a reason, we turn to alternative systems approaches to interpret the data on the antigen processing and presentation pathway shown in Fig. 2 and Table 2.
In panel a of Fig. 3, several known phagosome maturation markers including Lamp2, Ctss, Atp6v0c, H2-Ab1, and H2-Eb1 show higher amounts on Mφ KO phagosomes as well as Mφ BCG phagosomes. The enrichment pattern of these proteins on the Mφ phagosomes indicates that both KO and BCG phagosomes in Mφs are more mature than their DC counterparts and the H37Rv phagosomes. Mφ KO phagosomes appear to proceed further into maturation as suggested by the enrichment of additional phagosome maturation or antigen presentation markers.
The clustering of the six phagosome samples further suggests that all DC phagosomes are less mature than the KO and BCG phagosomes in Mφs. H37Rv is known to arrest phagosomes maturation. The DC phagosomes, especially the H37Rv and BCG ones, are more like the H37Rv phagosome in Mφs. In the clustering tree, the DC KO phagosome is located closer to the KO and BCG phagosomes in Mφs, suggesting that the KO phagosome might also be more mature than H37Rv and BCG phagosomes in DCs.
The upregulation of H2-Ab1 and H2-Eb1, along with Ctss and Lamp2 on KO and BCG phagosomes in Mφs indicate that MHCII antigen presentation pathway is upregulated in the KO and BCG phagosomes in Mφs. The upregulation of M6pr on the Mφ KO phagosome suggests that KO phagosomes proceed further into maturation. Thus, the Mφ KO phagosome is probably the most mature out of the six phagosomes (Fig. 3).
A similar pattern of clustering tree among the six phagosomes is observed in panel b when only the proteins in the antigen processing and presentation pathway were used to classify the samples. The results from the GSEA leading edge analysis shown in panel b lead to the identification of the proteins responsible for upregulation of the antigen presentation pathway in the Mφ phagosomes. These proteins include H2-L, Hspa8, Ctss, Tap1, Tap2, H2-Eb1, and H2-Ab1.
DCs are generally more efficient stimulators of naive, memory and alloreactive T cells than Mφs. The stronger ability of DCs to present antigens to T cells could be ascribed to the expression of higher levels of MHC and costimulatory molecules on DCs than on Mφs. The proteomic results obtained here indicate that mycobacteria are capable of excluding MHC molecules, such as the MHCII molecules H2-Ab1 and H2-Eb1, from DC phagosomes. The suppression of MHC enrichment on DC phagosomes at least in part explains how Mtb evades the host immune surveillance.
Although the data presented in Fig. 3 indicate that both KO and BCG phagosomes in Mφs are enriched with higher amounts of several known phagosome maturation markers including Lamp2, Ctss, and Atp6v0c, the degree of the phagosome maturation might still be limited compared to a fully mature phagosome. Indeed, BCG phagosomes are near neutral in pH due to the exclusion of vacuolar-type H+-ATPase enzyme, and as a consequence, intra-phagosomal cathepsin-D is inactive. The upregulation of Coro1a on BCG phagosomes versus KO or H37Rv phagosomes in either Mφs or DCs also suggests that BCG phagosomes are restricted from maturation. As mentioned before, we find that Mφs and DCs show a decrease in antigen processing and presentation by using a T-cell hybridoma specific for a major secreted antigen from BCG antigen 85B. Furthermore, the upregulation of Coro1a in DC phagosomes versus Mφ phagosomes coincides with the observation that the antigen processing and presentation pathway is downregulated in DC phagosomes. These results provide some basis for further investigation of how the BCG vaccine incur large variations in efficacy despite being the only Mtb vaccine used world wide to date.
Conclusion
We have demonstrated the first global comparison of mycobacterial phagosome proteomes from two important types of professional APCs, i.e. Mφs and DCs under three different infection conditions. Using label-free proteomics with peptide cross-reference algorithm, we substantially improved the phagosome proteome coverage even without sample fractionation prior to LC/MS analysis. The proteomic approach will be suitable for large-scale analysis of multiple samples under different infection conditions at multiple time points to generate datasets suitable for systems analysis.
With several systems biology informatics tools including functional annotation, gene set enrichment, gene ontology, and clustering analyses, we assessed the pathways in mycobacterial phagosomes with an emphasis on the regulation of phagosome maturation, antigen processing and antigen presentation pathways. We showed that a systems approach was needed to extract biological information from a relatively complex and noisy proteomic dataset, which otherwise would have been difficult to analyze with conventional protein-by-protein approaches.
The study indicates that antigen presentation pathways are repressed more in DCs than in Mφs, which in part explains how mycobacteria evade host immune surveillance. The upregulation of antigen presentation pathways in Mφs versus in DCs also suggest that Mtb might co-opt the host immune system to stimulate granuloma formation for persistence. Meanwhile, the downregulation of antigen processing and presentation in DCs helps Mtb to minimize the antimicrobial immune response to enhance Mtb survival.
The studies on phagosomal proteomes have also shown the promise to discover new antigen presentation mechanisms that a professional antigen presentation cell might use to overcome the mycobacterial blockade of conventional antigen presentation pathways. To further characterize the antigen presentation pathways in mycobacterial phagosomes, we will still need in-depth proteomic analysis of the phagosome at both a global and a targeted level. The work reported here also stresses the need of systems biology approaches for data processing to better recapitulate the antigen presentation pathways in professional antigen presentation cells modulated by the pathogen within a broader context of other biological processes in the phagosome. A better characterization of the phagosome maturation and antigen presentation pathways in Mφs and DCs will be important to improve vaccine strategies against tuberculosis disease.
Supplementary Material
Acknowledgements
Partial support for this work was provided by the NIH grants R03AI073469-01A1 (QL) and AI78420 (CJ), an NIH-NCI Howard Temin Pathway to Independence Award in Cancer Research (NDP), and an NSF Graduate Research Fellowship (SM). The LC/LTQ-FTMS system and the MassMatrix software utilized in this study were provided at the Proteomics and Informatics Services Facilities (PISF) at the Research Resources Center at University of Illinois at Chicago. The PISF was established by a grant from the Searle Funds at the Chicago Community Trust to the Chicago Biomedical Consortium.
Footnotes
Supporting Information Available
The supporting information includes a Word document and an Excel spreadsheet. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.Silverstein SC, Greenberg S, Di Virgilio F, Steinberg TH. Fundamentul Immunology. New York: Raven Press Ltd; 1989. [Google Scholar]
- 2.Kaufmann SH, Walker BD. Host-pathogen interactions. Curr Opin Immunol. 2006;18(4):371–373. doi: 10.1016/j.coi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenberg S, Burridge K, Silverstein SC. Colocalization of F-actin and talin during Fc receptor-mediated phagocytosis in mouse macrophages. J Exp Med. 1990;172(6):1853–1856. doi: 10.1084/jem.172.6.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helmy KY, Katschke KJ, Jr, Gorgani NN, Kljavin NM, Elliott JM, Diehl L, Scales SJ, Ghilardi N, van Lookeren Campagne M. CRIg: a macrophage complement receptor required for phagocytosis of circulating pathogens. Cell. 2006;124(5):915–927. doi: 10.1016/j.cell.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki K, Suda T, Naito T, Ide K, Chida K, Nakamura H. Impaired toll-like receptor 9 expression in alveolar macrophages with no sensitivity to CpG DNA. Am J Respir Crit Care Med. 2005;171(7):707–713. doi: 10.1164/rccm.200408-1078OC. [DOI] [PubMed] [Google Scholar]
- 6.Hazenbos WLW, Brown EJ. Phagocytosis: receptors and biology. In: Ernst JD, Stendahl O, editors. Phagocytosis of Bacteria and Bacterial Pathogenicity. Cambridge: Cambrige University Press; 2006. pp. 4–53. [Google Scholar]
- 7.Ramachandra L, Song R, Harding CV. Phagosomes are fully competent antigen-processing organelles that mediate the formation of peptide:class II MHC complexes. J Immunol. 1999;162(6):3263–3272. [PubMed] [Google Scholar]
- 8.Houde M, Bertholet S, Gagnon E, Brunet S, Goyette G, Laplante A, Princiotta MF, Thibault P, Sacks D, Desjardins M. Phagosomes are competent organelles for antigen cross-presentation. Nature. 2003;425(6956):402–406. doi: 10.1038/nature01912. [DOI] [PubMed] [Google Scholar]
- 9.Heath WR, Carbone FR. Cross-presentation in viral immunity and self-tolerance. Nat Rev Immunol. 2001;1(2):126–134. doi: 10.1038/35100512. [DOI] [PubMed] [Google Scholar]
- 10.Rogers LD, Foster LJ. Contributions of proteomics to understanding phagosome maturation. Cell Microbiol. 2008;10(7):1405–1412. doi: 10.1111/j.1462-5822.2008.01140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffiths G, Mayorga L. Phagosome proteomes open the way to a better understanding of phagosome function. Genome Biol. 2007;8(3):207. doi: 10.1186/gb-2007-8-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, Jagannath C, Rao PK, Singh CR, Lostumbo G. Analysis of phagosomal proteomes: From latex-bead to bacterial phagosomes. Proteomics. 2010;10(22):4098–4116. doi: 10.1002/pmic.201000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trost M, English L, Lemieux S, Courcelles M, Desjardins M, Thibault P. The phagosomal proteome in interferon-gamma-activated macrophages. Immunity. 2009;30(1):143–154. doi: 10.1016/j.immuni.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Jutras I, Laplante A, Boulais J, Brunet S, Thinakaran G, Desjardins M. Gamma-secretase is a functional component of phagosomes. J Biol Chem. 2005;280(43):36310–36317. doi: 10.1074/jbc.M504069200. [DOI] [PubMed] [Google Scholar]
- 15.Lee BY, Jethwaney D, Schilling B, Clemens DL, Gibson BW, Horwitz MA. The Mycobacterium bovis bacille Calmette-Guerin phagosome proteome. Mol Cell Proteomics. 2010;9(1):32–53. doi: 10.1074/mcp.M900396-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao PK, Singh CR, Jagannath C, Li Q. A systems biology approach to study the phagosomal proteome modulated by mycobacterial infections. Int J Clin Exp Med. 2009;2(3):233–247. [PMC free article] [PubMed] [Google Scholar]
- 17.Ullrich HJ, Beatty WL, Russell DG. Interaction of Mycobacterium avium-containing phagosomes with the antigen presentation pathway. J Immunol. 2000;165(11):6073–6080. doi: 10.4049/jimmunol.165.11.6073. [DOI] [PubMed] [Google Scholar]
- 18.Katti MK, Dai G, Armitige LY, Rivera Marrero C, Daniel S, Singh CR, Lindsey DR, Dhandayuthapani S, Hunter RL, Jagannath C. The Delta fbpA mutant derived from Mycobacterium tuberculosis H37Rv has an enhanced susceptibility to intracellular antimicrobial oxidative mechanisms, undergoes limited phagosome maturation and activates macrophages and dendritic cells. Cell Microbiol. 2008;10(6):1286–1303. doi: 10.1111/j.1462-5822.2008.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teitelbaum R, Cammer M, Maitland ML, Freitag NE, Condeelis J, Bloom BR. Mycobacterial infection of macrophages results in membrane-permeable phagosomes. Proc Natl Acad Sci U S A. 1999;96(26):15190–15195. doi: 10.1073/pnas.96.26.15190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moulton RA, Mashruwala MA, Smith AK, Lindsey DR, Wetsel RA, Haviland DL, Hunter RL, Jagannath C. Complement C5a anaphylatoxin is an innate determinant of dendritic cell-induced Th1 immunity to Mycobacterium bovis BCG infection in mice. J Leukoc Biol. 2007;82(4):956–967. doi: 10.1189/jlb.0206119. [DOI] [PubMed] [Google Scholar]
- 21.Copenhaver RH, Sepulveda E, Armitige LY, Actor JK, Wanger A, Norris SJ, Hunter RL, Jagannath C. A mutant of Mycobacterium tuberculosis H37Rv that lacks expression of antigen 85A is attenuated in mice but retains vaccinogenic potential. Infect Immun. 2004;72(12):7084–7095. doi: 10.1128/IAI.72.12.7084-7095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wisniewski JR, Nagaraj N, Zougman A, Gnad F, Mann M. Brain Phosphoproteome Obtained by a FASP-Based Method Reveals Plasma Membrane Protein Topology. J Proteome Res. 2010 doi: 10.1021/pr1002214. [DOI] [PubMed] [Google Scholar]
- 23.Wisniewski JR, Zougman A, Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res. 2009;8(12):5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- 24.Xu H, Freitas MA. MassMatrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9(6):1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q. Assigning significance in label-free quantitative proteomics to include single-peptide-hit proteins with low replicates. Int J Proteomics. 2010;2010(731582):15. doi: 10.1155/2010/731582. pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Roxas BA. An assessment of false discovery rates and statistical significance in label-free quantitative proteomics with combined filters. BMC Bioinformatics. 2009;10:43. doi: 10.1186/1471-2105-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreev VP, Li L, Cao L, Gu Y, Rejtar T, Wu SL, Karger BL. A new algorithm using cross-assignment for label-free quantitation with LC-LTQ-FT MS. J Proteome Res. 2007;6(6):2186–2194. doi: 10.1021/pr0606880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao PK, Rodriguez GM, Smith I, Li Q. Protein dynamics in iron-starved Mycobacterium tuberculosis revealed by turnover and abundance measurement using hybrid-linear ion trap-fourier transform mass spectrometry. Anal Chem. 2008;80(18):6860–6869. doi: 10.1021/ac800288t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38(Database issue):D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 35.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, Hanspers K, Isserlin R, Kelley R, Killcoyne S, Lotia S, Maere S, Morris J, Ono K, Pavlovic V, Pico AR, Vailaya A, Wang PL, Adler A, Conklin BR, Hood L, Kuiper M, Sander C, Schmulevich I, Schwikowski B, Warner GJ, Ideker T, Bader GD. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roxas BA, Li Q. Acid stress response of a mycobacterial proteome: insight from a gene ontology analysis. Int J Clin Exp Med. 2009;2(4):309–328. [PMC free article] [PubMed] [Google Scholar]
- 37.Shamir R, Maron-Katz A, Tanay A, Linhart C, Steinfeld I, Sharan R, Shiloh Y, Elkon R. EXPANDER--an integrative program suite for microarray data analysis. BMC Bioinformatics. 2005;6:232. doi: 10.1186/1471-2105-6-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulsamer AG, Wright PL, Wetzel MG, Korn ED. Plasma and phagosome membranes of Acanthamoeba castellanii. J Cell Biol. 1971;51(1):193–215. doi: 10.1083/jcb.51.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 40.Rogers LD, Foster LJ. The dynamic phagosomal proteome and the contribution of the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2007;104(47):18520–18525. doi: 10.1073/pnas.0705801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desjardins M, Celis JE, van Meer G, Dieplinger H, Jahraus A, Griffiths G, Huber LA. Molecular characterization of phagosomes. J Biol Chem. 1994;269(51):32194–32200. [PubMed] [Google Scholar]
- 42.Sturgill-Koszycki S, Haddix PL, Russell DG. The interaction between Mycobacterium and the macrophage analyzed by two-dimensional polyacrylamide gel electrophoresis. Electrophoresis. 1997;18(14):2558–2565. doi: 10.1002/elps.1150181411. [DOI] [PubMed] [Google Scholar]
- 43.Hasan Z, Schlax C, Kuhn L, Lefkovits I, Young D, Thole J, Pieters J. Isolation and characterization of the mycobacterial phagosome: segregation from the endosomal/lysosomal pathway. Mol Microbiol. 1997;24(3):545–553. doi: 10.1046/j.1365-2958.1997.3591731.x. [DOI] [PubMed] [Google Scholar]
- 44.Clemens DL, Horwitz MA. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J Exp Med. 1995;181(1):257–270. doi: 10.1084/jem.181.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kielian MC, Cohn ZA. Phagosome-lysosome fusion. Characterization of intracellular membrane fusion in mouse macrophages. J Cell Biol. 1980;85(3):754–765. doi: 10.1083/jcb.85.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vergne I, Chua J, Lee HH, Lucas M, Belisle J, Deretic V. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2005;102(11):4033–4038. doi: 10.1073/pnas.0409716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Loo KM, van Schijndel JE, van Zweeden M, van Manen D, Trip MD, Petersen DC, Schuitemaker H, Hayes VM, Martens GJ. Correlation between HIV-1 seropositivity and prevalence of a gamma-secretase polymorphism in two distinct ethnic populations. J Med Virol. 2009;81(11):1847–1851. doi: 10.1002/jmv.21601. [DOI] [PubMed] [Google Scholar]
- 48.Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3(3):285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- 49.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141(4):656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. Faseb J. 2001;15(12):2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 51.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450(7173):1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.