Summary
The Drosophila circadian oscillator is comprised of transcriptional feedback loops that are activated by CLOCK (CLK) and CYCLE (CYC) and repressed by PERIOD (PER) and TIMELESS (TIM) [1]. The timing of CLK-CYC activation and PER-TIM repression is regulated post-translationally, in part through rhythmic phosphorylation of CLK, PER and TIM [2–4]. Although kinases that control PER and TIM levels and subcellular localization have been identified [5–10], additional kinases are predicted to target PER, TIM and/or CLK to promote time-specific transcriptional repression. We screened for kinases that alter circadian behavior via clock cell directed RNA interference (RNAi) and identified the proline-directed kinase nemo (nmo) as a novel component of the circadian oscillator. Both nmo RNAi knock down and a nmo hypomorphic mutant shorten circadian period, whereas nmo overexpression lengthens circadian period. CLK levels increase when nmo expression is knocked down in clock cells, whereas CLK levels decrease and PER and TIM accumulation is delayed when nmo is overexpressed in clock cells. These data suggest that nmo slows the pace of the circadian oscillator by altering CLK, PER and TIM expression, thereby contributing to the generation of a ~24-hour circadian period.
Results and Discussion
nmo acts to lengthen circadian period
Because PER, TIM and CLK are maximally phosphorylated when CLK-CYC transcription is repressed [2–4], we sought to identify kinases that promote transcriptional repression. The loss of such kinases is predicted to increase CLK-CYC transcriptional activity, which is known to shorten circadian period [11]. In contrast, reducing the levels and/or activity of DOUBLETIME (DBT), SHAGGY (SGG) and CASEIN KINASE 2 (CK2) lengthens circadian period by slowing PER-TIM degradation or delaying PER-TIM nuclear localization [5, 8, 9, 12, 13]. To identify kinases that act to repress CLK-CYC transcription, we screened a series of kinase RNAi strains for short period rhythms. A tim-Gal4 driver was used to express 34 UAS-kinase RNAi lines in all clock cells, and one line (KK104885) targeting nemo (nmo) displayed a short period of ~22.0h in constant darkness (DD) (Table 1, compare rows 3–5; Fig. S1). Since the LNvs are key pacemaker cells that are sufficient to drive behavioral rhythms [14, 15], we predicted that expressing nmo RNAi only in LNvs would also shorten circadian period. Indeed, when the pdf-Gal4 driver was used to drive nmo RNAi in LNvs [16], the free-running period of locomotor activity rhythms shortened to ~22.5h (Table 1, compare rows 4, 6, 7).
Table 1.
Circadian locomotor activity of clock cell-specific nmo RNAi, nmo mutant, nmo mutant rescue and nmo overexpression flies.
| Row | Genotype | Na | % Rhythmicb | Period ± semc |
|---|---|---|---|---|
| 1 | per01 | 47 | 0 | NA |
| 2 | w1118 | 59 | 98.3 | 23.5 ± 0.03 |
| 3 | w1118;+/tim-Gal4 | 55 | 90.9 | 24.0 ± 0.07 |
| 4 | w1118;+/nmoRNAi-1* | 41 | 90.2 | 23.5 ± 0.05 |
| 5 | w1118;tim-Gal4/nmoRNAi-1d | 16 | 75 | 22.0 ± 0.17 |
| 6 | yw;;+/pdf-Gal4 | 11 | 100 | 24.3 ± 0.10 |
| 7 | yw;+/nmoRNAi-1;+/pdf-Gal4e | 16 | 87.5 | 22.6 ± 0.11 |
| 8 | yw;+/tim-Gal4 | 14 | 92.9 | 23.9 ± 0.08 |
| 9 | w1118;;+/pdf-Gal4 | 15 | 100.0 | 24.0 ± 0.10 |
| 10 | w1118;+/nmoRNAi-2**** | 35 | 97.1 | 23.6 ± 0.05 |
| 11 | w1118;tim-Gal4/nmoRNAi-2f | 16 | 75.0 | 22.1 ± 0.14 |
| 12 | w1118;+/nmoRNAi-2;+/pdf-Gal4g | 20 | 75.0 | 22.4 ± 0.14 |
| 13 | yw;;+/nmoP1** | 35 | 94.3 | 23.4 ± 0.03 |
| 14 | w1118;;+/nmoDf*** | 26 | 88.5 | 23.7 ± 0.05 |
| 15 | w1118;;nmoP1/nmoDfh | 74 | 36.5 | 21.4 ± 0.09 |
| 16 | w1118;tim-Gal4/+;nmoP1/nmoDf | 18 | 27.8 | 22.0 ± 0.22 |
| 17 | w1118;UAS-GFP-nmo/+;nmoP1/nmoDf | 14 | 42.9 | 21.3 ± 0.44 |
| 18 | w1118;UAS-GFP-nmo/tim-Gal4;nmoP1/nmoDfi | 25 | 56.0 | 25.4 ± 0.16 |
| 19 | w1118;UAS-GFP-nmo*****/+ | 20 | 100 | 23.7 ± 0.08 |
| 20 | w1118;UAS-GFP-nmo/tim-Gal4j | 12 | 100 | 25.1 ± 0.14 |
Number of animals tested;
Percentage of rhythmic animals.
Period of activity in constant darkness, given in hours ± sem.
nmoRNAi-1, VDRC# KK104885;
nmoP1, P[lacW]nmoP1;
nmoDf, Df(3L)Exel6279;
nmoRNAi-2, VDRC# KK101545;
UAS-GFP-nmo, UAS-GFP-nmoII. The genotypes in rows 8, 9, 11 and 12 contain +/UAS-Dcr-2 on chromosome 2 (rows 9 and 12) or chromosome 3 (rows 8 and 11) due to a very low number of rhythmic flies observed when nmoRNA-2 was crossed with the Gal4 drivers in rows 3 and 6.
Significantly shorter than w1118;+/tim-Gal4 and w1118;+/nmoRNAi-1 controls (p<10−8).
Significantly shorter than yw;;+/pdf-Gal4 and w1118;+/nmoRNAi-1 controls (p<10−8).
Significantly shorter than yw;+/tim-Gal4 and w1118;+/nmoRNAi-2 controls (p<10−10).
Significantly shorter than w1118;;+/pdf-Gal4 and w1118;+/nmoRNAi-2 controls (p<10−9).
Significantly shorter than yw;;+/nmoP1 and w1118;;+/nmoDf controls (p<10−25).
Significantly longer than w1118;tim-Gal4/+;nmoP1/nmoDf and w1118;UAS-GFP-nmo/+;nmoP1/nmoDf controls (p<10−8).
Significantly longer than w1118;+/tim-Gal4 and w1118;UAS-GFP-nmo controls (p<10−8).
To determine whether the short period phenotype was due to the action of nmo RNAi on nmo expression and not an off-target effect, nmo RNAi targeting a different portion of the nmo transcript (KK101545) was expressed in all clock cells or LNvs using the tim-Gal4 and pdf-Gal4 drivers, respectively. Expression of nmo RNAi KK101545 with either driver shortened circadian period to ~21.5h (Table 1, compare rows 8–12), thus confirming the specificity of nmo RNAi. To further demonstrate that loss of nmo function shortens circadian period, a severely hypomorphic nmo P[lacZ] insertion mutant (nmoP1) was tested for behavioral activity rhythms [17–19]. Heterozygous nmoP1/nmo deficiency (henceforth nmoP1/Df) flies shortened the period of circadian activity rhythms to ~21.5h, thus confirming that loss of nmo function shortens circadian period (Table 1, compare rows 13–15; Fig. S1). It is possible that loss of nmo function could shorten period by altering clock cell development. To test this possibility, we expressed nmo RNAi only in adults using the Gal80ts TARGET system [20]. At the permissive temperature (18°C) Gal80ts inhibits tim-Gal4 driven UAS-nmo RNAi and the circadian period is ~23.4h, whereas at restrictive temperature (30°C) nmo RNAi is expressed in clock cells and shortens circadian period by ~1h (Table S1, compare rows 5 and 11, and rows 7 and 13). Control flies that lack nmo RNAi expression at 30°C have 23.4h–24.4h behavioral activity rhythms, demonstrating that period shortening doesn’t result from increased temperature (Table S1, compare rows 3, 4 and 6 to rows 5 and 7). These results argue that period shortening is due to nmo function in differentiated oscillator cells.
Since nmo levels are severely reduced in nmoP1/Df flies [17–19], we reasoned that expressing nmo specifically in clock cells would rescue the short period rhythm phenotype. When tim-Gal4 was used to express UAS-GFP tagged NMO (UAS-GFP-nmo) in nmoP1/Df flies [21], short period behavioral rhythms were reverted to periods of ~25h (Table 1, compare rows 16–18;Fig. S1). Since the reverted period was ~1.5h longer than controls carrying only UAS-GFP-nmo, the period lengthening beyond a precise (e.g. ~23.5h period) rescue may be due to nmo overexpression. To determine if this was the case, GFP tagged nmo (GFP-NMO) was used to overexpress NMO in clock cells from wild-type flies, which resulted in long period (~25h) activity rhythms (Table 1, compare rows 3, 19, 20). The period shortening of nmo RNAi and nmoP1/Df mutants and the period lengthening of nmo overexpression flies suggested that NMO levels and/or activity are important determinants of circadian period. Though microarray and qPCR analysis demonstrate that nmo mRNA levels do not cycle in fly heads [22, 23] (data not shown), cycling of NMO protein levels could not be assessed because NMO antibodies are not available.
nmo interacts with core clock components and alters CLK and PER levels
Drosophila NMO is the founding member of the nemo-like kinase (Nlk) family of proline-directed protein kinases, which phosphorylate serine or threonine residues immediately preceding a proline residue [24]. During fly development, NMO acts in multiple signaling pathways (e.g. WNT, BMP) to regulate pattern formation in embryos [25], planar cell polarity [17, 19, 21], and apoptosis [18, 19]. NMO and its mammalian NLK counterpart are known to impart regulation by phosphorylating transcription factors such as Mad, TCF-4, and Eve [25–27].
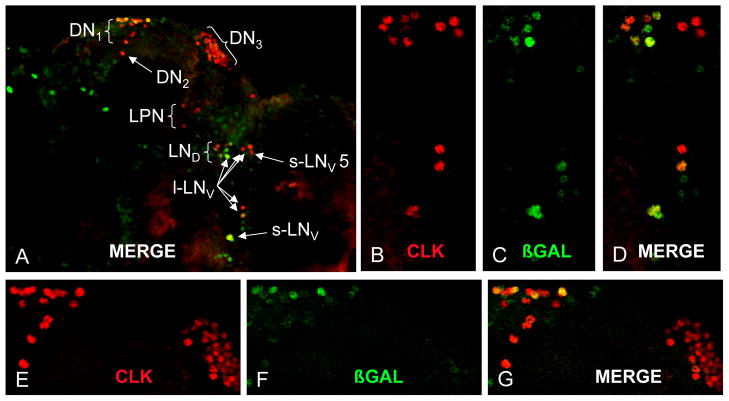
The short period phenotype produced by clock cell-specific nmo RNAi implies that nmo is expressed in clock cells. To determine the pattern of nmo expression in adults we employed the nmoP1 P[lacZ] enhancer trap insert, which was previously used to detect nmo spatial expression during development [17]. Brains from nmoP1/+ flies were dissected and co-immunostained with antisera against the oscillator cell marker CLK and the nmoP1 enhancer trap marker β-galactosidase. CLK positive neurons were assigned to cell groups according to size and position. β-galactosidase was co-localized with CLK in a subset of small ventrolateral neurons (sLNv), large ventrolateral neurons (lLNv), dorsolateral neurons (LNd), and dorsal neuron 1s (DN1), but not in dorsal neuron 2s (DN2) or dorsal neuron 3s (DN3) (Fig. 1). In addition, cells lacking CLK also expressed β-galactosidase (Fig. 1). These results demonstrate that nmo is expressed in a subset of oscillator neurons and in non-oscillator cells in the brain. Although we do not detect β-galactosidase expression in all brain oscillator neurons, it is likely that the nmoP1 insert does not trap all enhancers that drive expression in adults within the >70kb nmo gene.
Figure 1.
A nmo-lacZ enhancer trap is expressed in a subset of clock brain neurons. (A) Confocal image of brain hemisphere from w1118; nmoP1/+ flies coimmunostained with β-GAL (green) and CLK (red). DN1–3, dorsal neuron groups 1–3; l-LNV, large ventral lateral neurons; s-LNV, small ventral lateral neurons; LND, dorsal lateral neurons; LPN, lateral posterior neurons. (B–D) Magnification of the LN region from panel A. (E–G) Magnification of the DN region from panel A. Images are Z-stacks of 50μm thickness at 2μm per Z-image.
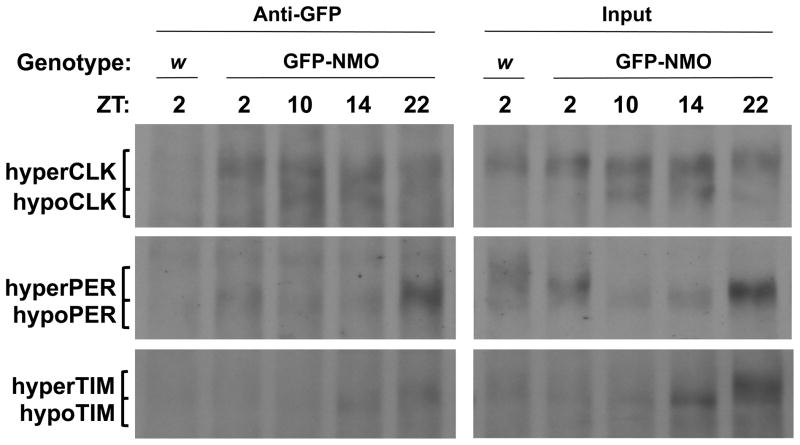
PER, TIM and CLK phosphorylation is maximal when they form a complex in the nucleus during the late night and early morning that represses transcription [2–4, 12]. If NMO phosphorylates one or more of these core clock components, we would expect NMO to be present in PER-TIM-CLK complexes when transcription is being repressed. To determine if this is the case, GFP antiserum was used to immunoprecipitate GFP-NMO complexes from flies expressing GFP-NMO in all clock cells and controls that lack GFP-NMO expression. During LD cycles, PER, TIM and CLK co-immunoprecipitate with GFP-NMO at ZT22, PER and CLK co-immunoprecipitate with GFP-NMO at ZT2, TIM co-immunoprecipitates with GFP-NMO at ZT14, and CLK co-immunoprecipitates with GFP-NMO at ZT10 and ZT14 (Fig. 2). Since TIM levels are low at ZT2 and ZT10 and PER levels are low at ZT10 and ZT14, we can’t rule out the possibility that these proteins interact with GFP-NMO at these times. These results demonstrate that NMO is present in circadian transcriptional repression complexes, and that NMO remains associated with CLK during times of transcriptional activation. Consistent with these co-immunoprecipitation results, GFP-NMO is present in the nucleus of LNvs when transcription is being repressed at ZT21 (Fig. S2).
Figure 2.
NMO interacts with PER, TIM and CLK. w1118;UAS-GFP-nmo/tim-Gal4 (GFP-NMO) and control w1118 (w) flies were entrained for three days in LD conditions and collected at the indicated times. Western blots of nuclear protein extracts of fly heads alone (Input) or after immunoprecipitation with GFP antiserum (Anti-GFP) were probed with CLK, PER and TIM antisera. Bands corresponding to hyperphosphorylated CLK, PER and TIM (hyperCLK, hyperPER and hyperTIM, respectively) and hypophosphorylated CLK, PER and TIM (hypoCLK, hypoPER, hypoTIM, respectively) are shown. See also Figure S2.
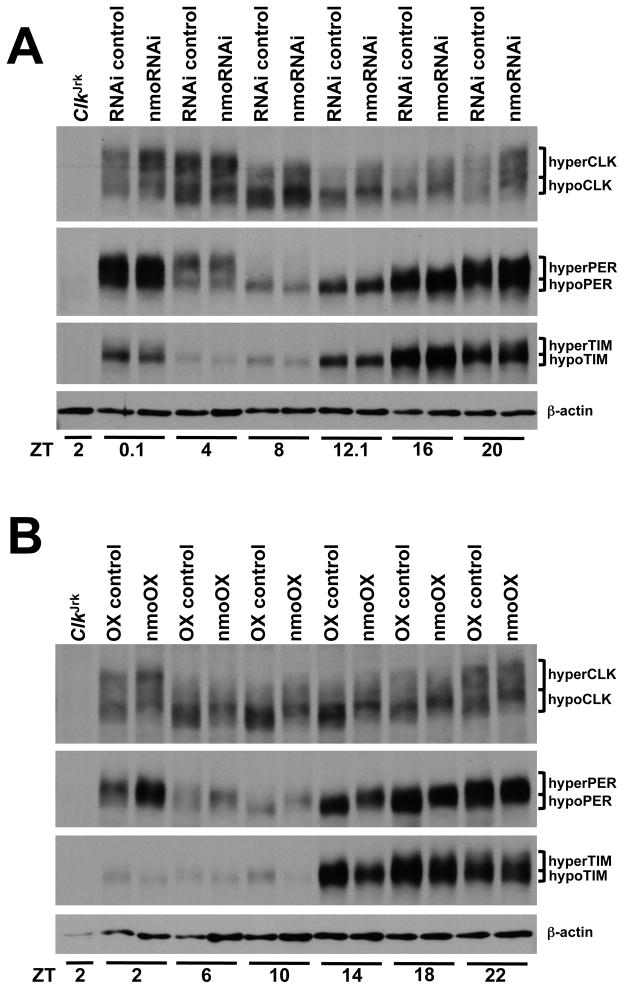
The presence of NMO in complexes with PER, TIM and CLK suggests that NMO may lengthen circadian period by phosphorylating one or more of these proteins, thereby altering their levels and/or activity. To determine if reducing NMO levels alters PER, TIM or CLK phosphorylation or abundance, antibodies against these proteins were used to probe western blots containing head extracts from nmo RNAi (tim-Gal4 driven UAS-nmo RNAi) and control (UAS-nmo RNAi/+) flies collected during LD. PER and TIM phosphorylation (as measured by the higher mobility forms) and levels were not noticeably different in tim-Gal4 driven nmo RNAi flies versus nmo RNAi/+ controls (Fig. 3a). In contrast, CLK phosphorylation and abundance were higher in tim-Gal4 driven nmo RNAi flies than in nmo RNAi/+ controls throughout the diurnal cycle (Fig. 3a; Fig. S3). This result is consistent with the progressive period shortening as Clk copy number increases [11], and suggests that NMO may act to destabilize CLK. It is also possible that knocking down nmo expression may increase CLK levels indirectly by boosting Clk transcription, but we favor the possibility that NMO destabilizes CLK because it is direct and there is ample precedent for phosphorylation-dependent degradation of clock proteins [28].
Figure 3.
NMO acts to stabilize CLK. Flies were entrained for three days under LD conditions and collected at the indicated times. (A) Western blot of head extracts from ClkJrk, w1118;+/UAS-nmoRNAi-2 (RNAi control) and w1118;UAS-nmoRNAi-2/tim-Gal4;+/UAS-Dcr-2 (nmoRNAi) flies probed with CLK, PER and TIM antisera. Bands corresponding to the different phosphorylation states of CLK, PER and TIM are marked as in Fig. 2. Actin serves as a loading control. (B) Western blot of head extracts from ClkJrk, w1118;UAS-GFP-nmo/+ (OX control) and w1118;UAS-GFP-nmo/tim-Gal4 (nmoOX) flies probed with CLK, PER and TIM antisera. Bands corresponding to the different phosphorylation states of CLK, PER and TIM are marked as above. Actin serves as a loading control. See also Fig. S3.
In contrast to the period shortening seen in nmo mutant and RNAi knock-down flies, overexpression of NMO in clock cells lengthens period by ~1.5h (Table 1). If NMO functions to destabilize CLK, then we would expect that CLK levels would be lower when NMO is overexpressed. To determine whether NMO alters the levels and/or phosphorylation of CLK and other core clock repression complex components, antibodies against CLK, PER and TIM were used to probe western blots containing head extracts from NMO overexpression (tim-Gal4 driven UAS-GFP-nmo) and control (UAS-GFP-nmo/+) flies collected during LD. CLK levels were generally lower and CLK phosphorylation was higher in NMO overexpression flies compared to controls (Fig. 3b; Fig. S3), consistent with a role for NMO in destabilizing CLK. Although nmo RNAi knockdown had little effect on PER levels and phosphorylation, NMO overexpression delayed PER accumulation and degradation and increased PER phosphorylation (Fig. 3b). TIM also accumulated in a delayed fashion (Fig. 3b), but is rapidly degraded upon exposure to light [29]. The delayed accumulation of PER and TIM and the lower levels of CLK in NMO overexpression flies are consistent with a lengthened circadian period. The delay in PER and TIM accumulation could be due to lower levels per and tim transcription, consistent with lower CLK levels in NMO overexpression flies. Alternatively, PER accumulates as a more highly phosphorylated form in NMO overexpression flies, suggesting that NMO may phosphorylate PER. If this phosphorylated form of PER is less stable it would delay PER accumulation. PER is not required for TIM stability [4, 30], thus a delay in PER accumulation due to NMO overexpression wouldn’t cause a delay in the accumulation of TIM. These results suggest that NMO phosphorylates more than one core clock component to slow the pace of the circadian oscillator.
PER, TIM and CLK each have a small number of consensus proline-directed phosphorylation sites, and could thus serve as NMO substrates. Although the identity of TIM and CLK phosphorylation sites have not been determined, at least six consensus proline-directed phosphorylation sites are phosphorylated when PER is expressed in S2 cells [12]. Eliminating a proline-directed phosphorylation site at S661 in PER delays nuclear localization and lengthens period by ~1.5h [31]. Another proline-directed phosphorylation site is situated in the “short period domain” of PER, a region spanning amino acids S585 to Y601 that contains multiple mutations that shorten circadian period including perS and perT [32–36]. When the only proline-directed phosphorylation site within the short period domain is mutated by replacing P597 with alanine, the period shortens to ~22h [33]. Replacing S596 with a non-phosphorylatable residue also shortens circadian period, and analysis using an antibody that detects phosphorylated S596 shows that this residue is phosphorylated by NMO (Chiu and Edery, personal communication). It is possible that loss of NMO phosphorylation only at PER S596 could account for the nmo mutant and nmo RNAi short period phenotype. For instance, loss of a phosphorylation site that destabilizes PER would advance feedback repression by PER complexes and shorten circadian period. Alternatively, NMO may phosphorylate multiple components of the PER repression complex to shorten circadian period. Knocking down nmo expression in clock cells via RNAi increased CLK levels, but did not detectably alter PER and TIM phosphorylation or abundance. In contrast, NMO overexpression decreased CLK levels, delayed PER and TIM accumulation and increased PER and CLK phosphorylation. These results suggest that NMO may directly phosphorylate CLK, which reduces CLK levels to maintain a 24h period. In this scenario, reduced nmo expression would increase CLK levels, thereby prematurely terminating PER repression and shortening period, whereas increased nmo expression would reduce CLK levels, thereby prolonging PER repression and lengthening period. These changes in CLK abundance likely work in concert with NMO-induced delays in PER accumulation to set circadian period.
We have identified nmo as a new component of the Drosophila circadian oscillator. The short period behavioral rhythms of nmoP1/Df and nmo RNAi flies indicates that NMO acts to slow the pace of the circadian oscillator, consistent with the lengthening of circadian period when nmo is overexpressed in clock cells. A nmo P[lacZ] enhancer trap line reveals nmo expression in the sLNv and other brain clock cells, consistent with the short period behavioral rhythms that result from expressing nmo RNAi in LNvs. NMO is present in complexes with PER, TIM and CLK when CLK-CYC transcription is repressed, and alters PER, TIM and CLK levels and/or phosphorylation state. Our results suggest that NMO acts within PER-TIM-CLK regulatory complexes to lengthen circadian period. These results are likely relevant to the mammalian circadian oscillator since mPER and CLOCK are also highly phosphorylated when transcription is repressed [37], and a single nmo ortholog, Nemo-like kinase (NLK), is present in mice and humans [38, 39].
Supplementary Material
Supplemental Figure 1, related to Table 1. Locomotor activity analysis of nmo RNAi, nmo mutant, nmo mutant revertant and control flies. nmo RNAi, nmo mutant, nmo mutant revertant and control flies of the indicated genotypes were entrained in LD cycles and then placed in DD (asterisk) for at least seven days. Representative examples of actograms and periodograms for single flies of each genotype are shown. Data collected in DD for the fly shown in the actogram was used for periodogram analysis. The period in hours is indicated in the periodogram.
Supplemental Figure 2, related to Figure 2. GFP-NMO is present in the nucleus during transcriptional repression. w1118; UAS-GFP-nmo/tim-Gal4; nmoP1/nmoDf flies were entrained for three days in LD conditions and collected at ZT21. Brains were dissected, immunostained with GFP (green) and PDF (red) antisera, and a 2 micron section through sLNvs (S) and lLNvs (L) was imaged via confocal microscopy.
Supplemental Figure 3, related to Figure 3. Quantification of CLK levels in nmo RNAi and nmo overexpression flies. (A) Quantification of CLK signals from three western blots containing independent collections of w1118; nmoRNAi-2/+ (RNAi control) and w1118; tim-Gal4/nmoRNAi-2; UAS-Dcr-2/+ (nmoRNAi) flies at the indicated timepoints. (B) Quantification of CLK signals from three western blots containing independent collections of w1118; UAS-GFP-nmo/+ (OX control) and w1118; tim-Gal4/UAS-GFP-nmo (nmoOX) flies at the indicated timepoints. Quantification was carried out as described in Supplemental Procedures. Relative CLK levels (mean +/− SEM) are shown.
Acknowledgments
We thank the Vienna Drosophila RNAi Center for providing the nmo RNAi flies, Dr. E. Verheyen for sending UAS-GFP-nmo flies, Dr. Isaac Edery for sharing unpublished results, and the Bloomington Drosophila Stock Center for providing the P{lacW}nmoP1, Df(3L)Exel6279, P{UAS-Dcr-2.D} and tubGal80ts flies. This work was supported by NIH grant NS052854.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci U S A. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 5.Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR. A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci. 2003;6:251–257. doi: 10.1038/nn1007. [DOI] [PubMed] [Google Scholar]
- 6.Akten B, Tangredi MM, Jauch E, Roberts MA, Ng F, Raabe T, Jackson FR. Ribosomal s6 kinase cooperates with casein kinase 2 to modulate the Drosophila circadian molecular oscillator. J Neurosci. 2009;29:466–475. doi: 10.1523/JNEUROSCI.4034-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 8.Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature. 2002;420:816–820. doi: 10.1038/nature01235. [DOI] [PubMed] [Google Scholar]
- 9.Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- 10.Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 11.Kadener S, Menet JS, Schoer R, Rosbash M. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 2008;6:e119. doi: 10.1371/journal.pbio.0060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu JC, Vanselow JT, Kramer A, Edery I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 2008;22:1758–1772. doi: 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muskus MJ, Preuss F, Fan JY, Bjes ES, Price JL. Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol Cell Biol. 2007;27:8049–8064. doi: 10.1128/MCB.00680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- 15.Helfrich-Forster C. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J Comp Physiol [A] 1998;182:435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- 16.Park JH, Hall JC. Isolation and chronobiological analysis of a neuropeptide pigment-dispersing factor gene in Drosophila melanogaster. J Biol Rhythms. 1998;13:219–228. doi: 10.1177/074873098129000066. [DOI] [PubMed] [Google Scholar]
- 17.Choi KW, Benzer S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell. 1994;78:125–136. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- 18.Mirkovic I, Charish K, Gorski SM, McKnight K, Verheyen EM. Drosophila nemo is an essential gene involved in the regulation of programmed cell death. Mech Dev. 2002;119:9–20. doi: 10.1016/s0925-4773(02)00289-7. [DOI] [PubMed] [Google Scholar]
- 19.Verheyen EM, Mirkovic I, MacLean SJ, Langmann C, Andrews BC, MacKinnon C. The tissue polarity gene nemo carries out multiple roles in patterning during Drosophila development. Mech Dev. 2001;101:119–132. doi: 10.1016/s0925-4773(00)00574-8. [DOI] [PubMed] [Google Scholar]
- 20.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 21.Fiehler RW, Wolff T. Nemo is required in a subset of photoreceptors to regulate the speed of ommatidial rotation. Dev Biol. 2008;313:533–544. doi: 10.1016/j.ydbio.2007.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keegan KP, Pradhan S, Wang JP, Allada R. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput Biol. 2007;3:e208. doi: 10.1371/journal.pcbi.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. Control of Daily Transcript Oscillations in Drosophila by Light and the Circadian Clock. PLoS Genet. 2006;2:e39. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyata Y, Nishida E. Distantly related cousins of MAP kinase: biochemical properties and possible physiological functions. Biochem Biophys Res Commun. 1999;266:291–295. doi: 10.1006/bbrc.1999.1705. [DOI] [PubMed] [Google Scholar]
- 25.Braid LR, Lee W, Uetrecht AC, Swarup S, Papaianni G, Heiler A, Verheyen EM. Nemo phosphorylates Even-skipped and promotes Eve-mediated repression of odd-skipped in even parasegments during Drosophila embryogenesis. Dev Biol. 343:178–189. doi: 10.1016/j.ydbio.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Ishitani T, Ninomiya-Tsuji J, Nagai S, Nishita M, Meneghini M, Barker N, Waterman M, Bowerman B, Clevers H, Shibuya H, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- 27.Zeng YA, Verheyen EM. Nemo is an inducible antagonist of Wingless signaling during Drosophila wing development. Development. 2004;131:2911–2920. doi: 10.1242/dev.01177. [DOI] [PubMed] [Google Scholar]
- 28.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 29.Ashmore LJ, Sehgal A. A fly’s eye view of circadian entrainment. J Biol Rhythms. 2003;18:206–216. doi: 10.1177/0748730403018003003. [DOI] [PubMed] [Google Scholar]
- 30.Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 31.Ko HW, Kim EY, Chiu J, Vanselow JT, Kramer A, Edery I. A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3beta/SGG in circadian clocks. J Neurosci. 2010;30:12664–12675. doi: 10.1523/JNEUROSCI.1586-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baylies MK, Bargiello TA, Jackson FR, Young MW. Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature. 1987;326:390–392. doi: 10.1038/326390a0. [DOI] [PubMed] [Google Scholar]
- 33.Baylies MK, Vosshall LB, Sehgal A, Young MW. New short period mutations of the Drosophila clock gene per. Neuron. 1992;9:575–581. doi: 10.1016/0896-6273(92)90194-i. [DOI] [PubMed] [Google Scholar]
- 34.Konopka RJ, Hamblen-Coyle MJ, Jamison CF, Hall JC. An ultrashort clock mutation at the period locus of Drosophila melanogaster that reveals some new features of the fly’s circadian system. J Biol Rhythms. 1994;9:189–216. doi: 10.1177/074873049400900303. [DOI] [PubMed] [Google Scholar]
- 35.Rutila JE, Edery I, Hall JC, Rosbash M. The analysis of new short-period circadian rhythm mutants suggests features of D. melanogaster period gene function. J Neurogenet. 1992;8:101–113. doi: 10.3109/01677069209084155. [DOI] [PubMed] [Google Scholar]
- 36.Yu Q, Jacquier AC, Citri Y, Hamblen M, Hall JC, Rosbash M. Molecular mapping of point mutations in the period gene that stop or speed up biological clocks in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1987;84:784–788. doi: 10.1073/pnas.84.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 38.Brott BK, Pinsky BA, Erikson RL. Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc Natl Acad Sci U S A. 1998;95:963–968. doi: 10.1073/pnas.95.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harada H, Yoshida S, Nobe Y, Ezura Y, Atake T, Koguchi T, Emi M. Genomic structure of the human NLK (nemo-like kinase) gene and analysis of its promoter region. Gene. 2002;285:175–182. doi: 10.1016/s0378-1119(02)00412-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1, related to Table 1. Locomotor activity analysis of nmo RNAi, nmo mutant, nmo mutant revertant and control flies. nmo RNAi, nmo mutant, nmo mutant revertant and control flies of the indicated genotypes were entrained in LD cycles and then placed in DD (asterisk) for at least seven days. Representative examples of actograms and periodograms for single flies of each genotype are shown. Data collected in DD for the fly shown in the actogram was used for periodogram analysis. The period in hours is indicated in the periodogram.
Supplemental Figure 2, related to Figure 2. GFP-NMO is present in the nucleus during transcriptional repression. w1118; UAS-GFP-nmo/tim-Gal4; nmoP1/nmoDf flies were entrained for three days in LD conditions and collected at ZT21. Brains were dissected, immunostained with GFP (green) and PDF (red) antisera, and a 2 micron section through sLNvs (S) and lLNvs (L) was imaged via confocal microscopy.
Supplemental Figure 3, related to Figure 3. Quantification of CLK levels in nmo RNAi and nmo overexpression flies. (A) Quantification of CLK signals from three western blots containing independent collections of w1118; nmoRNAi-2/+ (RNAi control) and w1118; tim-Gal4/nmoRNAi-2; UAS-Dcr-2/+ (nmoRNAi) flies at the indicated timepoints. (B) Quantification of CLK signals from three western blots containing independent collections of w1118; UAS-GFP-nmo/+ (OX control) and w1118; tim-Gal4/UAS-GFP-nmo (nmoOX) flies at the indicated timepoints. Quantification was carried out as described in Supplemental Procedures. Relative CLK levels (mean +/− SEM) are shown.