Summary
Studies in the yeast Saccharomyces cerevisiae have validated the major features of the double-strand break repair (DSBR) model as an accurate representation of the pathway through which meiotic crossovers are produced. This success has led to this model being invoked to explain double-strand break (DSB) repair in other contexts. However, most non-crossover recombinants generated during S. cerevisiae meiosis do not arise via a DSBR pathway. Furthermore, and it is becoming increasing clear that DSBR is a minor pathway for recombinational repair of DSBs that occur in mitotically proliferating cells; rather, the synthesis-dependent strand annealing (SDSA) model appears to describe mitotic DSB repair more accurately. Fundamental dissimilarities between meiotic and mitotic recombination are not unexpected, since meiotic recombination serves a very different purpose (accurate chromosome segregation, which requires crossovers) than mitotic recombination (repair of DNA damage, which typically generates non-crossovers).
Keywords: meiotic recombination, mitotic recombination, double-strand break repair
Introduction
The existence of DNA recombination was revealed by the behavior of segregating traits long before DNA was identified as the bearer of genetic information. At the start of the 20th century, pioneering Drosophila geneticists studied the behavior of chromosomal “factors” that determined traits such as eye color, wing shape, and bristle length. In 1910 Thomas Hunt Morgan published the observation that the linkage relationships of these factors were shuffled during meiosis [1]. Building on this discovery, in 1913 A. H. Sturtevant used linkage analysis to determine the order of factors (genes) on a chromosome, thus simultaneously establishing that genes are located at discrete physical locations along chromosomes as well as originating the classic tool of genetic mapping [2]. The revelation that somatic cells also experience recombination did not occur until some years after the discovery of meiotic recombination, when in 1936 that Stern proposed crossovers (COs) between homologous chromosomes to explain patches of mosaicism in D. melanogaster [3].
In the 1950’s several breakthroughs furthered our understanding of recombination. Research on meiotic recombination benefitted from analysis of asci from the fungi Saccharomyces cerevisiae and Neurospora crassa [4]. The recovery of all the products of a single meiosis within a single ascus allowed the observation of non-crossover (NCO) recombination such as gene conversion (GC). Further, the determination of the structure of DNA enabled geneticists to develop models of how the physical behavior of double-stranded DNA translates into the genetic properties of recombination. The models posited by Robin Holliday [5] and subsequent researchers sought to illustrate the DNA transactions that occur during meiosis and DNA repair that give rise to COs, GC, and in some cases, post-meiotic segregation (PMS, see below).
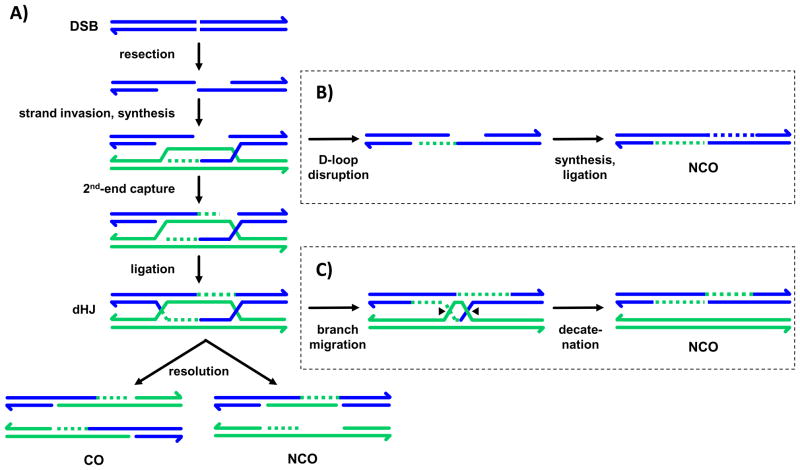
Current models for meiotic recombination are based on the double-strand break repair (DSBR) model outlined by Szostak and colleagues in the 1980s [6]. The central features of this model are: initiation by a double-strand break (DSB), formation of a double Holliday junction (dHJ) intermediate, and resolution of the dHJ by nicking two strands at each HJ (Fig. 1A). Although several modifications have been made to the original model, these core features are well supported for meiotic recombination in the S. cerevisiae, the model organism workhorse in which this process has been most extensively studied.
Figure 1.
Models for double-strand break repair. A: In the canonical double-strand break repair (DSBR) model proposed by Szostak et al. [6], an initiating DSB is processed to produce 3′ overhangs. One or both of these overhangs can invade a homologous duplex, usually a sister chromatid or homologous chromosome. A D-loop is displaced by the invading strand. The free 3′ end of the invading strand primes synthesis using the homologous sequence as a template. The other resected end of the break anneals to the D-loop, in a process called second-end capture. Additional synthesis and ligation of nicks produces a dHJ intermediate. To resolve the dHJ, two strands are nicked at each of the HJs. If two strands are each nicked twice (once at each junction), NCO repair products are produced. If different strands are cut at each junction (so that each of the four strands is nicked once), CO repair products are produced. B: In synthesis-dependent strand annealing (SDSA), the invading strand is displaced from the D-loop structure and its newly synthesized sequence anneals to the other side of the break. This yields a NCO repair product. C: In dHJ-dissolution, the two HJs are branch-migrated together and then decatenated to produce a NCO.
Because mitotic recombination is comparatively rare and more difficult to detect, characterization of this process has lagged. (Note: We use the term “mitotic recombination” to refer to COs and/or GC that takes place in cells that are proliferating mitotically, whether or not the process occurs during the mitotic phase of the cell cycle.) One consequence of meiotic recombination being more readily studied than mitotic recombination is that models for mitotic recombination have historically been strongly influenced by, and dependent on, models of meiotic recombination. Indeed, the strong evidence for the DSBR model in meiotic recombination in S. cerevisiae has frequently led to the assumption that this model also applies to mitotic DSB repair. However, as we discuss in this review, there is little evidence to support the direct application of the canonical DSBR model to mitotic DSB repair, and other models appear to describe mitotic DSB repair more accurately.
Meiotic vs. mitotic recombination
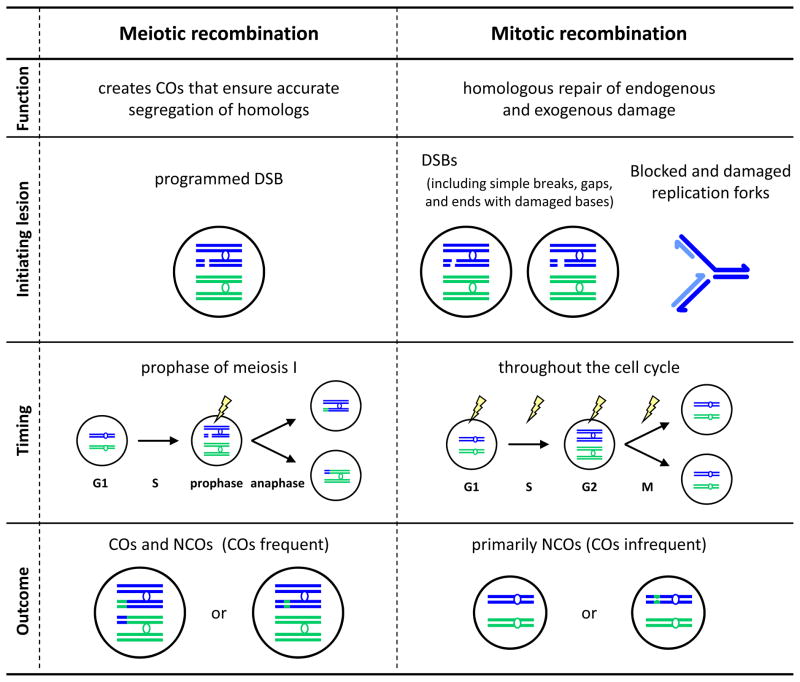
Mitotic and meiotic recombination differ fundamentally in purpose (Fig. 2). Meiotic recombination is actively promoted and highly regulated because it is crucial for accurate chromosome segregation. Meiotic COs create physical links (chiasmata) between homologous chromosomes, thereby facilitating their proper alignment at the metaphase plate and their subsequent disjunction at anaphase of meiosis I. Disruption of meiotic COs therefore leads to increased chromosome non-disjunction and sterility. In contrast, mitotic recombination is used in the homologous repair of spontaneous and induced DNA damage. Although there are well-studied examples of programmed mitotic recombination, including mating-type switching in fungi and mammalian V(D)J recombination, the discussion below focuses on spontaneous and induced mitotic recombination, especially during DSB repair.
Figure 2.
Meiotic and mitotic recombination are fundamentally different in several aspects, including function, initiating lesions, timing during the cell cycle, and outcome. In this figure, a single pair of homologous chromosomes is shown in each nucleus. Both DNA strands of each chromatid are shown.
The different functions of recombination are reflected in the ratios of COs to NCOs after DSB repair in meiotic and mitotic cells. In most eukaryotes, meiotic recombination frequently results in COs, consistent with the requirement for COs to direct chromosome segregation. For example, in mouse meiosis there is approximately one NCO for every two COs [7], in Drosophila it’s been estimated that there are three NCOs for every CO [8], and a genome-wide measurement in S. cerevisiae found an equal ratio of NCOs to COs [9]. In contrast, COs are usually rare in mitotic DSB repair [10–13]. COs are likely suppressed during mitotic DSB repair due to their potential for negative consequences, such as loss of heterozygosity distal to CO sites, or chromosome rearrangements that occur when non-allelic sequences (e.g., dispersed repetitive sequences) recombine with one another.
These functional differences are reflected in the differing genetic requirements for meiotic and mitotic recombination, which may stem from the use of different recombination mechanisms and/or differences in types of initiating damage, choice of repair template, or cell cycle stage. In meiosis, a DNA nuclease produces a simple DSB, whereas damage incurred by mitotic cells may include single-stranded and double-stranded gaps, breaks ending with damaged bases, one-ended DSBs, and other more complex arrangements. Furthermore, mitotic DNA damage is not limited to strand breaks, but also includes deleterious structures that can arise during DNA metabolism, such as stalled or blocked replication forks; repair of such structures adds another level of complexity to mitotic recombination.
Thus, while both meiotic and mitotic recombination are crucial for maintaining stable genomes, there are essential differences in the execution of each. Meiotic recombination is a programmed process that frequently generates COs used to accurately segregate homologous chromosomes from one another, whereas the scope and diversity of mitotic recombination are broader, and COs are a rare outcome.
Meiotic recombination in Saccharomyces cerevisiae
The studies that led to the proposal of the original DSBR model, as well as subsequent modifications, have been done predominantly in S. cerevisiae. This organism has several features that facilitate study of meiotic recombination, including hotspots for DSB formation, a cell cycle that is easily synchronized across a population, recovery of all four products of meiosis, and a relatively easily manipulated genome. Thus, our introduction to the DSBR model will focus on meiotic recombination research done in S. cerevisiae.
The DSBR model (Fig. 1A) was put forward to resolve discrepancies between predictions made by earlier models and subsequent experimental observations. Experiments performed since the proposal of the DSBR model have likewise required that some modifications be made, but there is still strong support for the central features of the canonical DSBR model, at least for the generation of COs. These central features include initiating DSBs and dHJ intermediates.
DSBs initiate meiotic recombination
Initial support for the DSBR model came from the finding that DSBs occur at hotspots for meiotic recombination and induced DSBs can initiate recombination [14]. The enzyme that catalyzes DSB formation was determined to be Spo11, a nuclease related to type II topoisomerases [15]. Other factors are also important for production of DSBs, including the MRX complex, comprising Mre11, Rad50, and Xrs2, and several proteins that are less well-conserved in primary sequence and whose precise functions are not yet known [16].
After the DSB is made, the ends are processed to produce long 3′ single-stranded overhangs. This feature of the model is supported by the physical detection of 3′ overhangs at hotspots [17]. The process of resection is still not completely understood, but recent evidence indicates that the MRX complex and the protein Sae2 are important for removing covalently bound Spo11, thereby allowing processing by the nucleases Exo1 and Dna1 [18].
After resection, one 3′ end invades the homologous template. This single-ended invasion intermediate has been detected in physical assays at recombination hot spots [19]. Mutations in genes in the rad52 epistasis group, which encode proteins that catalyze or facilitate strand invasion, disrupt meiotic recombination. An important byproduct of strand invasion is the creation of heteroduplex DNA (hDNA), which may contain mismatches or insertion/deletion loops between the two strands if there are heterologies between the homologous chromosomes. Both genetic and physical studies have detected hDNA in S. cerevisiae meiosis [20,21]. Heterologies within hDNA are usually repaired, leading either to GC or to restoration of the original sequence. If hDNA is unrepaired, the heterologies between homologous chromosomes undergo post-meiotic segregation (PMS). PMS is increased in mismatch repair (MMR) mutants, supporting a role for MMR proteins in repairing hDNA produced during meiotic recombination [22].
Resolvases cut dHJ intermediates to yield crossovers
Other than recombination initiation by a DSB, the most fundamental feature of the DSBR model is the dHJ. Consequently, the strongest support for the DSBR model came with detection and isolation of “joint molecules” formed during recombination in S. cerevisiae [23,24]. These joint molecules have the predicted properties of dHJ intermediates: They can be resolved into CO and NCO products by RuvC, an E. coli nuclease with high specificity for HJs [25], and all four strands are thought to be continuous (i.e., there are no unligated nicks). Additionally, all four strands of the joint molecules have the parental arrangement of flanking markers [24]. This finding is consistent with an intermediate with two HJs; in an intermediate a single HJ, two strands would have a parental arrangement of markers, and two would have a recombinant arrangement. The in vitro RuvC experiment also supports the postulate that dHJs can produce both COs and NCOs, as predicted by the DSBR model.
The final step of the DSBR model involves cutting of the dHJ by one or more HJ resolvases. Despite intensive searches, it is unclear what resolvase carries out this process in S. cerevisiae. One candidate is the nuclease Mus81. In vitro, the Mus81-Mms4/Eme1 dimer from several organisms cleave HJs; however, these enzymes have higher activity on other structures, such as D-loops and nicked HJs [26–28]. S. cerevisiae mus81 and mms4 mutants lack a subset of meiotic COs that do not exhibit CO interference; however, Mus81-Mms4 is not required to produce the COs that participate in crossover interference, which are the majority of COs in S. cerevisiae. Based on the robust activity of Mus81-Mms4 on nicked HJs, it has been suggested that these COs arise not from fully ligated dHJs, but from some other intermediate [29].
COs that exhibit interference require a set of proteins that includes the mismatch repair-related MutS homologs Msh4 and Msh5 [30]. It is unclear what role Msh4-Msh5 plays in promoting crossing over, but one important function seems to be to bind to dHJs to stabilize them and/or block NCO-promoting proteins [31,32]. The enzyme that then cuts these stabilized dHJs remains elusive. The nuclease Yen1 was recently found to have robust HJ resolvase activity in vitro [33]; however, Yen1 does not seem to have an important function in meiotic recombination. Rather, it is partially redundant with Mus81, and appears to be secondary to Mus81 in the resolution of recombination intermediates [34].
Meiotic non-crossovers come from SDSA
Despite strong support for many features of the DSBR model, additional data inconsistent with the canonical model have required an important modification. In a study of the timing of appearance and disappearance of meiotic recombination intermediates in S. cerevisiae, Allers and Lichten made the surprising finding that NCOs appear earlier than dHJs or COs, challenging the prediction that COs and NCOs are both produced from dHJs [35]. These authors also cited previous characterization of several mutants that have decreased frequencies of COs but not of NCOs, consistent with the possibility that these two types of products do not both arise from the same intermediate. They hypothesized that NCOs arise instead from synthesis-dependent strand annealing (SDSA) (Fig. 1B). In SDSA, repair is initiated as in the DSBR model, but before second-end capture the invading strand is dissociated from the D-loop and the newly synthesized sequences anneals to the complementary single strand on the other side of the break. SDSA does not involve a dHJ intermediate. Support for SDSA in meiosis has come from the analysis of hDNA found in recombination products that exhibit PMS. In S. cerevisiae, hDNA tracts are frequently restricted to one side of the break, as predicted by the SDSA model [36,37]. In a recent test of the SDSA model, McMahill et al. [38] found that a class of GCs best explained by the SDSA model comprised a high percentage of the NCO products recovered.
Another mechanism that has been suggested for generation of NCOs is dHJ dissolution [36,39]. Dissolution occurs when the two HJs are branch migrated toward one another, and the remaining catenation is removed by a type I topoisomerase (Fig. 1C). In vitro studies have shown that BLM helicase and TOP3α topoisomerase can catalyze this reaction efficiently [40]. After dHJ dissolution, the chromatid that received the DSB will have hDNA with sequence from the homologous chromosome on one strand to one side of the break and on the other strand to the other side of the break – the trans configuration (Fig. 1C). Trans hDNA has been detected in S. cerevisiae, but the frequency is extremely low, suggesting that some or all of the instances noted may actually result from the occurrence of overlapping recombination events at two nearby DSB sites [36,39,37]. Furthermore, dHJ dissolution cannot account for the appearance of NCOs prior to the appearance of dHJs. Based on these arguments, dHJ dissolution does not seem to be a major source of NCOs in S. cerevisiae meiosis.
Meiotic recombination in other eukaryotes
Meiotic recombination in S. cerevisiae is perhaps best explained by a model that unites the DSBR and SDSA models. The extent to which this compound model is applicable to other organisms is still in question. The key initiation event – formation of a DSB – appears to be universal. Orthologs of Spo11 have been found to be essential for meiotic recombination in S. pombe, Arabidopsis, C. elegans, D. melanogaster, and mouse [41–45]. Indeed, the relationship between Spo11 and meiotic recombination initiation is so well established that the presence of a gene orthologous to SPO11 has been taken as evidence for a meiotic cell cycle in a species not known to reproduce sexually [46]. Removal of covalently bound Spo11 and the production of single-stranded 3′ ends is also well conserved, with the MRE1 RAD50 NBS1 (MRN) complex (analgous to the yeast MRX complex) being important to this process in many organisms [47]. Similarly, the families of proteins required for strand invasion are well conserved, and homologs of the canonical strand invasion protein RecA are required for fertility in S. pombe, Arabidopsis, C. elegans, D. melanogaster, and mouse [48–52].
Although these early steps in recombination appear to be very similar across eukaryotic meiosis, the degree to which the later steps are conserved is unclear. As noted above, some COs in S. cerevisiae require the Mus81-Mms4 nuclease, whereas others require Msh4-Msh5. This is also true in Arabidopsis [53]. In S. pombe, however, there are no orthologs of Msh4 or Msh5, and most or all COs require Mus81 [54]. Recent work has revealed that most meiotic recombination in S. pombe involves an intermediate with a single HJ rather than a dHJ [55]. This suggests that orthologous proteins (e.g., Mus81) may act on different intermediates in different species.
Conversely, a common intermediate, such as a dHJ, may be acted upon by different proteins to produce COs in different species. In C. elegans, all COs seem to require MSH-4– MSH-5, suggesting they go through a dHJ intermediate [56]. Some of these COs are dependent on HIM-18– SLX-1, which is orthologous to the BTBD12–SLX1 HJ resolvase [57]. Similarly, meiotic COs in D. melanogaster have been suggested to go through a dHJ intermediate, despite the absence of Msh4 and Msh5 orthologs [58,59]. Most of these COs are generated by a complex that contains the MEI-9–ERCC1 endonuclease, which is orthologous to S. cerevisiae Rad1–Rad10 and mammalian XPF–ERCC1, and the proteins MUS312 [60,61] and HDM [62]. Genetic studies are consistent with the hypothesis that this complex resolves dHJs to produce COs [59].
Production of meiotic NCOs in other organisms is even less well understood. It has been suggested that meiotic NCOs in D. melanogaster are generated through SDSA, based on the observation that most NCO GC tracts in mei-9 mutants are indistinguishable from those in wild-type flies [59]. In C. elegans, COs are elevated in mutants that lack RTEL-1, a helicase that efficiently disrupts D-loops, suggesting that RTEL-1 promotes meiotic NCOs via SDSA [63].
Taken together, the data from different model organisms indicates that the initiation of meiotic recombination is conserved across eukaryotes, but that there are multiple ways to turn a DSB into a CO (or NCO). The DSBR model appears to be an accurate description of CO formation in S. cerevisiae and probably some other organisms, but it doesn’t seem to be deployed universally.
The DSBR model and mitotic DSB repair
Though intended to describe meiotic recombination, the classic DSBR model drew on evidence from studies of mitotic recombination. Prior studies on plasmid-chromosome recombination in yeast provided evidence that DSBs are recombinogenic, and suggested that GC could be produced by the repair of double-strand gaps [64]. These earlier studies provided the basis for the original formulation of the DSBR model [6]. Some of this interchange of ideas between meiotic and mitotic recombination models explains why the DSBR model is frequently co-opted in attempts to describe the mechanism of mitotic recombination. However, the application of the DSBR model to DSB repair in mitotic cells should be cautioned, because, as discussed above, meiotically and mitotically dividing cells have very different recombination requirements and outcomes.
The early steps of meiotic and mitotic recombination are similar
While the purpose and environment of recombination is different in mitotic cells, the early steps of the DSBR model are consistent with what is known about mitotic DSBR. There is, for example, no question that DSBs can induce COs in a mitotic context. It has long been known that treatments that produce DSBs, such as X-ray irradiation, can produce breaks and induce somatic COs [65]. Treating cells with agents that produce other types of damage, such as alkylation of bases, can also yield recombination, but it is generally thought that this is the result of a DSB formed as secondary damage [66]. For instance, the induction of genome rearrangements by crosslinking agents is DNA synthesis-dependent [67,68], suggesting that the recombinogenic damage ultimately results from secondary damage produced by replication forks encountering the primary interstrand crosslink damage. There is also evidence that mitotic recombination can result from single-stranded breaks or gaps as well [69]; for comparison to meiotic recombination, we restrict the discussion below to mitotic DSB repair.
Mutations that disrupt the early steps of meiotic DSBR have similar effects on mitotic recombination, which suggests that these steps are similar. A prime example is Rad51: In S. cerevisiae, rad51 mutants are defective in spontaneous and induced mitotic recombination and mating-type switching, in addition to the their defects in meiotic recombination [70]. Conversely, there are some notable differences in the early-acting genetic requirements for meiotic vs. mitotic recombination. There are meiosis-specific recombination factors, such as Dmc1, a Rad51 paralog found in many eukaryotes. Like rad51 mutants, dmc1 mutants are deficient in meiotic recombination [71] but not mitotic recombination [72]. There are also factors that vary in their relative importance or specific role. The Rad54 paralog Rdh54/Tid1 seems to be more important for meiotic interaction between homologs, whereas Rad54 is more important for recombinational repair between sister chromatids [73]. These differences, however, are indicative of redundancies of function and varying requirements due to differences in cell cycle and type of damage, and do not by themselves suggest different repair mechanisms.
Meiotic and mitotic recombination diverge in later steps
Although the early steps of mitotic DSB repair are similar to those of meiotic DSB repair, less is known about the later steps. In principle, mitotic DSB repair may be biased toward NCOs by any combination of the mechanisms described above for meiotic recombination: SDSA, dHJ dissolution, or dHJ resolution that is biased to produce NCOs. Some evidence favors SDSA being the primary NCO mechanism in mitotic DSB repair in S. cerevisiae. In a study of repair of a single DSB associated with a repetitive sequence, the repeat underwent expansion and contraction in the product, but the template was unaltered; sometimes more than one donor templates was used, suggesting repeated rounds of strand invasion and synthesis [74]. Bzymek et al. recently measured formation of joint molecules (dHJ intermediates) at sites of DSBs induced in mitotically dividing diploid S. cerevisiae and found them to be reduced in frequency by at least a factor of ten compared to meiotic DSBs, indicating that DSBR is not a primary DSB repair pathway [75]. Differences observed in the hDNA of CO vs NCO repair products of a gapped plasmid assay also support the use of SDSA as the primary DSB repair mechanism [76].
Studies of mitotic gap repair in D. melanogaster have also yielded strong evidence for SDSA. In gap repair, the chromosome that receives the break fills the gap by copying information from a template, without alteration of the template [77]. Furthermore, the two ends of the gap can use different templates independently [77]. In experiments in which the gap spans a direct repeat, one of the most common products has collapse of the repeat to a single copy, with loss of intervening sequences [78,79]. These findings are not compatible with repair by DSBR, but are readily accommodated by the SDSA model.
Studies of the genetic requirements of NCO repair have also provided important insights into the primary repair mechanisms. A key player in the process of actively promoting NCO repair and blocking CO-associated repair is the RecQ helicase BLM. Cells lacking the BLM helicase have elevated spontaneous COs, primarily between sister chromatids, but also between homologous and heterologous chromosomes [80,81]. Discussions of the anti-crossover functions of BLM are most often based on the hypothesis that the primary anti-crossover activity is in dHJ dissolution [40]. This hypothesis has been driven by a combination of genetic and biochemical studies.
S. cerevisiae top3 mutants grow slowly, but the slow growth is suppressed by mutations in SGS1 (slow growth suppressor 1), which encodes the only RecQ helicase in this species. One interpretation of this finding is that Sgs1 produces an intermediate that is toxic in the absence of Top3 activity [82]. The identity of the toxic intermediate is suggested by the in vitro dHJ dissolution activity of BLM and TOP3α. In vitro, BLM and TOP3α (together with accessory proteins) catalyze dHJ dissolution efficiently, with BLM migrating the HJs together so they can be decatenated by TOP3α [83–85]. Thus, unresolved, branch-migrated dHJs may be the toxic intermediates in top3 mutants. However, direct evidence that dHJ dissolution occurs during mitotic DSB repair in vivo is lacking; indeed, it is now clear that dHJs are a minor intermediate in break repair in S. cerevisiae [75]. The dissolvase activity of BLM – TOP3α may be important in preventing mitotic COs that arise from other types of spontaneous damage. For example, it has been hypothesized that repair of gaps that occur when replication is blocked on the lagging strand might involve formation of dHJ intermediates that do not originate with a DSB [86].
Regardless of its roles in replication fork repair, Sgs1 and the Drosophila ortholog, DmBLM, do have important anti-crossover functions during DSB repair [79,87,88]. Interestingly, mutants lacking DmBLM have severe defects in gap repair, suggesting that the primary role of DmBLM in preventing COs during break repair may be in promoting SDSA rather than in dissolving dHJs [79]. It is possible that the genetic interaction between BLM and TOP3α reflects a heretofore uncharacterized requirement for TOP3α in SDSA. Perhaps TOP3α is required to relax supercoiling produced by D-loop production/migration. Such a requirement is unlikely to be revealed with the short substrates used in in vitro biochemical assays.
Although DmBLM appears to facilitate SDSA in Drosophila, the same role in S. cerevisiae is thought to be taken on by a different helicase, Srs2. As in sgs1 mutants, srs2 mutants have increased COs during repair of induced DSBs [89,90]. Also like Sgs1 and BLM, Srs2 is a 3′→5′ DNA helicase and has a D-loop disrupting activity in vitro, suggesting that it might play a role in SDSA [91,92]. Srs2 is also able to disrupt Rad51 filaments, so it might prevent COs by preventing strand invasion [92]. However, sgs1 srs2 double mutants have extremely slow growth, and this phenotype is dependent on the presence of Rad51 and other strand invasion proteins, suggesting that Srs2 and Sgs1 have partially redundant functions in removing some recombination intermediate, such as D-loops [93,94]. Srs2 orthologs have not been identified outside of fungi, but the functional analog in C. elegans appears to be RTEL-1. Like Srs2, RTEL-1 disrupts D-loops in vitro and is believed to promote SDSA in vivo [95].
Although NCO mechanisms are strongly promoted in mitotic cells, spontaneous mitotic COs do occur. It is generally believed that at least some of these COs arise from resolution of a dHJ intermediate, as in DSBR. Identification of the resolvase(s) responsible for generating mitotic COs would lend support to this hypothesis. In S. pombe, Mus81-Eme1, which is required for most or all meiotic COs, is required for a subset of mitotic COs associated with induced DSBs [96]. In contrast, spontaneous mitotic COs are not affected by loss of Mus81 in Arabidopsis [97], and in S. cerevisiae mus81 mutants, the frequency of spontaneous mitotic COs is slightly elevated [98]. Thus, the resolvases that are involved in production of mitotic COs remain unknown.
Conclusions
A joint DSBR/SDSA model currently provides the best description of meiotic recombination in several of the model organisms in which it has been most extensively studied, including S. cerevisiae, D. melanogaster, and C. elegans. However, this picture is not yet complete. For example, the model does not satisfactorily account for the differing biochemical activities of the nucleases and MMR-related proteins required for different subsets of COs in a single species, or the different requirements between species. For mitotic DSB repair, the DSBR model is less well supported; rather, the SDSA model seems to be a more accurate description of the primary recombinational repair mechanism. Several proteins, including S. cerevisiae Srs2, Drosophila DmBLM, and C. elegans RTEL-1, have been implicated in promoting SDSA and thereby preventing mitotic COs. Thus, while many parallels have been found between meiotic and mitotic recombination, we must remain mindful that the differing functions for recombination may have prompted the development of different solutions to meet different needs in meiotic and mitotic contexts.
Acknowledgments
Research in the Sekelsky lab is supported by grants from the NIH: R01GM061252 and RO1GM088606.
Abbreviations
- CO
crossover
- dHJ
double Holliday junction
- DSB
double-strand break
- DSBR
double-strand break repair
- GC
gene conversion
- NCO
non-crossover
- PMS
post-meiotic segregation
- SDSA
synthesis-dependent strand annealing
References
- 1.Morgan TH. The method of inheritance of two sex-linked characters in the same animal. Proc Soc Exp Biol Med. 1910;8:17. [Google Scholar]
- 2.Sturtevant AH. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J Exp Zool. 1913;14:43–59. [Google Scholar]
- 3.Stern C. Somatic crossing over and Segregation in Drosophila melanogaster. Genetics. 1936;21:625–730. doi: 10.1093/genetics/21.6.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindegren CC. Non-Mendelian segregation in a single Tetrad of Saccharomyces ascribed to gene conversion. Science. 1955;121:605–7. doi: 10.1126/science.121.3147.605. [DOI] [PubMed] [Google Scholar]
- 5.Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;78:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 6.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 7.Guillon H, Baudat F, Grey C, Liskay RM, et al. Crossover and noncrossover pathways in mouse meiosis. Mol Cell. 2005;20:563–73. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 8.Mehrotra S, McKim KS. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2006;2:e200. doi: 10.1371/journal.pgen.0020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–95. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haber JE, Hearn M. Rad52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics. 1985;111:7–22. doi: 10.1093/genetics/111.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virgin JB, Bailey JP, Hasteh F, Neville J, et al. Crossing over is rarely associated with mitotic intragenic recombination in Schizosaccharomyces pombe. Genetics. 2001;157:63–77. doi: 10.1093/genetics/157.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stark JM, Jasin M. Extensive loss of heterozygosity is suppressed duringhomologous repair of chromosomal breaks. Mol Cell Biol. 2003;23:733–43. doi: 10.1128/MCB.23.2.733-743.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nickoloff JA, Brenneman MA. Analysis of recombinational repair of DNA double-strand breaks in mammalian cells with I-SceI nuclease. Methods Mol Biol. 2004;262:35–52. doi: 10.1385/1-59259-761-0:035. [DOI] [PubMed] [Google Scholar]
- 14.Kolodkin AL, Klar AJ, Stahl FW. Double-strand breaks can initiate meiotic recombination in S. cerevisiae. Cell. 1986;46:733–40. doi: 10.1016/0092-8674(86)90349-1. [DOI] [PubMed] [Google Scholar]
- 15.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–84. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 16.Keeney S, Neale MJ. Initiation of meiotic recombination by formation of DNA double-strand breaks: mechanism and regulation. Biochem Soc Trans. 2006;34:523–5. doi: 10.1042/BST0340523. [DOI] [PubMed] [Google Scholar]
- 17.Sun H, Treco D, Szostak JW. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–61. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 18.Manfrini N, Guerini I, Citterio A, Lucchini G, et al. Processing of meiotic DNA double strand breaks requires cyclin-dependent kinase and multiple nucleases. J Biol Chem. 2010;285:11628–37. doi: 10.1074/jbc.M110.104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 20.Goyon C, Lichten M. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol Cell Biol. 1993;13:373–82. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nag DK, Petes TD. Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2324–31. doi: 10.1128/mcb.13.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramer B, Kramer W, Williamson MS, Fogel S. Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS genes. Mol Cell Biol. 1989;9:4432–40. doi: 10.1128/mcb.9.10.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins I, Newlon CS. Meiosis-specific formation of joint DNA molecules containing sequences from homologous chromosomes. Cell. 1994;76:65–75. doi: 10.1016/0092-8674(94)90173-2. [DOI] [PubMed] [Google Scholar]
- 24.Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 25.Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–91. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 26.Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J Biol Chem. 2002;277:32753–9. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- 27.Gaillard PH, Noguchi E, Shanahan P, Russell P. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell. 2003;12:747–59. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- 28.Ehmsen KT, Heyer WD. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 2008;36:2182–95. doi: 10.1093/nar/gkm1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitby MC. Making crossovers during meiosis. Biochem Soc Trans. 2005;33:1451–5. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- 30.de los Santos T, Hunter N, Lee C, Larkin B, et al. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snowden T, Acharya S, Butz C, Berardini M, et al. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell. 2004;15:437–51. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 32.Jessop L, Rockmill B, Roeder GS, Lichten M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2006;2:e155. doi: 10.1371/journal.pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ip SC, Rass U, Blanco MG, Flynn HR, et al. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–61. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 34.Blanco MG, Matos J, Rass U, Ip SC, et al. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair (Amst) 2010;9:394–402. doi: 10.1016/j.dnarep.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 35.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 36.Gilbertson LA, Stahl FW. A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1996;144:27–41. doi: 10.1093/genetics/144.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merker JD, Dominska M, Petes TD. Patterns of heteroduplex formation associated with the initiation of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics. 2003;165:47–63. doi: 10.1093/genetics/165.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMahill MS, Sham CW, Bishop DK. Synthesis-dependent strand annealing in meiosis. PLoS Biol. 2007;5:e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stahl F. Meiotic recombination in yeast: coronation of the double-strand-break repair model. Cell. 1996;87:965–8. doi: 10.1016/s0092-8674(00)81791-2. [DOI] [PubMed] [Google Scholar]
- 40.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–4. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 41.Dernburg AF, McDonald K, Moulder G, Barstead R, et al. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–98. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- 42.McKim KS, Hayashi-Hagihara A. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 1998;12:2932–42. doi: 10.1101/gad.12.18.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell. 2000;6:975–87. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 44.Grelon M, Vezon D, Gendrot G, Pelletier G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001;20:589–600. doi: 10.1093/emboj/20.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharif WD, Glick GG, Davidson MK, Wahls WP. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome. 2002;1:1. doi: 10.1186/1475-9268-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malik SB, Ramesh MA, Hulstrand AM, Logsdon JM., Jr Protist homologs of the meiotic Spo11 gene and topoisomerase VI reveal an evolutionary history of gene duplication and lineage-specific loss. Mol Biol Evol. 2007;24:2827–41. doi: 10.1093/molbev/msm217. [DOI] [PubMed] [Google Scholar]
- 47.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–95. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muris DF, Vreeken K, Schmidt H, Ostermann K, et al. Homologous recombination in the fission yeast Schizosaccharomyces pombe: different requirements for the rhp51+, rhp54+ and rad22+ genes. Curr Genet. 1997;31:248–54. doi: 10.1007/s002940050202. [DOI] [PubMed] [Google Scholar]
- 49.Ghabrial A, Ray RP, Schupbach T. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 1998;12:2711–23. doi: 10.1101/gad.12.17.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pittman DL, Cobb J, Schimenti KJ, Wilson LA, et al. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 51.Rinaldo C, Bazzicalupo P, Ederle S, Hilliard M, et al. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics. 2002;160:471–9. doi: 10.1093/genetics/160.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bleuyard JY, White CI. The Arabidopsis homologue of Xrcc3 plays an essential role in meiosis. EMBO J. 2004;23:439–49. doi: 10.1038/sj.emboj.7600055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 2007;3:e132. doi: 10.1371/journal.pgen.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boddy MN, Gaillard PH, McDonald WH, Shanahan P, et al. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–48. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 55.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, et al. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–78. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zalevsky J, MacQueen AJ, Duffy JB, Kemphues KJ, et al. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 1999;153:1271–83. doi: 10.1093/genetics/153.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saito TT, Youds JL, Boulton SJ, Colaiacovo MP. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 2009;5:e1000735. doi: 10.1371/journal.pgen.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sekelsky J, Brodsky MH, Burtis KC. DNA repair in Drosophila. Insights from the Drosophila genome sequence. J Cell Biol. 2000;150:F31–F36. doi: 10.1083/jcb.150.2.f31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Radford SJ, McMahan S, Blanton HL, Sekelsky J. Heteroduplex DNA in meiotic recombination in Drosophila mei-9 mutants. Genetics. 2007;176:63–72. doi: 10.1534/genetics.107.070557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sekelsky JJ, McKim KS, Chin GM, Hawley RS. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics. 1995;141:619–27. doi: 10.1093/genetics/141.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yildiz O, Majumder S, Kramer B, Sekelsky JJ. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol Cell. 2002;10:1503–9. doi: 10.1016/s1097-2765(02)00782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joyce EF, Tanneti SN, McKim KS. Drosophila hold’em is required for a subset of meiotic crossovers and interacts with the dna repair endonuclease complex subunits MEI-9 and ERCC1. Genetics. 2009;181:335–40. doi: 10.1534/genetics.108.093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Youds JL, Mets DG, McIlwraith MJ, Martin JS, et al. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327:1254–8. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orr-Weaver TL, Szostak JW, Rothstein RJ. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–45. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- 65.Bauer H, Demerec M, Kaufmann BP. X-Ray induced chromosomal alterations in Drosophila melanogaster. Genetics. 1938;23:610–30. doi: 10.1093/genetics/23.6.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaina B. Mechanisms and consequences of methylating agent-induced SCEs and chromosomal aberrations: a long road traveled and still a far way to go. Cytogenet Genome Res. 2004;104:77–86. doi: 10.1159/000077469. [DOI] [PubMed] [Google Scholar]
- 67.Akkari YM, Bateman RL, Reifsteck CA, Olson SB, et al. DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol. 2000;20:8283–9. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barber LJ, Ward TA, Hartley JA, McHugh PJ. DNA interstrand cross-link repair in the Saccharomyces cerevisiae cell cycle: overlapping roles for PSO2 (SNM1) with MutS factors and EXO1 during S phase. Mol Cell Biol. 2005;25:2297–309. doi: 10.1128/MCB.25.6.2297-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Branzei D, Foiani M. The DNA damage response during DNA replication. Curr Opin Cell Biol. 2005;17:568–75. doi: 10.1016/j.ceb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 70.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–70. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 71.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–56. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 72.Fukushima K, Tanaka Y, Nabeshima K, Yoneki T, et al. Dmc1 of Schizosaccharomyces pombe plays a role in meiotic recombination. Nucleic Acids Res. 2000;28:2709–16. doi: 10.1093/nar/28.14.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shinohara M, Shita-Yamaguchi E, Buerstedde JM, Shinagawa H, et al. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–56. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paques F, Leung WY, Haber JE. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol. 1998;18:2045–54. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bzymek M, Thayer NH, Oh SD, Kleckner N, et al. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–41. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mitchel K, Zhang H, Welz-Voegele C, Jinks-Robertson S. Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol Cell. 2010;38:211–22. doi: 10.1016/j.molcel.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nassif N, Penney J, Pal S, Engels WR, et al. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–25. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurkulos M, Weinberg JM, Roy D, Mount SM. P element-mediated in vivo deletion analysis of white-apricot: deletions between direct repeats are strongly favored. Genetics. 1994;136:1001–11. doi: 10.1093/genetics/136.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adams MD, McVey M, Sekelsky JJ. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–7. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 80.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc Natl Acad Sci USA. 1974;71:4508–12. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ellis NA, Groden J, Ye TZ, Straughen J, et al. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–66. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 82.Gangloff S, McDonald JP, Bendixen C, Arthur L, et al. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol Cell. 1999;3:611–20. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 84.Harmon FG, Brockman JP, Kowalczykowski SC. RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. J Biol Chem. 2003;278:42668–78. doi: 10.1074/jbc.M302994200. [DOI] [PubMed] [Google Scholar]
- 85.Plank JL, Wu J, Hsieh TS. Topoisomerase IIIalpha and Bloom’s helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci USA. 2006;103:11118–23. doi: 10.1073/pnas.0604873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu L. Role of the BLM helicase in replication fork management. DNA Repair (Amst) 2007;6:936–44. doi: 10.1016/j.dnarep.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 87.Ira G, Malkova A, Liberi G, Foiani M, et al. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–11. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson-Schlitz D, Engels WR. Template disruptions and failure of double Holliday junction dissolution during double-strand break repair in Drosophila BLM mutants. Proc Natl Acad Sci USA. 2006;103:16840–5. doi: 10.1073/pnas.0607904103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aguilera A, Klein HL. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–90. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rong L, Palladino F, Aguilera A, Klein HL. The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics. 1991;127:75–85. doi: 10.1093/genetics/127.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rong L, Klein HL. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268:1252–9. [PubMed] [Google Scholar]
- 92.Dupaigne P, Le Breton C, Fabre F, Gangloff S, et al. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: Implications for crossover incidence during mitotic recombination. Mol Cell. 2008;29:243–54. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 93.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–4. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 94.McVey M, Kaeberlein M, Tissenbaum HA, Guarente L. The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics. 2001;157:1531–42. doi: 10.1093/genetics/157.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–71. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hope JC, Cruzata LD, Duvshani A, Mitsumoto J, et al. Mus81-Eme1-dependent and -independent crossovers form in mitotic cells during double-strand break repair in Schizosaccharomyces pombe. Mol Cell Biol. 2007;27:3828–38. doi: 10.1128/MCB.01596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hartung F, Suer S, Bergmann T, Puchta H. The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 2006;34:4438–48. doi: 10.1093/nar/gkl576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Robert T, Dervins D, Fabre F, Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25:2837–46. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]