Abstract
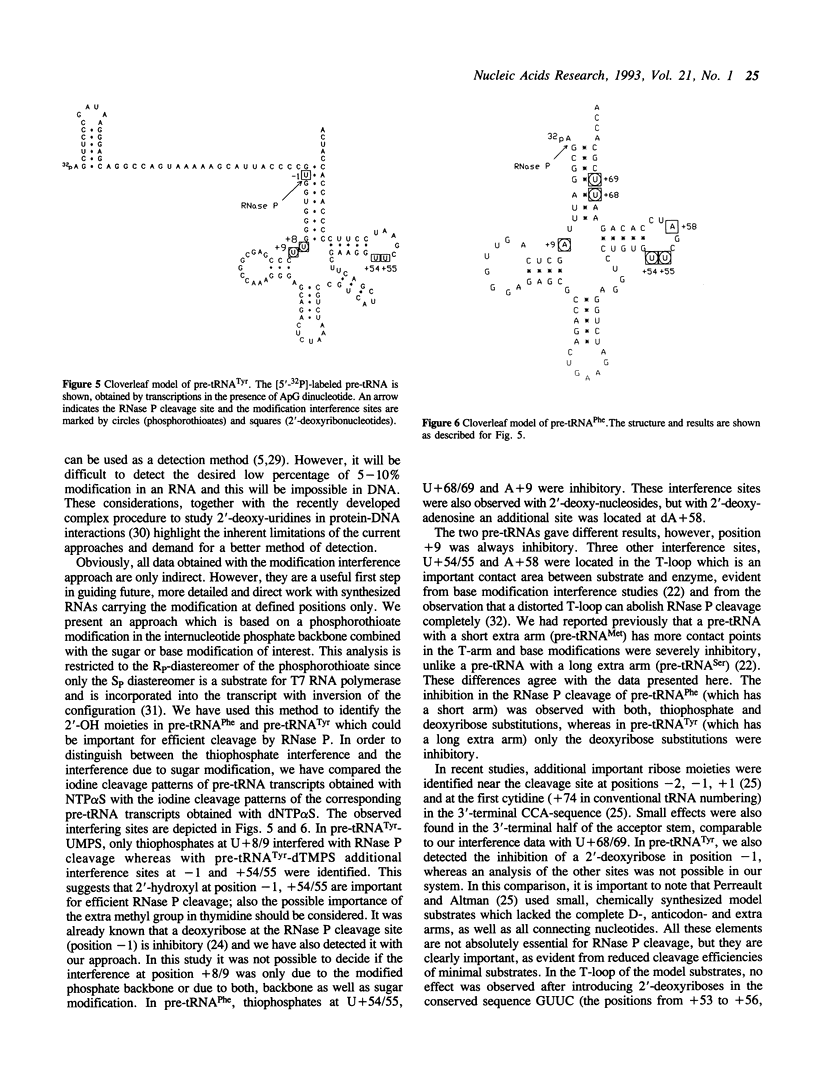
A new approach for modification interference studies is presented. It involves the use of phosphorothioates as a handle to analyze any desired base or sugar modification. This method was applied to identify ribose and phosphate moieties which could be important in the pre-tRNA recognition of E. coli RNase P RNA (M1 RNA). The utility of this technique was confirmed by detecting the inhibitory effect of a deoxyribose in the 5'-flank (position-1). This site was already known to interfere with RNase P cleavage, if modified. We have analyzed pre-tRNA(Tyr) and pre-tRNA(Phe) and found different interference patterns for both tRNAs. Two unpaired regions were involved in both pre-tRNAs. Phosphorothioates interfered at the transition between acceptor- and D-arms. The results with deoxythymidines in the T-loop indicated that deoxyribose moieties or the extra methyl group in thymidine could interfere with RNAse P cleavage. These data suggest that even in complete pre-tRNAs, only a few intact ribonucleotides are important in the substrate recognition by RNase P. We have demonstrated the potential of this new approach which offers many future applications in all fields involving nucleic acids, for example RNA processing, action of ribozymes, tRNA charging and studies related to DNA promoter recognition.
Full text
PDF
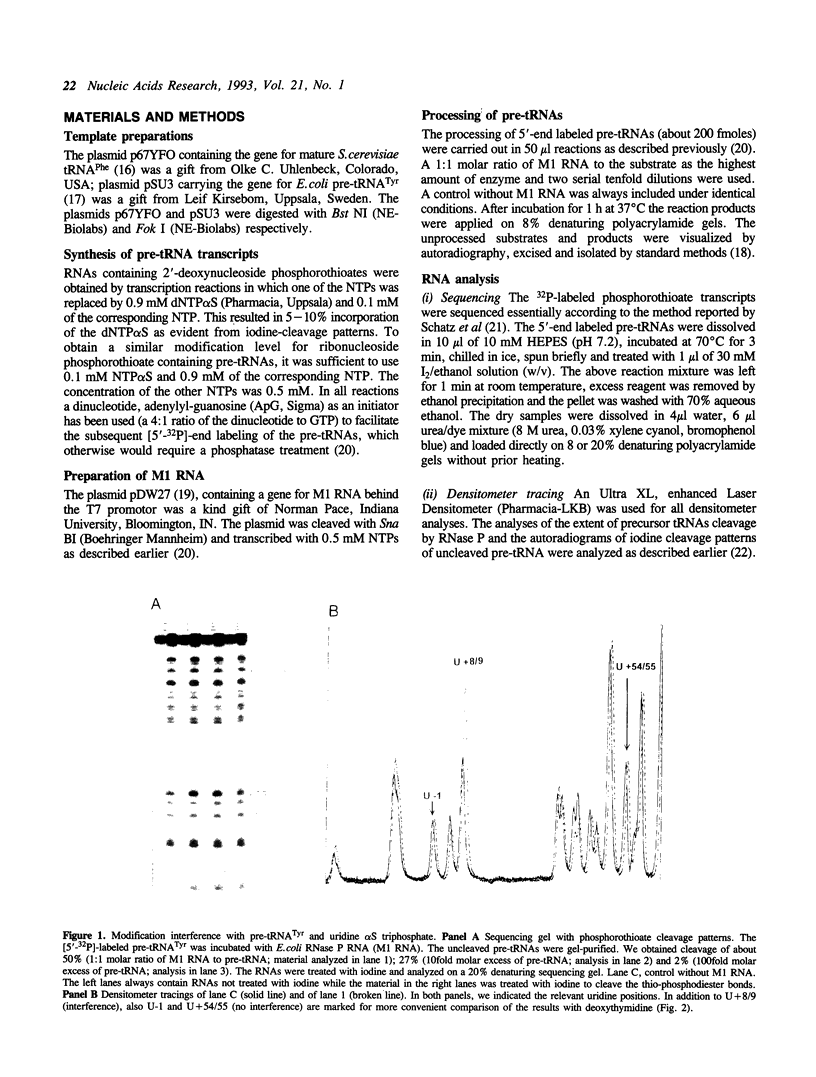
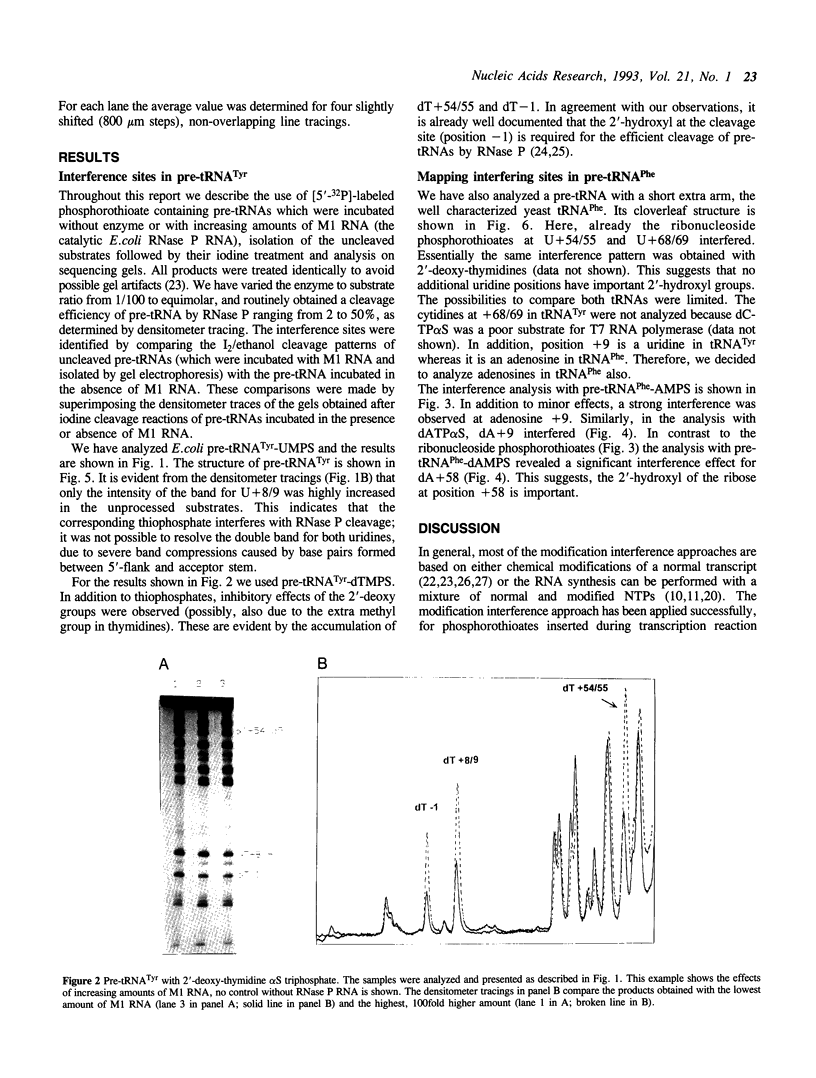
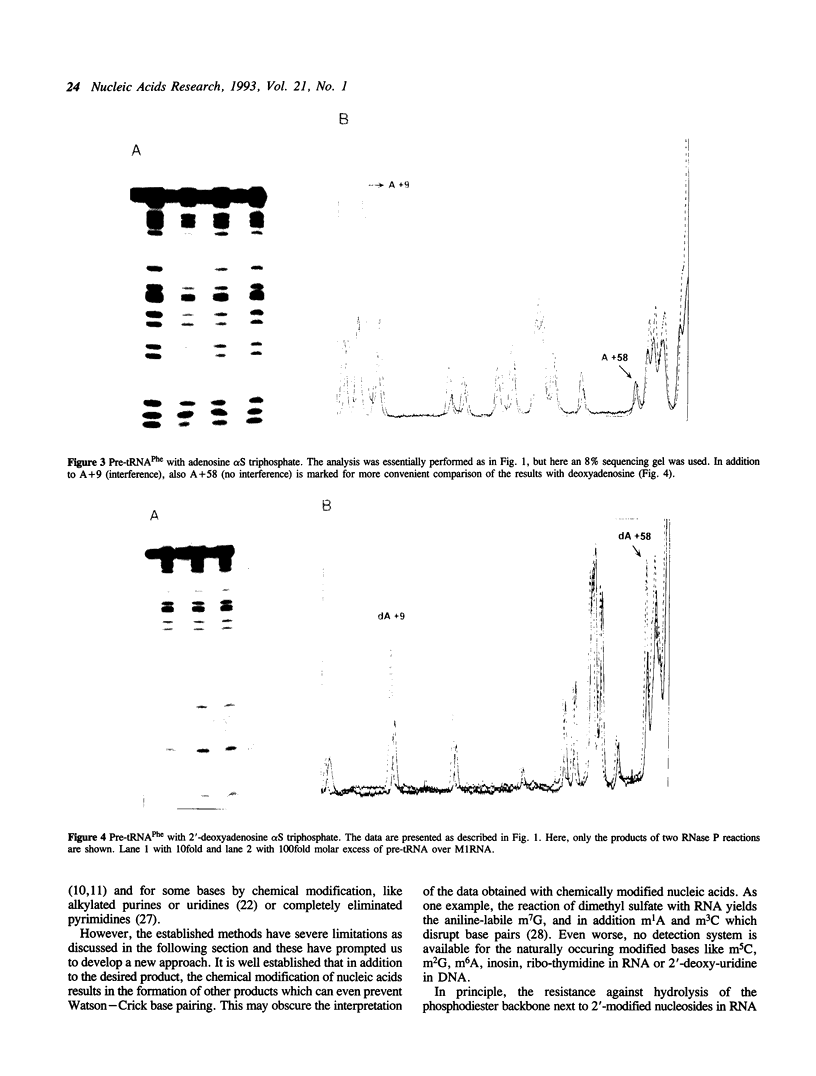

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Buzayan J. M., van Tol H., Feldstein P. A., Bruening G. Identification of a non-junction phosphodiester that influences an autolytic processing reaction of RNA. Nucleic Acids Res. 1990 Aug 11;18(15):4447–4451. doi: 10.1093/nar/18.15.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowrira B. M., Burke J. M. Extensive phosphorothioate substitution yields highly active and nuclease-resistant hairpin ribozymes. Nucleic Acids Res. 1992 Jun 11;20(11):2835–2840. doi: 10.1093/nar/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway L., Wickens M. Analysis of mRNA 3' end formation by modification interference: the only modifications which prevent processing lie in AAUAAA and the poly(A) site. EMBO J. 1987 Dec 20;6(13):4177–4184. doi: 10.1002/j.1460-2075.1987.tb02764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein F. Nucleoside phosphorothioates. Annu Rev Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Altman S. External guide sequences for an RNA enzyme. Science. 1990 Aug 17;249(4970):783–786. doi: 10.1126/science.1697102. [DOI] [PubMed] [Google Scholar]
- Gardiner K. J., Marsh T. L., Pace N. R. Ion dependence of the Bacillus subtilis RNase P reaction. J Biol Chem. 1985 May 10;260(9):5415–5419. [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hegg L. A., Thurlow D. L. Residual tRNA secondary structure in 'denaturing' 8M urea/TBE polyacrylamide gels: effects on electrophoretic mobility and dependency on prior chemical modification of the tRNA. Nucleic Acids Res. 1990 May 25;18(10):2993–3000. doi: 10.1093/nar/18.10.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle D., Wehmeyer U., Char S., Krupp G. The methylation of one specific guanosine in a pre-tRNA prevents cleavage by RNase P and by the catalytic M1 RNA. Nucleic Acids Res. 1990 Feb 25;18(4):837–844. doi: 10.1093/nar/18.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle D., Wehmeyer U., Krupp G. Substrate recognition by RNase P and by the catalytic M1 RNA: identification of possible contact points in pre-tRNAs. EMBO J. 1990 Jun;9(6):1929–1937. doi: 10.1002/j.1460-2075.1990.tb08320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom L. A., Baer M. F., Altman S. Differential effects of mutations in the protein and RNA moieties of RNase P on the efficiency of suppression by various tRNA suppressors. J Mol Biol. 1988 Dec 20;204(4):879–888. doi: 10.1016/0022-2836(88)90048-4. [DOI] [PubMed] [Google Scholar]
- Lang K. M., Keller W. Sequence requirements in different steps of the pre-mRNA splicing reaction: analysis by the RNA modification-exclusion technique. Mol Cell Biol. 1990 Sep;10(9):4942–4947. doi: 10.1128/mcb.10.9.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Determination of RNA-protein contacts using thiophosphate substitutions. Biochemistry. 1989 Apr 4;28(7):2849–2855. doi: 10.1021/bi00433a016. [DOI] [PubMed] [Google Scholar]
- Paolella G., Sproat B. S., Lamond A. I. Nuclease resistant ribozymes with high catalytic activity. EMBO J. 1992 May;11(5):1913–1919. doi: 10.1002/j.1460-2075.1992.tb05244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault J. P., Altman S. Important 2'-hydroxyl groups in model substrates for M1 RNA, the catalytic RNA subunit of RNase P from Escherichia coli. J Mol Biol. 1992 Jul 20;226(2):399–409. doi: 10.1016/0022-2836(92)90955-j. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Labuda D., Usman N., Yang J. H., Cedergren R. Relationship between 2'-hydroxyls and magnesium binding in the hammerhead RNA domain: a model for ribozyme catalysis. Biochemistry. 1991 Apr 23;30(16):4020–4025. doi: 10.1021/bi00230a029. [DOI] [PubMed] [Google Scholar]
- Perreault J. P., Wu T. F., Cousineau B., Ogilvie K. K., Cedergren R. Mixed deoxyribo- and ribo-oligonucleotides with catalytic activity. Nature. 1990 Apr 5;344(6266):565–567. doi: 10.1038/344565a0. [DOI] [PubMed] [Google Scholar]
- Pu W. T., Struhl K. Uracil interference, a rapid and general method for defining protein-DNA interactions involving the 5-methyl group of thymines: the GCN4-DNA complex. Nucleic Acids Res. 1992 Feb 25;20(4):771–775. doi: 10.1093/nar/20.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly R. M., RajBhandary U. L. A single mutation in loop IV of Escherichia coli SuIII tRNA blocks processing at both 5'- and 3'-ends of the precursor tRNA. J Biol Chem. 1986 Feb 25;261(6):2928–2935. [PubMed] [Google Scholar]
- Ruffner D. E., Uhlenbeck O. C. Thiophosphate interference experiments locate phosphates important for the hammerhead RNA self-cleavage reaction. Nucleic Acids Res. 1990 Oct 25;18(20):6025–6029. doi: 10.1093/nar/18.20.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D., Leberman R., Eckstein F. Interaction of Escherichia coli tRNA(Ser) with its cognate aminoacyl-tRNA synthetase as determined by footprinting with phosphorothioate-containing tRNA transcripts. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6132–6136. doi: 10.1073/pnas.88.14.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark B. C., Kole R., Bowman E. J., Altman S. Ribonuclease P: an enzyme with an essential RNA component. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3717–3721. doi: 10.1073/pnas.75.8.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow D. L., Shilowski D., Marsh T. L. Nucleotides in precursor tRNAs that are required intact for catalysis by RNase P RNAs. Nucleic Acids Res. 1991 Feb 25;19(4):885–891. doi: 10.1093/nar/19.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh D. S., Green C. J., Pace N. R. The design and catalytic properties of a simplified ribonuclease P RNA. Science. 1989 Jun 30;244(4912):1569–1571. doi: 10.1126/science.2472671. [DOI] [PubMed] [Google Scholar]
- Williams D. M., Pieken W. A., Eckstein F. Function of specific 2'-hydroxyl groups of guanosines in a hammerhead ribozyme probed by 2' modifications. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):918–921. doi: 10.1073/pnas.89.3.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. H., Perreault J. P., Labuda D., Usman N., Cedergren R. Mixed DNA/RNA polymers are cleaved by the hammerhead ribozyme. Biochemistry. 1990 Dec 25;29(51):11156–11160. doi: 10.1021/bi00503a002. [DOI] [PubMed] [Google Scholar]
- Yang J. H., Usman N., Chartrand P., Cedergren R. Minimum ribonucleotide requirement for catalysis by the RNA hammerhead domain. Biochemistry. 1992 Jun 2;31(21):5005–5009. doi: 10.1021/bi00136a013. [DOI] [PubMed] [Google Scholar]