Abstract
Autism spectrum disorders (ASDs) comprise a range of disorders that share a core of neurobehavioural deficits characterized by widespread abnormalities in social interactions, deficits in communication as well as restricted interests and repetitive behaviours. The neurological basis and circuitry mechanisms underlying these abnormal behaviours are poorly understood. Shank3 is a postsynaptic protein, whose disruption at the genetic level is thought to be responsible for development of 22q13 deletion syndrome (Phelan-McDermid Syndrome) and other non-syndromic ASDs. Here we show that mice with Shank3 gene deletions exhibit self-injurious repetitive grooming and deficits in social interaction. Cellular, electrophysiological and biochemical analyses uncovered defects at striatal synapses and cortico-striatal circuits in Shank3 mutant mice. Our findings demonstrate a critical role for Shank3 in the normal development of neuronal connectivity and establish causality between a disruption in the Shank3 gene and the genesis of autistic like-behaviours in mice.
Introduction
Autism and autism spectrum disorders (ASDs) are neurodevelopmental disorders diagnosed based on a triad of criteria: deficits in communication, impaired social interaction, and repetitive or restricted interests and behaviours1. ASDs are highly heritable disorders with concordance rates as high as 90% for monozygotic twins2. Recent genetic and genomic studies have identified a large number of candidate genes for ASDs3, many of which encode synaptic proteins4–6, suggesting synaptic dysfunction may play a critical role in ASDs7,8. One of the most promising ASD candidate genes is Shank3, which codes for a key postsynaptic density (PSD) protein at glutamatergic synapses. Disruption of Shank3 is thought to be the cause of core neurodevelopmental and neurobehavioural deficits in the 22q13 deletion syndrome (Phelan-McDermid syndrome), an autism spectrum disorder9–11. Furthermore, recent genetic screens have identified several mutations/rare variants of the Shank3 gene in ASD patients outside of diagnosed 22q13 deletion syndrome12,13.
Shank family proteins (Shank1–3) directly bind SAPAP to form the PSD95/SAPAP/Shank complex14,15. This core of proteins is thought to function as a scaffold, orchestrating the assembly of the macromolecular postsynaptic signaling complex at glutamatergic synapses. Currently, however, little is known about the in vivo function of Shank3 at the synapse and how a disruption of Shank3 may contribute to ASDs. Here we demonstrate that genetic disruption of Shank3 in mice leads to compulsive/repetitive behaviour and impaired social interaction, resembling two of the cardinal features of ASD. Biochemical, morphological and electrophysiological studies revealed synaptic dysfunction at cortico-striatal synapses, part of the neural circuits strongly implicated as dysfunctional in ASDs. Our studies provide a synaptic and circuitry mechanism underlying Shank3 disruption and ASD-like behaviours.
Shank3B−/− mice display repetitive grooming
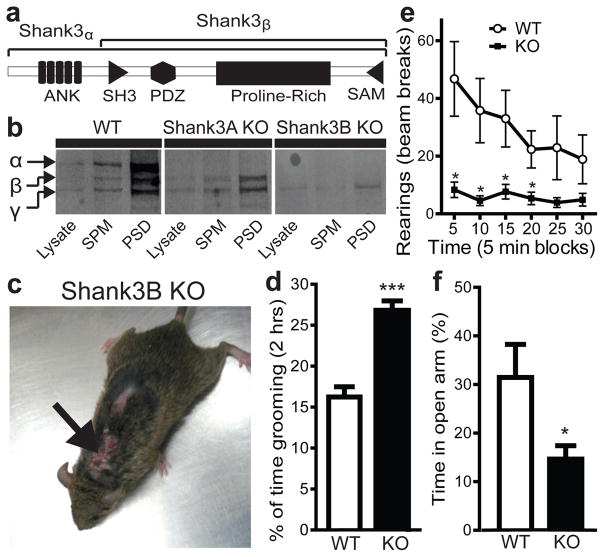
The Shank3 gene codes for large proteins with multiple protein-protein interaction domains (Fig. 1a). We generated two different alleles of Shank3 mutant mice. In Shank3A mutant mice, we targeted a portion of the gene encoding the ankyrin repeats (Supplementary Fig. 1b). This resulted in a complete elimination of Shank3α, the longest Shank3 isoform (Fig. 1b). However, the other two isoforms were not affected (here named Shank3β and Shank3γ). In Shank3B mutants, we targeted the fragment encoding the PDZ domain (Supplementary Fig. 1c). This led to the complete elimination of both Shank3α and Shank3β isoforms and a significant reduction of the putative Shank3γ isoform at the PSD (−42.12% ± 9.27% of control, n=3, p<0.05) (Fig. 1b). Our analysis is mainly focused on the Shank3B−/− mutants due to their more pronounced behavioural and physiological defects.
Figure 1. Excessive grooming, skin lesions and anxiety-like behaviour in Shank3B−/− mice.
a, Shank3 protein structure. b, Western blot showing a pan-Shank3 antibody staining in brain lysate, synaptosomal plasma membrane (SPM) and PSD 2T fraction in wildtype (WT), Shank3A−/− and Shank3B−/− mice. c, Four month old Shank3B−/− mice display neck and head lesions (arrows). d, Pre-lesion Shank3B−/− (KO) mice spent more time in self-grooming than WT. e, In the open field test, Shank3B−/− mice, when compared to controls, display decreased rearing activity. f, In the zero maze test, Shank3B−/− mice spent less time in the open area than wildtype controls. * p<0.05, *** p<0.001, two-tailed t-test for d and f, Two-way repeated measures ANOVA with post hoc two-tailed t-test for e; all data are presented as means ± s.e.m. from 6–9 mice per genotype.
We used mice with a hybrid genetic background to avoid the potential contribution to behavioral phenotypes of homozygous genetic variants on a pure inbred background7,16. Initially, F1 hybrids from heterozygous x heterozygous matings were generated and homozygous mice were born at an expected Mendelian rate. However, homozygous knockout mice from this type of mating are smaller than their wildtype littermates, presumably due to inadequate competition for resources during early postnatal days leading to different developmental trajectories. We postulated that this size difference would influence our behavioral tests. To alleviate this confound, heterozygous animals were crossed in direct brother-sister matings for 5 generations from which we derived F5 isogenic hybrids in a mixed background. These isogenic animals were then used to generate time-mated homozygous x homozygous breeding pairs to obtain wildtype and mutant animals used in the experiments. F5 Shank3A and F5 Shank3B knockouts from these matings are reared to weaning age with body weights similar to those from F5 control animals.
Shank3B−/− mice did not display any gross anatomical or histological brain abnormality, but on rare occasion exhibited seizures during handling in routine husbandry procedures. However, spontaneous seizures were never observed. By the age of 3–6 months, Shank3B−/− mice developed pronounced skin lesions with varying degrees of phenotypical penetrance: approximately 35% in the general holding colony (Fishers exact test, p<0.0001), and 100% in mating females that have produced 4–6 litters. The lesions tend to appear first on the back of the neck or on the face (Fig. 1c) and usually progressed bilaterally to cover large areas of the body. The lesions were self-inflicted, as they were present in animals socially isolated at weaning age, and not due to excessive allogrooming, as no lesions were found in wildtype or Shank3B+/− mice housed from birth with Shank3B−/− animals. Furthermore, 24 h videotaping in pre-lesion animals revealed that Shank3B−/−mice showed an increase in time spent grooming when compared to wildtype controls (Fig. 1d). These observations indicate that Shank3B−/− mice display excessive grooming and self-injurious behaviour.
We further characterized the animals in a battery of behavioural tests. In the rotarod motor test, Shank3B−/− and control animals performed at similar levels (Supplementary Fig. 2). In the open field test, when compared to controls, Shank3B−/− mice showed similar levels of activity and thigmotaxis (Supplementary Fig. 2). However, rearing, which is a form of vertical exploration considered to be anxiogenic for mice, was significantly reduced in the mutants (Fig. 1e). In the elevated zero maze, the Shank3B−/− mice spent less time exploring the open arms of the maze versus the closed arms (Fig. 1f). In the light-dark emergence test, the Shank3B−/− mice displayed an increased latency to cross into the brightly lit area, although the time spent in each side of the box was similar between mutant animals and controls (Supplementary Fig. 2). Thus, the Shank3B−/− mice display an anxiety-like behaviour and excessive, self-injurious grooming. In contrast, Shank3A−/−mice displayed no lesions or anxiety-like behaviour (Supplementary Fig. 3).
Social interaction deficits in Shank3B−/− mice
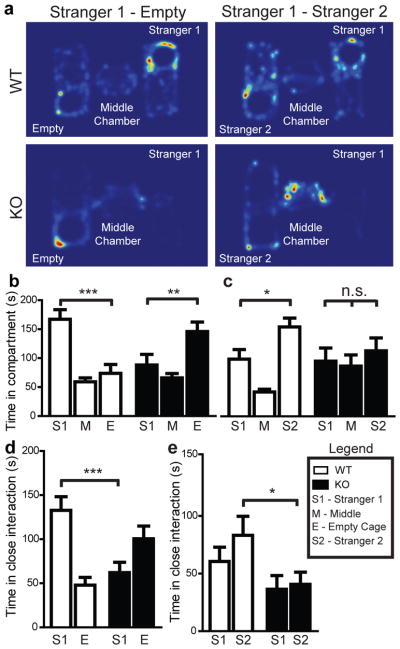
Deficits in social interaction are the most recognizable manifestation of autistic behaviours in humans. We used a modified version of a three-chamber social arena17 to probe animals for their voluntary initiation of social interaction and their ability to discriminate social novelty. Initially, the test animal was left to explore and initiate social contact with a partner (“Stranger 1”) held inside a wired cage or an identical but empty wired cage (“Empty Cage”). In this test, the Shank3B−/− mice displayed dysfunctional social interaction behaviour, as measured by observing both time spent in the compartment containing the social partner (Fig. 2a, b) or in close interaction (Fig. 2d). Notably, Shank3B−/− mice exhibited a clear preference for interacting with the empty cage rather than with the social partner (Fig. 2a, d). In a subsequent trial, a novel social partner (“Stranger 2”) was introduced into the previously empty wired cage. Wildtype mice displayed a preference for the novel animal, as shown by the increase in time spent in the compartment containing “Stranger 2”. The Shank3B−/−mutants markedly spent more time in the center chamber (Fig. 2c) and a reduced amount of time closely interacting with either social partner (Fig. 2e). In an identical test, the Shank3A−/− mice displayed normal initiation of social interaction, but perturbed recognition of social novelty (Supplementary Fig 4).
Figure 2. Reduced social interaction and abnormal social novelty recognition in Shank3B−/−mice.
a, Representative heat map analysis from “Stranger 1 Empty” and “Stranger 1 Stranger 2” trials from Shank3B−/− mice and controls. b, In the social interaction test, Shank3B−/− mice (closed bars) spent less time in the chamber containing the social partner (Stranger 1) and more time in the chamber containing the empty wire cage (Empty Cage) when compared to controls (open bars). c, In the social novelty test, Shank3B−/− mice do not display a preference for the novel social partner (Stranger 2), and spent more time in the middle chamber. d, e, When analyzing social interaction by close proximity (within 5 cm) to either “Stranger 1”, “Empty Cage” (d), or “Stranger 1”, “Stranger 2” (e), Shank3B−/− mice displayed a clear reduction in social interaction when compared to controls (d); while under a social novelty paradigm (e), Shank3B−/− mice displayed a clear reduction in time spent with “Stranger 2 * p<0.05, ** p<0.01, *** p<0.0001; One-way ANOVA, with Bonferroni post hoc t-test for b–e; all data presented as means ± s.e.m; 12–14 mice per group.
Additionally, in an open arena test, freely interacting dyadic pairs of wildtype-Shank3B−/−mice displayed less time spent in reciprocal interaction, a lower frequency of nose-to-nose interaction and anogenital sniffing when compared to wildtype-wildtype pairs (Supplementary Fig. 5). Thus, data from both social interaction tests indicate that Shank3B−/− mice display abnormal social interaction as well as deficits in discriminating social novelty.
In our breeding scheme, Shank3B−/− mice and wildtype mice were nurtured by Shank3B−/− and wildtype dams, respectively. To assess the impact of maternal rearing on the observed sociability defects, we performed time-mated cross-fostering of Shank3B−/− mice and controls. Cross-fostering of Shank3B−/− neonatal pups with wildtype dams (KOcf) revealed qualitatively equivalent social defects in the mutant mice as compared to those observed in mutant mice from homozygous x homozygous matings. Additionally, rearing wildtype neonatal pups by Shank3B−/− dams (WTcf) did not perturb normal sociability in wildtype animals (Supplementary Fig. 6). These data further suggest a genetic origin of the abnormal social behaviors in the Shank3 mutant mice.
Altered PSD composition in the striatum
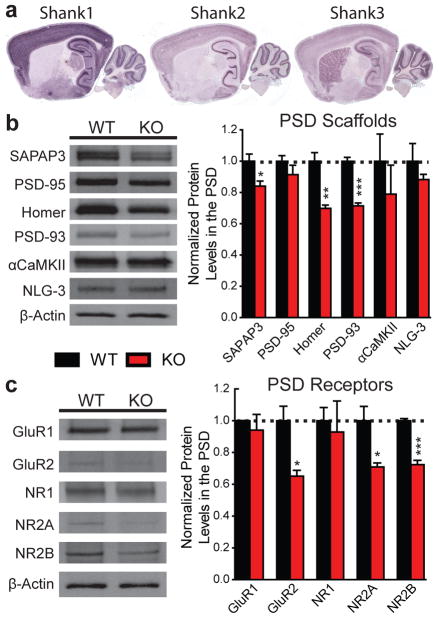
The basal ganglia are one of the brain regions implicated as dysfunctional in ASD. The repetitive/compulsive grooming behaviour in Shank3B−/− mice also suggests defects in cortico-striatal function. Furthermore, Shank3, but not Shank1 or Shank2, is highly expressed in the striatum (Fig. 3a) (Supplementary Fig. 7 and Supplementary Table 1). Therefore, we focused our analyses on striatal neurons and cortico-striatal synapses.
Figure 3. Biochemical changes in striatal synapses of Shank3B−/− mice.
a, Only Shank3 mRNA is highly expressed in the striatum. b, Protein levels of the scaffolding proteins SAPAP3, Homer and PSD93 are reduced in striatal PSD fractions from Shank3B−/− mice. c, Protein levels of glutamate receptor subunits GluR2, NR2A and NR2B are reduced in striatal PSD fractions from Shank3B−/− mice. Each lane was loaded with 3 μg of protein with β-Actin as loading control and normalized to wildtype levels. * p<0.05, ** p<0.01, *** p<0.001, two-tailed t-test; all data are presented as means ± s.e.m; n=3 samples per group, with each sample being a combined pool of striatal tissue from 3 animals.
Shank family members have been proposed as key regulators of the PSD at glutamatergic synapses18. To determine how the disruption of Shank3 may affect the PSD protein network, we used biochemically purified PSDs from the striatum of wildtype and Shank3B−/− mice and performed semi-quantitative Western blotting for several scaffolding proteins (Fig. 3b) and glutamate receptor subunits (Fig. 3c). At the PSD level, we observed reduced levels of SAPAP3, Homer-1b/c and PSD93 (Fig. 3b) as well as a reduction in the glutamate receptor subunits GluR2, NR2A and NR2B (Fig. 3c). These results suggest an altered molecular composition of postsynaptic machinery in the striatum and a possible disruption of glutamatergic signalling.
Morphological defects of medium spiny neurons
To test whether disruption of Shank3 affects neuronal morphology, we traced Golgi stained striatal MSNs and their dendrites to investigate the cellular morphology and complexity of these cells. Sholl analysis revealed neuronal hypertrophy as measured by an increase in complexity of dendritic arbors (Fig. 4a), total dendritic length (Fig. 4b) and also an increase in surface area (Fig. 4c) in Shank3B−/− MSNs.
Figure 4. Morphological and ultrastructural neuronal abnormalities in Shank3B−/− mice.
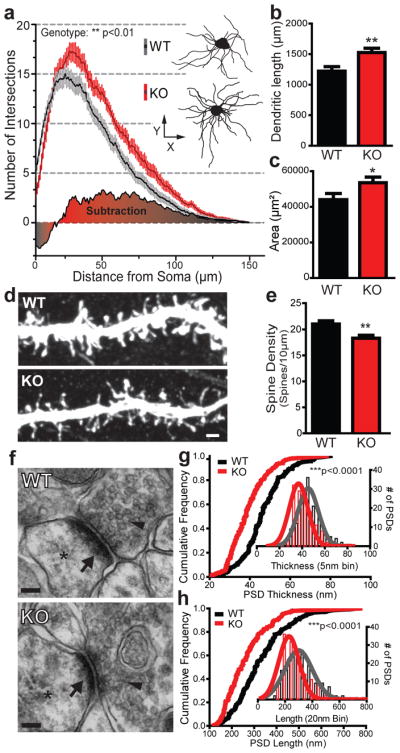
a, Sholl analysis reveals an increased neuronal complexity of Shank3B−/− MSNs (red) when compared to MSNs from wildtype mice (grey); example neurons are shown as insets (WT top; KO bottom). b, c, MSNs from Shank3B−/− mice show an increase in total dendritic length (b) and surface area (c) when compared to controls. d, Representative confocal stacks of dye-filled MSNs from KO and WT mice; scale bar 1μm. e, Spine density in MSNs from Shank3B−/− mice is lower than that of wildtype MSNs. f, Examples of electron micrographs depicting the synaptic contacts with presynaptic vesicles (arrowheads), postsynaptic densities (arrow) and dendritic spine (asterisk); scale bar 100 nm. g, Shank3B−/− PSDs are thinner than wildtype PSDs. h, Shank3B−/− PSDs are shorter than wildtype PSDs. * p<0.05, ** p<0.01, *** p<0.0001; Two-Way repeated measures ANOVA for a; two-tailed t-test for b, c and e; Two-sample Kolmogorov-Smirnov test for g and h. Data in g and h are presented as cumulative frequency plot with histogram distribution and Gaussian curve fit for the insets. Data from b, c and e are presented as means ± s.e.m; n= 36 from 3 wildtype mice and n= 36 from 3 Shank3B−/− mice for a–c; n=41 dendritic segments from 3 wildtype mice and n=36 dendritic segments from 3 Shank3B−/− mice for e; n=144 PSDs from 3 wildtype mice and n= 140 PSDs from 3 Shank3B−/− mice g, h.
Next, we performed patch-assisted Lucifer Yellow cell filling of MSNs and measured spine density in control and Shank3B−/− mice. Shank3B−/− mice displayed a significant reduction in spine density (Fig 4d, e). We did not observe significant changes in spine length or head diameter, however, the neck width of Shank3B−/− MSN spines was slightly larger than that of controls (Supplementary Fig. 8).
Finally we analyzed PSD morphology by electron microscopy (Fig. 4f). We found a significant reduction in mean thickness (Fig. 4g) of PSDs from Shank3B−/− mice relative to controls. Additionally, PSD length was also significantly reduced in the Shank3B−/− mice (Fig. 4h). Taken together, these results highlight a critical in vivo role for Shank3 in the normal development of medium spiny neurons and striatal glutamatergic synapses.
Striatal hypertrophy in Shank3B−/− mice
Even though there is no clear correlation between brain size or neuronal hypertrophy specifically for Shank3 disruptions in humans, a potential link between enlarged brain size, neuronal hypertrophy and autism has been previously suggested19. In particular, increased caudate volume in autism patients has been proposed to be linked to repetitive behaviors20,21. We measured striatal volume using 3-dimensional magnetic resonance imaging in the intact brain of Shank3B−/− and control mice. We found that there was no significant difference in overall brain size between the genotypes. However, measurement of caudate volume in the same animals revealed a small but significant volumetric enlargement of this structure in Shank3B−/− mice (Supplementary Fig. 9). These data suggest a correlation between neuronal hypertrophy and brain volume, consistent with studies from other mouse models of ASD22,23.
Perturbation of striatal postsynaptic function
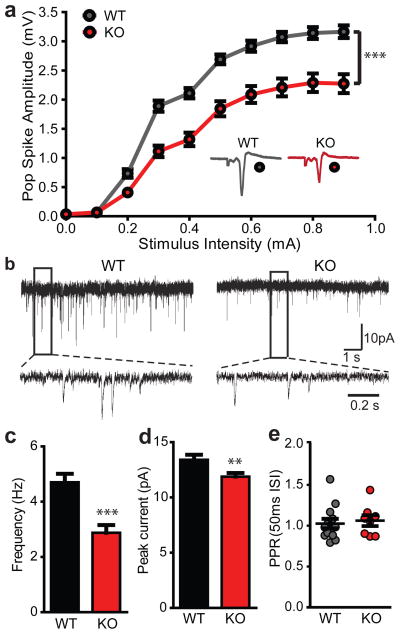
To elucidate the functional consequences of a disruption in Shank3 on synaptic function, we performed recordings of cortico-striatal synaptic circuitry in acute brain slices of 6–7 week old animals. We found that field population spikes were significantly reduced in Shank3B−/− mice when compared with controls (Fig. 5a). Presynaptic function was not altered, as indicated by the relationship of stimulation intensity to the amplitude of the action potential component of the response termed negative peak 1 (NP1) and the paired-pulse ratio (PPR; Supplementary Fig. 10). These results indicate that the reduction in total field responses was most likely due to a postsynaptic impairment in synaptic function and/or a reduction in the number of functional synapses. Consistent with their mild behavioural phenotypes Shank3A−/− mice displayed minimal disruption at cortico-striatal synapses (Supplementary Fig. 11).
Figure 5. Reduced cortico-striatal synaptic transmission in Shank3B−/− MSNs.
a, Cortico-striatal pop spike amplitude is decreased in Shank3B−/− mice (red trace) as measured by extracellular field recordings. Inset, example traces for Shank3B−/− (KO) and wildtype (WT). b, mEPSC example traces from wildtype and Shank3B−/− MSNs recorded with whole-cell voltage clamp. c, d, Reduced mEPSC frequency (c) and amplitude (d) in Shank3B−/− MSNs when compared to wildtype. e, PPR is unaltered in Shank3B−/− MSNs. ** p<0.01, *** p<0.001; Two-way repeated measures ANOVA, with Bonferroni post hoc test for a; two-tailed t-test for c, d; all data presented as means ± s.e.m. For field recordings, n=13 slices from 4 mice per group; for mEPSCs, n=29 MSNs from wildtype mice, n=32 MSNs from Shank3B−/− mice.
We next performed whole-cell voltage clamp recordings of AMPAR-mEPSCs in dorsolateral striatal MSNs. We found that the frequency of mEPSCs was significantly reduced in Shank3B−/− MSNs (Fig. 5b, c), suggesting a reduction in the number of functional synapses in Shank3B−/− MSNs since we did not observe defects on presynaptic function by measuring PPR (Fig. 5e). We also found a significant reduction of peak mEPSC amplitude in Shank3B−/− MSNs (Fig. 5b, d), indicating a reduction in the postsynaptic response from the available synapses. We did not observe significant differences in NMDA/AMPA ratio in Shank3B−/− neurons (Supplementary Fig. 12). Finally, similar defects in mEPSC frequency and amplitude were observed in Shank3B−/− and wildtype littermate mice obtained from heterozygous matings (Supplementary Fig. 13). Together, these data demonstrate a critical role for Shank3 in postsynaptic function in cortico-striatal circuitry.
To assess if the defects arising from Shank3 dysfunction were specific to striatal circuitry or due to a more broad CNS perturbation, we performed a Morris Water Maze task for hippocampal-dependent learning and memory. We found that Shank3B−/− mice performed at the same levels as controls in both learning and probe trials (Supplementary Fig. 14a–c). Reversal learning and probe trials, again demonstrated similar levels of performance between Shank3B−/− mice and controls (Supplementary Fig. 14d–f). Concomitantly, we performed electrophysiological recordings from the hippocampal CA1 subregion and found no obvious difference in field recording of population spikes or PPR between genotypes (Supplementary Fig 15a–c). In addition, we found no significant differences in mEPSC frequency, or mEPSC amplitude (Supplementary Fig 15d–f). These data suggest that the observed behavioral and synaptic defects are specific to discrete brain regions and are not part of an overall CNS dysfunction.
Discussion
Despite recent advances in the understanding of autism spectrum disorder genetics, the underlying neurobiological substrates and neural circuits involved in these disorders remain largely unknown. The Shank3 gene has become the focus of substantial interest, with an increasing body of evidences suggesting Shank3 as the causative gene of the major neurological symptoms in the 22q13 deletion syndrome9,11–13,24. Our present study with Shank3 mutant mice not only sheds light on a critical in vivo role for Shank3 in striatal glutamatergic synaptic structure and function, but also demonstrates causality between a disruption in this gene and the development of autistic-like behaviours in mice.
In this study, we generated two mutant alleles for the Shank3 gene. These two lines of mice displayed different levels of severity in synaptic defects and phenotypes. In humans, multiple mutations/variants of Shank3 gene have been identified to coalesce at the Ankyrin repeats and downstream of PDZ domain9,13. Our data indicate that disruptions of different locations of the Shank3 gene can lead to varying degrees of functional defects, which may in part contribute to phenotypic heterogeneity in Shank3-related ASDs. We should note that, in clinical conditions, the 22q13 deletions and the autism-associated Shank3 mutations are heterozygous, whereas in our current study, we used homozygous mutant mice to get a clear understanding of the physiological role of the Shank3 gene and the underlying functional consequences of its disruption. Further studies will be needed to elucidate potential functional deficits resulting from Shank3 haploinsufficiency in Shank3B−/− mice.
PSD95-SAPAP-Shank proteins form a key postsynaptic scaffold at glutamatergic synapses which interacts with many synaptic proteins including the Neurexin-Neuroligin complex25. In addition to Shank39, it is worth noting that Shank26,8, SAPAP26, Neurexin-126, Neuroligin-3 and −427 have all been implicated in human ASDs. Therefore, the dysfunction of Neurexin/Neuroligin/PSD95/SAPAP/Shank complex could underlie a common synaptic mechanism for a subset of autistic spectrum disorders.
The precise circuitry defects involved in autistic behaviours are poorly understood. Neuroimaging studies provide evidence that caudate and frontal-striatal circuitries are dysfunctional areas in ASD28–30. Cortico-striatal circuitry dysfunction has also been strongly implicated in repetitive/compulsive behaviours in OCD31–33. We previously found that deletion of SAPAP3, which directly interacts with Shank3 and is highly expressed in the striatum, leads to cortico-striatal circuitry dysfunction in mice and OCD-like behaviours including repetitive/compulsive grooming31. Repetitive behaviours are also often seen in autistic patients and in some mouse models of ASDs34–36. Shank3 is the most abundant Shank family member expressed in the striatum and Shank3B−/−mice exhibit excessive/repetitive grooming leading to skin lesions. Our data support the hypothesis that repetitive behaviours in OCD and ASD may share a common circuitry mechanism.
The regulation of social behaviours and social interaction is thought to be controlled by several brain regions and circuits37. Similarly, genetic makeup is thought to play a key role in the phenotypical manifestation of social behaviours38. The robust social interaction deficits in Shank3B mutant mice demonstrate a casual role for the disruption of this gene in the genesis of social dysfunction and provide a valuable experimental system for future genetic dissection of the neuronal basis of social behaviour.
Methods Summary
Behavioural analysis
Young adult mice 5–6 week old were used for all behavioural analyses except lesion scores which were performed in 4–5 months old mice. All experiments were done blind to genotypes. All experimental procedures were reviewed and approved by the Duke University Institutional Animal Care and Use Committee and the MIT Committee on Animal Cares.
Statistical Analysis
Analyses were performed using Prism (GraphPad Software, La Jolla, CA) and MATLAB (MathWorks, Natick, Massachusetts). Details on particular tests used are described in the main text and in the methods section; a summary of statistical analysis for the behavioral data is presented in Supplementary Table 2.
Full Methods
Mice
Shank3 mutant mice were generated by homologous recombination in R1 embryonic stem cells and implanted in C57 blastocysts using standard procedures. One targeting vector (Shank3A) was designed to replace exon 4–7 (containing the ankyrin repeat domains) and another vector (Shank3B) was designed to replace exon 13–16 (containing the PDZ domain) of the Shank3 gene with a NEO cassette. Genotypes were determined by PCR of mouse tail DNA, using: for Shank3A; primer F1a (GGTTGAGGATGAGCAAGCTAG) and R1a (GGGACATAAGTGAAGGTTAGG) for the wildtype allele (318 base pairs), and F1a and R2 (TCAGGGTTATTGTCTCATGAGC; in the neo cassette) for the mutant allele (361 base pairs); for Shank3B, primer F1b (GAGCTCTACTCCCTTAGGACTT) and R1b (TCCCCCTTTCACTGGACACCC) for the wildtype allele (316 base pairs), and F1b and R2 (TCAGGGTTATTGTCTCATGAGC; in the neo cassette) for the mutant allele (360 base pairs). The NEO cassette was not removed.
Chimeric mice were crossed to C57 females (Jackson Labs). Initially, F1 hybrids from het x het matings were generated. However, homozygous knockouts mice from this type of mating are smaller than their wildtype littermates, presumably due to an inadequate competition for resources during early postnatal days leading to different developmental trajectories. We postulated that this size difference would influence our behavioral tests. To alleviate this confound, heterozygous animals were crossed in direct brother-sister matings for 5 generations from which we derived F5 isogenic hybrids in a mixed background. These isogenic animals were then used to generate time-mated homozygous x homozygous breeding pairs to obtain wildtype and mutant animals used in the experiments. F5 Shank3A and F5 Shank3B knockouts from these matings are reared to weaning age with weights similar to those from F5 control animals.
Animals were housed at a constant 23oC in a 12h light/dark cycle (lights off at 19:00), with food and water available ad libidum. Mice were housed 3–5 by genotype per cage with the exception of the animals individually house for grooming measurements. Only aged-matched male mice were used for behavioral experiments, all other tests included age-matched males and females in proportional contribution across groups. Unless otherwise noted, all tests were conducted with naive cohorts of mice. All procedures were conducted in compliance with the Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals policies on animal welfare and approved by the Duke University IACUC.
Grooming behaviour31
Young adult male mice 5–6 weeks old were used for analysis of grooming behavior. Habituated, individually housed animals were video-taped for 24 hours under 700 lux (day, 12 h) and ~2 lux (red light at night, 12 h) illumination. Grooming behaviours were coded from 1900–2100 hours (i.e. 2 hours beginning at the initiation of the dark cycle); this segment was analyzed using Noldus Observer software (Leesburg, VA) and the total amount of time in the 2-hour segment spent grooming was determined. Grooming included all sequences of face-wiping, scratching/rubbing of head and ears, and full-body grooming. The observer was blinded to genotype during the scoring of the videotapes.
In situ hybridization
mRNA in situ hybridization was performed as described elsewhere39. Briefly, reactions were performed with 20 μm cryosections from freshly frozen 5 week old brain mouse tissue using digoxygenin (DIG)-labelled riboprobes against mouse SHANK1 cDNA (NM_001034115; bp 4107–4924), SHANK2 cDNA (NM_001081370; bp 2063–2876) and SHANK3 (NM_021423; bp 3159–3959). The complementary DNAs used were all verified by sequencing compared to the following sequences GenBank accession numbers: (SHANK1: NM_001034115), (SHANK2: NM_001081370) and (SHANK3: NM_021423). The hybridization signal was detected using an alkaline phosphatase (AP)-conjugated anti-DIG antibody (Roche) and developed using 5-bromo-4-cloro-indolylphosphate/nitroblue tetrazolium (BCIP/NBT; Roche).
Motor and Anxiety-like behaviours31
Zero maze
An elevated zero maze was indirectly illuminated at 100 lux. Testing commenced with an animal being introduced into a closed area of the maze. Behaviour was video-taped for 5 min and subsequently scored by a trained observer using Noldus Observer software (Leesburg, VA). Anxiety-like behaviour was deduced based upon the percent time spent in the open areas. The observer was blinded to genotype. The animals used in the zero maze test, both Shank3A−/−, Shank3B−/− and respective controls were previously tested in the open field test with a 2 day period in between tasks.
Open field
Spontaneous locomotor activity was evaluated over 30 min in an automated Omnitech Digiscan apparatus (AccuScan Instruments, Columbus, OH) as described31. Locomotor activity was assessed as total distance traveled (m). Anxiety-like behaviour was defined by number of rearings and time spent in the center as compared to time spent in the perimeter (thigmotaxis) of the open field.
Dark-Light Emergence Test
Mice were habituated in an adjacent room to low light conditions (~40 lux) and the test room was initially under similar illumination. Testing was conducted in a two-chambered test apparatus (Med Associates, St. Albans, VT), with one side draped in black cloth (i.e. dark-chamber) and the other illuminated at ~1000 lux (i.e. light-chamber) with a high intensity house light and overhead fluorescent lamps. Upon placing the mice into the dark chamber, the light chamber was illuminated and the door between the two chambers was opened. The mice were allowed to freely explore the apparatus for 5 min. The latency to emerge from the darkened into the lighted chamber and the percentage of time spent in the illuminated chamber were used as indices of anxiety-like behaviours.
Social Interaction Paradigm
Three-chamber social test
Sociability and response to social novelty test was performed as previously described17 with minor modifications. Briefly, 5–6 week old male animals were used across all tests. Target subjects (Stranger 1 and Stranger 2) were 5–6 week old males habituated to being placed inside wire cages for 5 days prior to beginning of testing. Test mice were habituated to the testing room for at least 45 min prior to the start of behavioural tasks. The social test apparatus consisted of a transparent acrylic box with removable floor and partitions dividing the box into three chambers. Here, the middle chamber (20 cm × 17.5 cm) is half the width of Chamber 1 (20 cm × 35 cm) and Chamber 2 (20 cm × 35 cm) with the overall dimensions of the box being: 60 cm (length) × 35 cm (width) with 5 cm openings between each chamber which can be closed or open with a lever operated door. The wire cages used to contain the stranger mice were cylindrical, 11 cm in height, a bottom diameter of 10.5 cm with the bars spaced 1 cm apart (Galaxy Cup, Spectrum Diversified Designs, Inc., Streetsboro, OH). An inverted transparent cup was placed on the top of the cage to prevent the test mice from climbing on the top of the wire cage.
For the sociability test, the test animal was introduced to the middle chamber and left to habituate for 5 min, after which an unfamiliar mouse (Stranger 1) is introduced into a wire cage in one of the side-chambers and an empty wire cage on the other side-chamber. The dividers are then raised and the test animal is allowed to freely explore all 3 chambers over a 5 min session. Following the 5 min session, the animal remains in the chamber for an extra 5 min (post-test) to better acquire the identification cues from Stranger 1 animal. Following this, a novel stranger mouse (Stranger 2) is inserted in the wire cage previously empty and again the test animal is left to explore for a 5 min session. Time spent in each chamber, time spent in close proximity and heat maps were calculated using the automated software Noldus Ethovison software (Leesburg, VA). The release of the animals and relative position of social and inanimate targets was counterbalanced. However, for each individual test animal the location of S1 was maintained during S1-E and S1–S2 testing of the social behavior.
Dyadic social interaction
Animals were acclimatized to the test room for at least 1 hour prior to the experiment. Target mice were wildtype and Shank3B−/− of 6 weeks of age. Stimulus mice were conspecific age-matched wildtype mice socially naive to the target mice. At least 3 hours prior to the beginning of the test, stimulus mice were given identifiable markings on the tails using a black marker pen. A pair of target and stimulus mice were introduced in a transparent plexiglass arena (40cm × 40cm × 30cm) covered with fresh bedding and the session recorded for 10 minutes. Quantification of social behaviors was performed using Noldus Observer software (Leesburg, VA) by a researcher blinded to the genotype of the target animals. Quantifications included: reciprocal social interaction, as determined by any sequence or combination of sequences involving close huddling, sniffing (e.g. nose-to-nose, anogenital sniffing) or allogroming by the target and stimulus mouse; the frequency of nose-to-nose sniffing; and the frequency of anogenital sniffing initiated by the target animal towards the stimulus mouse. Statistical tests were performed using unpaired two-tailed t-test.
Rotarod
Motor coordination was assessed in an accelerating rotarod test (4–40 rpm). Briefly, animals were introduced in the apparatus (Med Associates, St. Albans, VT) and the latency to fall was determined. Animals were tested for 3 trials in a single day with an inter-trial interval of 10 minutes.
Morris Water Maze
Morris Water Maze testing was conducted as describe elsewhere40 with minor modifications. 4–5 week old male mice selected for the test were individually handled daily for 5 days prior to beginning the experiment. Testing pool was 120 cm in diameter and the platform 8 cm in diameter. The platform was submerged 1 cm below the water surface. Pool water was maintained at 23.0 ± 0.5 °C and made opaque by mixing-in white non-toxic tempera paint. During training, 90 s duration trials were used, if the animals did not find the platform within 90 s the experimenter guided the animal to the platform. After reaching the platform the animals were left for 15s on top of the platform before being removed. Trials were administered for 5 days with 4 trials per animal per day with the platform located in the SW quadrant. On the sixth day a 60 s probe trial was performed. On the seventh day, the reversal training commenced with the platform in the NE quadrant, and proceeded as described above. The experimenter followed the animals’ progress using tracking software outside of the testing room. Tracking and analysis were performed using the Noldus Ethovison software (Leesburg, VA).
Golgi Staining and Sholl Analysis
All brains and collected sections were coded in order to blind the experimenter of the genotype until after all data was collected and analyzed. Brains from 5 week old, gender matched Shank3B−/− and controls mice were prepared using standard Golgi-Cox impregnation technique using the FD Rapid GolgiStain Kit (NeuroTechnologies, Ellicott City, MD). Serial coronal sections of 100μm were collected from controls and Shank3B mutant animals. A total of 12 cells per animal were traced across the dorsal striatum as to sample representatively from this structure for a final number of 36 cells per genotype. For each animal, sections were selected to be between rostral-caudal bregma 1.18mm and 0.86mm. Criteria to identify medium spiny neurons was, a) presence within the caudate putamen; b) full impregnation of the neuron along the entire length of the dendritic arbor; c) relative non-overlap with surrounding neurons and isolation from astrocytes and blood vessels and d) morphologically, by the presence of high number of spines and relatively short neuronal arbors as characteristic of MSNs. For each selected neuron the entire neuronal arbor was reconstructed under a 100x oil lens in a motorized microscope with a digital CCD camera connected to a computer running Neurolucida Software (MBF Biosciences, USA). The 3D analysis of the reconstructed neurons was performed using NeuroExplorer software (MBF Biosciences, USA) and data from branch length, number of branches and neuronal complexity was measured and analyzed in Prism (Graph Pad, USA). Two-way repeated measures ANOVA was used for Sholl analysis. Statistical significance was accepted when: * P < 0.05, **P < 0.01 and ***P < 0.0001.
Cortico-striatal electrophysiology
Brain Slice preparation for extracellular field recording
Acute brain slices were prepared from 6–7 week old mice. Slices were prepared from one WT and one KO pair each day and the experimenter was blind to the genotype. The mice were deeply anesthetized by intra-peritoneal injection of avertin and then transcardially perfused with carbogenated ice-cold protective cutting artificial cerebrospinal fluid (aCSF) with the composition (in mM): 119 glycerol, 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3, 25 glucose, 2 thiourea, 5 L-ascorbic acid, 3 Na-pyruvate, 0.5 CaCl2.4H2O, 10 MgSO4.7H2O. Mice were then decapitated and the brains were removed into ice-cold cutting solution for an additional 1 minute. The brains were then rapidly blocked for coronal sectioning at 300uM thickness on a VF200 model compresstome (Precisionary Instruments) using either a sapphire or zirconium ceramic injector style blade. Slices containing the dorsal striatum were initially recovered for 30 minutes at room temperature (23–25°C) in a carbogenated protective recovery aCSF (same composition as the cutting aCSF except that glycerol was replaced with NMDG-Cl as a substitute for NaCl to prevent initial excitotoxic swelling during re-warming). After this initial 30 minute period the slices were transferred into a holding chamber containing carbogenated normal aCSF of the composition (in mM): 119 NaCl, 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3, 12.5 glucose, 2 CaCl2.4H2O, 1 MgSO4.7H2O. The holding aCSF was supplemented with (in mM): 2 thiourea, 1 L-ascorbic acid, 3 Na-pyruvate to improve slice health and longevity, and slices were stored for 1–6 hrs prior to transfer to the recording chamber for use. The osmolarity of all solutions was measured at 300–310 mOsm and the pH was maintained at ~7.3 after equilibration under constant carbogenation.
Supplemental Fig 11 shows summary data for corticostriatal field recordings from acute coronal brain slices of Shank3A mutant versus WT mice. The method of slice preparation differed significantly in these earlier experiments. Mice were transcardially perfused with carbogenated ice-cold protective sucrose aCSF with the composition (in mM): 185 sucrose, 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3, 25 glucose, 0.5 CaCl2.4H2O, 4 MgSO4.7H2O (pH 7.3, 300–310 mOsm) without supplementation of antioxidants. Slices were immediately transferred into a holding chamber containing carbogenated normal aCSF of the composition (in mM): 119 NaCl, 2.5 KCl, 1.25 NaH2PO4, 26 NaHCO3, 12.5 glucose, 2 CaCl2.4H2O, 1 MgSO4.7H2O (pH 7.3, 300–310 mOsm) without supplementation of antioxidants, and slices were stored for 1–4 hrs prior to transfer to the recording chamber. The absence of the initial 30 min recovery period in “protective” aCSF in addition to the absence of antioxidant supplementation in the cutting aCSF and in the aCSF in the holding chamber results in more rapid deterioration of slice health and smaller evoked population spike amplitudes on average, indicating reduced overall slice viability compared to slices prepared with a 30 min NMDG aCSF recovery protocol described above. However, WT and KO brain slices were always subjected to identical procedures on any given day of recording and the procedures were always standardized for each discrete experimental data set so that these factors would not introduce any potential confounds.
Extracellular field recording
A platinum iridium concentric bipolar stimulating electrode (CBAPC75, 25uM inner pole diameter; FHC) was placed on the inner border of the corpus callosum between the cortex and dorsolateral striatum. This electrode position was chosen to predominantly activate cortical axons within the corpus callosum which heavily converge upon striatal MSNs to form excitatory corticostriatal synaptic connections. Though there is ample evidence on which to base our assertion that stimulation of the corpus callosum predominantly results in activation of cortical axons41,42, we are unable to exclude the possibility of a relatively smaller contribution arising from activation of thalamostriatal axons that have distal terminals in dorsolateral striatum nearby to the stimulated region. Thus, although we refer to our measurements as primarily reflecting corticostriatal transmission, our measurements are not “pure” corticostriatal responses. Borosilicate glass recording electrodes filled with 2M NaCl were placed in the dorsolateral striatum approximately 400–450 μm away from the stimulating electrode. Corticostriatal field population spikes were evoked with 0.15 ms step depolarizations at 0.5 mA intensity at a frequency of 0.05–0.1 Hz. Paired pulses were evoked with a 100 ms inter-stimulus interval. Baseline responses were monitored to ensure stable population spike amplitude for a minimum of 5 minutes. Input-output functions were then determined for the negative peak 1 (NP1; presynaptic fiber volley) and population spike amplitude by 3 consecutive rounds of stimulation from 0–1.0 mA in 0.1 mA increments. All recordings were performed at room temperature and acquired using pCLAMP 10 software (Axon Instruments/Molecular Devices). Data analysis was performed blind of genotype in Clamp fit (Axon Instruments/Molecular Devices). Population spike amplitude was measured as the average of the early peak positivity to the peak negativity and from the peak negativity to the late peak positivity. This standard method takes into account the fact that the downward population spike is superimposed on an upward field excitatory postsynaptic potential (fEPSP). Paired pulse ratio was calculated as the ratio of the 2nd population spike amplitude to the 1st population spike amplitude for responses to paired pulse stimulation at 0.5 mA fixed intensity with a 100 msec interstimulus interval for the pair.
Extracellular field recordings and whole-cell mEPSC recordings in the hippocampal CA1 region were conducted in 300 μM thick acute brain slices from 6–9 week old WT and Shank3B mutant mice. For measurement of hippocampal CA1 population spikes, a concentric bipolar stimulating electrode was placed in the stratum radiatum to stimulate the Schaffer collateral pathway, and a boroscilate glass recording electrode (~2–3 MOhms) filled with recording aCSF was placed in the CA1 pyramidal cell layer approximately 400 μm from the stimulation site. The recording electrode was placed at the depth in the slice that gave the largest population spike amplitude, and a stable baseline was established for < 10 min. Input-output recordings were conducted by increasing the stimulation intensity from 0 to 160 μA in 20 μA increments. Three successive rounds were collected and values at each intensity represent the average of the three measurements. CA1 population spike amplitude was quantified exactly as described previously for cortico-striatal population spikes. For CA1 pyramidal neuron whole-cell recordings, pyramidal neurons in CA1 were identified under IR-DIC visualization. Cells were patched with a CsGluconate based internal solution containing (in mM): 110 CsGluconate, 15 KCl, 4 NaCl, 5 TEA-Cl, 20 HEPES, 0.2 EGTA, 5 Lidocaine N-ethyl chloride, 4 ATP magnesium salt, and 0.3 GTP sodium salt. The pH was adjusted to 7.25 with D-gluconic acid and osmolarity was adjusted to 290–300 mOsm with sucrose as necessary. The recording aCSF contained 1 μM TTX, 100 μM picrotoxin, 5 μM CGP55845, and 50 μM D-APV to isolate pure AMPAR-mediated mEPSCs. CA1 neurons were voltage clamped at −80 mV to amplify the smallest spontaneous miniature synaptic events that might otherwise escape detection. Criteria for acceptance were uncompensated stable Ra < 25 MOhms and holding current < −300 pA. mEPSCs were detected using MiniAnalysis software (Synaptosoft Inc., USA) as described for striatal MSNs. All recordings were carried out at room temperature (23–25°C). Slices were prepared in a 20–30 degree off-horizontal cutting angle (optimal for CA1 region) from one WT and one KO pair each day and the experimenter was blind to the genotypes of the animals.
Striatal slice preparation for whole-cell recording
Mice 5–6 week old were used for all whole-cell electrophysiology procedures by an experimentalist blinded to genotype. Acute coronal striatal slices were prepared as follows. Briefly, mice were anesthetized with Avertin solution (20 mg/ml, 0.5 mg g body weight) and perfused through the heart with a small volume (about 20 ml) of ice-cold and oxygenated (95% O2, 5% CO2) cutting solution containing (mM): 105 NMDG (N-Methyl-D-glucamine), 105 HCl, 2.5 KCl, 1.2 NaH2PO4, 26 NaHCO3, 25 Glucose, 10 MgSO4, 0.5 CaCl2, 5 L-Ascorbic Acid, 3 Sodium Pyruvate, 2 Thiourea (pH was around 7.4, with osmolarity of 295–305 mOsm). The brains were rapidly removed and placed in ice-cold and oxygenated cutting solution. The coronal slices (300 μm) were prepared using a slicer (Vibratome 1000 Plus, Lecia Microsystems, USA) and then transferred to an incubation chamber (BSK4, Scientific System Design Inc., USA) at 32 °C with oxygenated cutting solution, which was gradually replaced with artificial cerebrospinal fluid (aCSF) in 30 min through a peristaltic pump (Rainin, RP-1) allowing a precise regulation of flowing rates. The slices were then kept in the aCSF that contained (mM): 119 NaCl, 2.3 KCl, 1.0 NaH2PO4, 26 NaHCO3, 11Glucose, 1.3 MgSO4, 2.5 CaCl2 (pH was adjusted to 7.4 with HCl, with osmolarity of 295–305 mOsm)at room temperature for at least 30 min.
Whole-cell patch-clamp
The slice was placed in a recording chamber (RC-27L, Warner Instruments, USA) and constantly perfused with oxygenated aCSF at 24 °C (TC-324B, Warner Instruments, USA) at a rate of 1.5–2.0 ml/min. The striatum and individual medium spiny neurons (MSNs) were visualized and identified with a microscope equipped with Nomarski optics and infrared illumination (BX-51WI, Olympus, Japan) by location, shape and size (ovoid cell body with major axis of 10 to 14 μm). Two additional measures were used to distinguish them from similar sized GABAergic interneurons. First, GABAergic interneurons show smaller membrane capacitance (Cm) and membrane time constant (τm) (at least 2 times less) when compared to that of MSNs. In the case of recordings done with Cs+ internal, these membrane properties were measured immediately after membrane rupture when the Cs+ internal has not been dialyzed and taken effect yet. Second, AMPA receptor mediated mEPSCs showed much faster kinetics (including both rise time and decay time constant, τ decay) in GABAergic interneurons. Whole-cell patch-clamp recordings were obtained from MSNs using recording pipettes (King Precision Glass, Inc, Glass type 8250) pulled in a horizontal pipette puller (P-87, Sutter Instruments, USA) to a resistance of 3–4 MΩ, when filled with the internal solution containing (in mM): 107 CsMeSO3, 10 CsCl, 3.7 NaCl, 5 TEA-Cl, 20 HEPES, 0.2 EGTA, 5 Lidocaine N-ethyl chloride, 4 ATP magnesium salt, and 0.3 GTP sodium salt. PH was adjusted to 7.3 with KOH and osmolarity was adjusted to 298–300 mOsm with 15mM K2SO4.
To record AMPA miniature excitatory postsynaptic currents (mEPSCs), the cells were held in voltage clamp at −70 mV in the presence of 50 μM APV (DL-2-amino-5-phosphono-valeric acid), 25 μM BMR (1(S),9(R)-(−)-Bicuculline methbromide,), 10μM D-serine and 1μM TTX (all from Tocris). The miniature events were not recorded until 5 min after entering whole cell patch clamp recording mode to allow the dialysis of Cs+ internal solution for a relatively complete block of the potassium channels in the MSNs. The mEPSCs were detected and analyzed with MiniAnalysis (Synaptosoft Inc., USA).
For paired-pulse stimulation experiments, AMPAR mediated excitatory post-synaptic currents (EPSCs) were evoked by a local concentric bipolar stimulating electrode (CBARC75, FHC, USA) that was placed in the inner edge of corpus callosum within the dorso-lateral region of the striatum. Recordings were made in the presence of Picrotoxin (100 μM) and APV (50 μM) to block activation of GABAA receptors and NMDA receptors. Stimulation was current-controlled (ISO-Flex, A.M.P.I., Israel). The stimulus intensity was set at a level that could evoke 300–400 pA AMPAR-mediated response for all the cells measured and delivered with an inter-stimulus interval of 50 ms. The paired-pulse measurements were obtained for 15–20 consecutive traces and only those traces with stable evoked first current response were used for data analysis. The paired-pulse ratio (PPR) was calculated with the peak current response to the second pulse divided by that of the first response.
NMDAR- and AMPAR- mediated synaptic current ratio (NMDA/AMPA ratio) was recorded in the presence of picrotoxin at holding potentials of +40mV and −70 mV, respectively. The NMDA/AMPA ratios were measured according to previously described methods43. Briefly, the stimulus intensity was set at a level that could evoke 300–400 pA AMPAR-mediated response with a holding potential at −70 mV. Each evoked response was repeated for 15–20 times with an inter-stimulus interval of 20s for all the cells measured, The time point of the peak current at −70 mV, considered to be fully mediated by AMPARs, was used to establish the time window for measuring the AMPA peak at +40 mV. The decay to baseline of the AMPA current at −70 mV was used to select a time window for measurement of the NMDA current; a 10-ms measurement window beginning 40 ms after the stimulus artifact was used. This current amplitude at this point was designated as the NMDAR mediated synaptic current response. (INMDA at +40mV/IAMPA at −70mV) was taken as the NMDA/AMPA ratio.
Data acquisition and analysis
A Multiclamp 700B amplifier (Molecular Devices Corporation, Sunnyvale, CA, USA) and digidata 1440A were used to acquire whole cell signals. The signals were acquired at 20KHz and filter at 2KHz. The series-resistance was < 20 MΩ. Values are expressed as means ± s.e.m. Data were tested for significance using either an unpaired t-test or a Two-way repeated measures ANOVA.
Cell Filling
Mice were assigned a code previous to dissection, as to maintain a blinded genotype across all procedures, including dissection, cell filling, imaging and quantification. Mice were deeply anesthetized with an overdose of isofluorane and transcardially perfused with PBS (pH 7.4) followed by ice-cold 4% paraformaldehyde/PBS (PFA) (pH 7.4). The brains were removed and post-fixed overnight in PFA 4%. After post-fixation, the brain was sliced at 200μm thickness coronal sections in a vibratome and kept in PBS at 4oC. For cell filling injections, selected brain slices immersed in PBS were mounted in a tissue stage. Dorsal striatal medium cells were targeted with post hoc confirmation of being medium spiny neurons (morphology and spine density). Using a micromanipulator, micropipettes loaded with Lucifer Yellow dye (Sigma L-0259, 8% solution in 0.05M Tris buffer, pH 7.4) were used to impale the cell body. A micropipette containing a solution of 0.1M LiCl was used to deliver the dye with a continuous 10 nA current for 5 min. Following cell filling, a post-staining was used to amplify the fluorescent signal, briefly, sections were transferred to blocking solution (5% sucrose, 2% BSA, and 1% triton x-100 in PBS) containing 1:500 rabbit anti-lucifer yellow antibody (Invitrogen A5750) and incubated gently for 3 days at 4oC. Sections were washed 3 times for 5 min in blocking solution and incubated 2 hours at room temperature with 1:400 biotinylated goat anti-rabbit antibody (Vector Laboratories BA-1000). Next, sections were washed 3 times for 5 min in PBS. A tertiary incubation was performed by incubating sections for 2 hours at room temperature in streptavidin-conjugated Alexa 488 (Invitrogen S11223) diluted 1:1000 in PBS. Finally, sections were washed 3 times in PBS, mounted on slides using Fluoro-Gel (EMS, 17985-10) and imaged by confocal microscopy. Spine density was calculated automatically using Neuron Studio (Mount Sinai School of Medicine, NY) and manually curated by an observer using a 3D analysis of the dendritic image stack. All spine counts began 30um away from the outer edge of the soma and extended for an additional 10 μm-60 μm away from the starting point. The data from spine density passed the Lilliefors Normality Test and D’Agostino & Pearson Omnibus Normality Test. Spine metrics relating to spine length, spine neck diameter and spine neck width were collect using ImageJ (NIH, Bethesda, Maryland). All analysis of spine metrics were performed by observers that were blinded to the genotypes of the animals.
Electron microscopy
Mice were assigned a code previous to dissection, as to maintain a blinded genotype across all procedures, including dissection, sample processing, imaging and quantification. Mice were deeply anesthetized with an overdose of isofluorane and transcardially perfused with PBS (pH 7.4) followed by ice-cold 4% paraformaldehyde (PFA) in phosphate buffer (pH 7.4). The brains were removed, the striatum dissected and post-fixed overnight in PFA 4%, then transferred into a 4% glutaraldehyde solution and kept at 4°C for 3 days. The samples were washed twice, 20 minutes each, in 7.5% sucrose, 0.1M sodium cacodylate buffer, then post-fixed in 1% Osmium tetroxide for 2 hours with initial microwave treatment for 6 minutes. Next, the samples were washed twice in 0.11M Veronal acetate buffer for 20 minutes each. Following en block staining in 1% Uranyl acetate in distilled (DI) water for 1 hour the samples were washed twice in 0.11M Veronal acetate buffer for 20 minutes each. Samples were dehydrated using serial dilutions of ethanol (70%, 95%, 2x 100%) for 20 minutes each, with initial microwave treatment of 2 minutes. Samples were then treated for 20 minutes twice with Propaline oxide and impregnated with 50:50 Propaline oxide:Epon resin overnight at 4°C, with initial microwave treatment for 3 minutes. Next, the samples were impregnated with 100% Epon resin, 3 changes of 2 hours each, with initial microwave treatment for 3 minutes each. Tissue samples were embedded in molds and incubated for 48 hours at 60°C. Afterwards, semi-thin sections (0.5μm) were cut on a Leica UltraCut S ultramicrotome and stained with Toluidine (0.8%) stain. From these, thin striatal sections (70nm) were cut on an UltraCut S, mounted on 200 mesh Metaxaform Copper Rhodium grids and poststained in 2% Uranyl acetate in DI water for 15 minutes and Sato’s Lead citrate stain for 7 minutes. Grids were examined on a Philips (FEI) CM 12 transmission electron microscope. Images were acquired at 40,000x magnification using an AMT 2Vue system, with an ORCA HR High resolution digital camera 7Mpix, a Hamamatsu DCAM board for acquisition and AMT Image Capture Engine software version 600.335f. Images were saved as 7.5 Mpixel 8 bit TIFF format files. PSD measurements were performed using ImageJ (NIH, Bethesda, Maryland) by an observer that was blinded to the genotype of the samples..
Magnetic Resonance Image Acquisition
Animals were assigned a blinding code, which was maintained during magnetic resonance (MR) data acquisition and analysis. MR mouse brain imaging was performed on a 7T Bruker Biospec 70/30 horizontal bore system (Billerica, MA). Animals were lightly anesthetized under isofluorane with continuous monitoring and maintenance of physiological parameters throughout the imaging session (~60 min for each animal). Axial 2D T2-weighted fast spin echo images (TURBO-RARE, TE/TR = 11/4200 ms with 1mm slice thick, matrix = 256 X 256 and FOV of 2.4cm X 2.4 cm, 5 averages, 0.0 mm interslice gap) images were first obtained for screening purposes and supplemental anatomic information. For directed striatal and brain volumetric analysis, 64 contiguous 500 μM thick 3D FSE proton density images (TURBO-RARE, TE/TR = 9/1500 ms, matrix = 256 X 256 X 64 and FOV 2.2 cm X 2.2 cm X 2.2 cm, 25 minutes duration) were acquired.
MR Volumetry Measurements
Volumetric analysis of MR data sets was performed in Osirix software, an open source image processing application developed and maintained by Pixmeo (Geneva, SUI). The left caudate-putamen was manually segmented in each animal by an investigator blinded to genotype. Each caudate-putamen was traced on contiguous axial slices from the 3D volume acquisition with reference to a high-resolution age-matched mouse brain atlas (The Mouse Brain Atlas, The Mouse Brain Library @ www.mbl.org. Rosen GD et al., Int. Mouse Genome Conference 14: 166 (2000). Selected areas were reviewed for consistency on coronal and sagittal representations, and cross-correlated with axial 2D FSE images. Volumes were computed within Osirix. Two separate striatal segmentations were obtained for each animal, with the average volume then taken. Intrarater reliability (kappa value) was = 0.97.
Volume Normalization
Unilateral caudate-putamen volumes were normalized to brain volume measurements obtained from the same 3D volume sets. Because of susceptibility distortions within the posterior fossa, and variable inclusion from animal to animal of posterior fossa contents at the caudal end of the 64 slice 3D volume set, ‘whole’ brain volumes for normalization were obtained in each animal from traces beginning rostrally at the olfactory bulbs and ending caudally through the cerebral aqueduct at the roof of the fourth ventricle. As with striatal volumes, brain volumes were computed in Osirix from the average of two segmentations. Intrarater reliability (kappa value) was > 0.99. Statistical analysis was performed with unpaired two-tailed t-test.
Supplementary Material
Acknowledgments
We thank C. Duarte, S. Chaterjee and A. Oliveira-Maia for helpful discussion; L. Kruger and Q. Liu for technical assistance; A. Hadiono for assistance in behavioural annotation; D. Bredt for the PSD-93 antibody; T. Boeckers for the anti-Shank3 antibody; S. Miller and P. Christopher for advice and assistance with EM techniques; J. Crawley for the demonstration of social behaviour tests; Y. Wan for advice on electrophysiology studies; A. Graybiel for critical comments of the manuscript; D. Wang and the other members of the Feng lab for their support. We thank The Poitras Center for Affective Disorders Research. This work was funded by a grant from NIMH/NIH (R01MH081201), a Hartwell Individual Biomedical Research Award from The Hartwell Foundation, and a Simons Foundation Autism Research Initiative (SFARI) grant Award to G.F; a NARSAD Young Investigator Award and NIH Ruth L. Kirschstein National Research Service Award (F32MH084460) to J.T.T.; a NIH (R03MH085224) grant to Z.F.; and doctoral fellowships from the Portuguese Foundation for Science and Technology to J.P. (SFRH/BD/15231/2004) and C.F. (SFRH/BD/15855/2005). C.F. would like to acknowledge the support from the “Programa Gulbenkian de Doutoramento em Biomedicina” (PGDB, Oeiras, Portugal) and J.P. the “Programa Doutoral em Biologia Experimental e Biomedicina” (CNC, Coimbra, Portugal).
Footnotes
Author Contributions: J.P., C.F., J.T., W.W., M.W., T.V., C.L. and Z.F. participated in the execution and analysis of experiments. J.P., C.F., J.T., C.L, Z.F. and G.F participated in the interpretation of the results. J.P., C.F. and G.F. designed the experiments and wrote the paper.
Author Information: The authors declare no competing financial interests.
References
- 1.American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-IV-TR. American Psychiatric Association; Washington, DC: 2000. Task Force on DSM-IV. [Google Scholar]
- 2.Rosenberg RE, et al. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch Pediatr Adolesc Med. 2009;163:907–914. doi: 10.1001/archpediatrics.2009.98. [DOI] [PubMed] [Google Scholar]
- 3.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgeron T. A synaptic trek to autism. Current Opinion in Neurobiology. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- 6.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabuchi K, et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkel S, et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010;42:489–491. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- 9.Durand CM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prasad C, et al. Genetic evaluation of pervasive developmental disorders: the terminal 22q13 deletion syndrome may represent a recognizable phenotype. Clin Genet. 2000;57:103–109. doi: 10.1034/j.1399-0004.2000.570203.x. [DOI] [PubMed] [Google Scholar]
- 11.Wilson HL, et al. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moessner R, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gauthier J, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- 14.Kim E, et al. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi M, et al. SAPAPs. A family of PSD-95/SAP90-associated proteins localized at postsynaptic density. J Biol Chem. 1997;272:11943–11951. doi: 10.1074/jbc.272.18.11943. [DOI] [PubMed] [Google Scholar]
- 16.Zoghbi HY, Warren ST. Neurogenetics: advancing the “next-generation” of brain research. Neuron. 2010;68:165–173. doi: 10.1016/j.neuron.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moy SS, et al. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- 18.Hung AY, et al. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci. 2008;28:1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redcay E, Courchesne E. When is the brain enlarged in autism? A meta-analysis of all brain size reports. Biol Psychiat. 2005;58:1–9. doi: 10.1016/j.biopsych.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 20.Langen M, et al. Changes in the developmental trajectories of striatum in autism. Biol Psychiatry. 2009;66:327–333. doi: 10.1016/j.biopsych.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Hollander E, et al. Striatal volume on magnetic resonance imaging and repetitive behaviors in autism. Biol Psychiatry. 2005;58:226–232. doi: 10.1016/j.biopsych.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 22.Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19:231–234. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Kwon CH, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonaglia MC, et al. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J Med Genet. 2006;43:822–828. doi: 10.1136/jmg.2005.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irie M, et al. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- 26.Kim HG, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jamain S, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silk TJ, et al. Visuospatial processing and the function of prefrontal-parietal networks in autism spectrum disorders: a functional MRI study. Am J Psychiat. 2006;163:1440–U1443. doi: 10.1176/ajp.2006.163.8.1440. [DOI] [PubMed] [Google Scholar]
- 29.Horwitz B, Rumsey JM, Grady CL, Rapoport SI. The cerebral metabolic landscape in autism. Intercorrelations of regional glucose utilization. Arch Neurol. 1988;45:749–755. doi: 10.1001/archneur.1988.00520310055018. [DOI] [PubMed] [Google Scholar]
- 30.Sears LL, et al. An MRI study of the basal ganglia in autism. Prog Neuropsychopharmacol Biol Psychiatry. 1999;23:613–624. doi: 10.1016/s0278-5846(99)00020-2. [DOI] [PubMed] [Google Scholar]
- 31.Welch JM, et al. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature. 2007;448:894–900. doi: 10.1038/nature06104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shmelkov SV, et al. Slitrk5 deficiency impairs corticostriatal circuitry and leads to obsessive-compulsive-like behaviors in mice. Nat Med. 2010;16:598–602. doi: 10.1038/nm.2125. 591p following 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graybiel AM. Habits, rituals, and the evaluative brain. Annu Rev Neurosci. 2008;31:359–387. doi: 10.1146/annurev.neuro.29.051605.112851. [DOI] [PubMed] [Google Scholar]
- 34.McFarlane HG, et al. Autism-like behavioral phenotypes in BTBR T+tf/J mice. Genes Brain Behav. 2008;7:152–163. doi: 10.1111/j.1601-183X.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- 35.Blundell J, et al. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010;30:2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci U S A. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Insel TR, Fernald RD. How the brain processes social information: searching for the social brain. Annu Rev Neurosci. 2004;27:697–722. doi: 10.1146/annurev.neuro.27.070203.144148. [DOI] [PubMed] [Google Scholar]
- 38.Ebstein RP, Israel S, Chew SH, Zhong S, Knafo A. Genetics of human social behavior. Neuron. 2010;65:831–844. doi: 10.1016/j.neuron.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 39.Welch JM, Wang D, Feng G. Differential mRNA expression and protein localization of the SAP90/PSD-95-associated proteins (SAPAPs) in the nervous system of the mouse. J Comp Neurol. 2004;472:24–39. doi: 10.1002/cne.20060. [DOI] [PubMed] [Google Scholar]
- 40.Vorhees CV, Williams MT. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat Protoc. 2006;1:848–858. doi: 10.1038/nprot.2006.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin HH, Davis MI, Ronesi JA, Lovinger DM. The role of protein synthesis in striatal long-term depression. J Neurosci. 2006;26:11811–11820. doi: 10.1523/JNEUROSCI.3196-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malenka RC, Kocsis JD. Presynaptic actions of carbachol and adenosine on corticostriatal synaptic transmission studied in vitro. J Neurosci. 1988;8:3750–3756. doi: 10.1523/JNEUROSCI.08-10-03750.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myme CI, Sugino K, Turrigiano GG, Nelson SB. The NMDA-to-AMPA ratio at synapses onto layer 2/3 pyramidal neurons is conserved across prefrontal and visual cortices. Journal of Neurophysiology. 2003;90:771–779. doi: 10.1152/jn.00070.2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.