Abstract
Transcription factor-based cellular reprogramming has opened the way to converting somatic cells to a pluripotent state, but has faced limitations resulting from the requirement for transcription factors and the relative inefficiency of the process. We show here that expression of the miR302/367 cluster rapidly and efficiently reprograms mouse and human somatic cells to an iPS state without a requirement for exogenous transcription factors. This miRNA-based reprogramming approach is two orders of magnitude more efficient than standard Oct4/Sox2/Klf4/Myc-mediated methods. Mouse and human miR302/367 iPS cells display similar characteristics to Oct4/Sox2/Klf4/Myc-iPS cells, including pluripotency marker expression, teratoma formation, and, for mouse cells, chimera contribution and germline contribution. We found that miR367 expression is required for miR302/367-mediated reprogramming and activates Oct4 gene expression, and that suppression of Hdac2 is also required. Thus, our data show that miRNA and Hdac-mediated pathways can co-operate in a powerful way to reprogram somatic cells to pluripotency.
INTRODUCTION
The transformation of differentiated cells to induced pluripotent stem (iPS) cells has revolutionized stem cell biology by providing a more tractable source of pluripotent cells for regenerative therapy. Although powerful, there are currently several limitations to iPS cell generation including the rather low efficiency of the process (0.2–1.0%) and the necessity of forced expression of at least one pluripotent stem cell transcription factor including Oct4, Nanog, Sox2, Klf4, and/or Myc. These limitations hamper the use of iPS technology in high throughput formats such as generation of human iPS clones from large patient populations.
The current standard strategy for iPS generation relies upon ectopic expression of Oct4, Sox2, Klf4 and Myc (OSKM) (Takahashi and Yamanaka, 2006). Although there are several alternatives to some of these factors including the use of other transcription factors, signaling factors, and pharmacological molecules, at least one pluripotent stem cell transcription factor, usually Oct4, is required for efficient iPS reprogramming (Huangfu et al., 2008a; Huangfu et al., 2008b; Judson et al., 2009; Melton et al., 2010; Yoshida et al., 2009). Recently, several microRNAs (miRNAs) have been shown to enhance iPS reprogramming when expressed along with combinations of the OSKM factors (Judson et al., 2009). These miRNAs belong to families of miRNAs that are expressed preferentially in embryonic stem cells and are thought to help maintain the ES cell phenotype (Babiarz et al., 2008; Wang et al., 2008; Wang and Blelloch, 2009; Wang et al., 2007). How these miRNAs enhance iPS reprogramming is unclear but may have to do with their ability to regulate the cell cycle (Judson et al., 2009).
Of the miRNAs expressed at high levels in ES and iPS cells, the miR302/367 cluster has been shown to be a direct target of Oct4 and Sox2 (Card et al., 2008), two of the critical factors required for iPS reprogramming. Levels of miR302/367 correlate with Oct4 transcripts in ES cells and early embryonic development, indicating an important role in ES cell homeostasis and maintenance of pluripotency (Card et al., 2008). Despite their ability to enhance iPS reprogramming in the presence of several of the OSKM factors (Judson et al., 2009), the ability of these miRNAs to directly reprogram somatic cells to an iPS phenotype is unclear. We show that expression of the miR302/367 cluster can directly reprogram mouse and human somatic cells to a pluripotent stem cell state in the absence of any of the previously described pluripotent stem cell transcription factors. Reprogramming by miR302/367 is up to two orders of magnitude more efficient than that with the OSKM factors. We also show that valproic acid (VPA) is required for reprogramming mouse fibroblasts by specifically degrading Hdac2 protein, a finding that is supported by the efficient reprogramming of Hdac2−/− fibroblasts in the absence of VPA. Thus, the expression of miR302/367 along with Hdac2 suppression allows for highly efficient iPS reprogramming without the expression of the known reprogramming factors.
RESULTS
miR302/367 reprograms fibroblasts to an iPS cell phenotype
Pervious studies have shown that the miR302/367 cluster is comprised of five miRNAs, four of which, miR302a/b/c/d, have an identical seed sequences (Card et al., 2008) and Fig. 1A). The miR302/367 cluster is located in intron 8 of the Larp7 gene on chromosome 3 and is transcribed as a single polycistronic primary transcript (Card et al., 2008). The sequence of the miR302/367 miRNAs are highly conserved across species (Card et al., 2008; Rosa et al., 2009). To determine whether expression of miR302/367 could reprogram somatic cells, we generated a lentiviral vector which expressed the 690 bp region encoding the mouse miR302/367 sequences and used it to transfect mouse embryonic fibroblasts (MEFs) derived from the Oct4-GFP mouse line ((Lengner et al., 2007) and Fig. 1B). We included the Hdac inhibitor VPA in these experiments as this has been shown to enhance iPS reprogramming (Huangfu et al., 2008a). Surprisingly, we observed clones derived from miR302/367 transduced MEFs within 6–8 days after the start of viral infection that had already assumed an ES cell like morphology (Fig. 1C and 3A). Most of these clones were Oct4-GFP positive and alkaline phosphatase positive (Fig. 1C and D). These clones also expressed Nanog, Sox2, and SSEA1 (Fig. 1E). In comparison, parallel expression of OSKM expressing viruses in addition to VPA did not result in any visible clones until at least 8–10 days after starting viral transduction (Fig. 3 and data not shown). Use of a polycistronic virus did not alter the timing or overall number of colonies generated by OSKM expression (data not shown and (Sommer et al., 2009). Moreover, in the absence of VPA, miR302/367 was unable to reprogram MEFs efficiently (see below and data not shown).
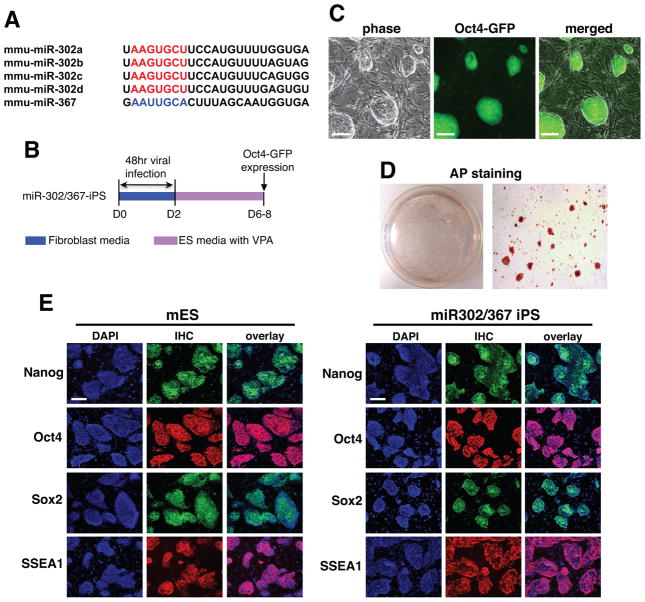
Figure 1. miR302/367 can reprogram mouse fibroblasts to a pluripotent stem cell phenotype.
(A) The sequences of the miR302/367 cluster showing the similarity between members of the miR302a/b/c/d subfamily. miR367 has a different seed sequence than miR302a/b/c/d. (B) Schematic of viral expression protocol for miR302/367 iPS reprogramming with VPA. Day 0 is the start of viral transduction. (C) Oct4-GFP positive miR302/367 clones at seven days after starting viral transduction. (D) AP staining of a primary induction plate of miR302/367 iPS clones at eight days after starting viral transduction. (E) Immunostaining for Nanog, Oct4, Sox2, and SSEA1 in both mouse ES and primary induction samples of miR302/367 iPS cells at day 10 showing expression of pluripotent genes. See also Figures S1 and S2. Scale bars=100 μm.
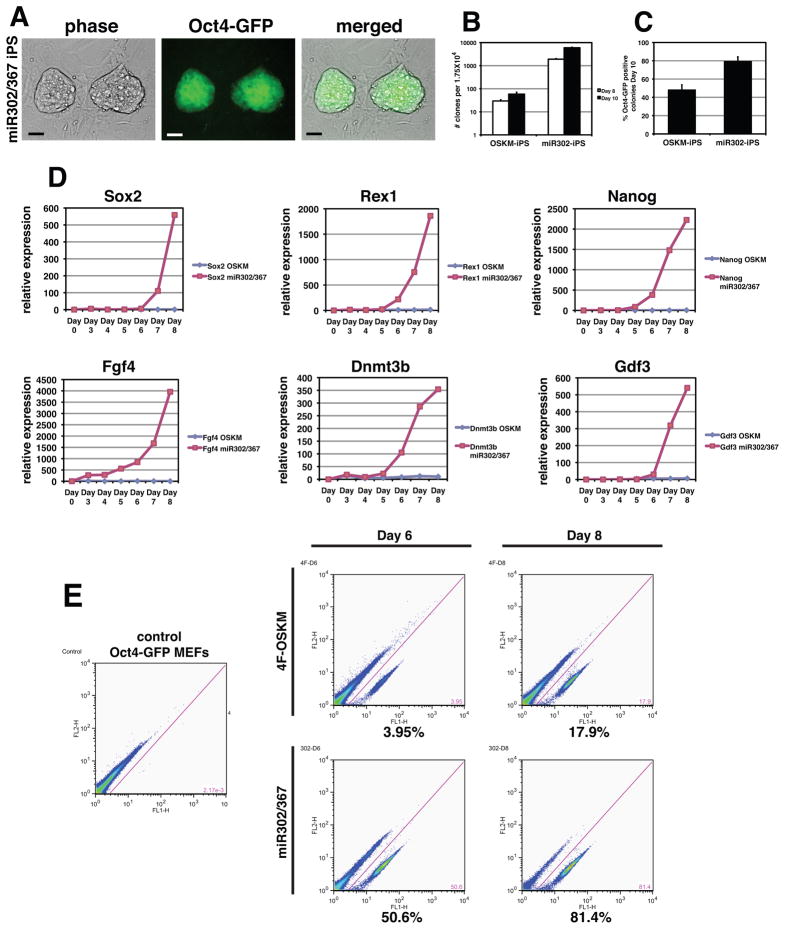
Figure 3. miR302/367 plus VPA is two orders of magnitude more efficient that OSKM factors in iPS reprogramming of mouse fibroblasts.
(A) miR302/367 iPS clones are readily observed 6–7 days after starting viral transduction and express high levels of Oct4-GFP while OSKM induced clones are not observed until 8–10 days, are very rare, and do not express significant levels of GFP from the Oct4 locus. (B) Counts of clones with ES like morphology from transduction of 1.75 × 104 Oct4-GFP MEFs with equivalent amounts of either OSKM or miR302/367 virus at eight and ten days after viral transduction. Data are the average of three assays ± S.E.M. (C) Percentage of Oct4-GFP positive clones ten days after viral transduction with OSKM or miR302/367. Data are the average of three assays ± S.E.M. (D) Q-PCR of the indicated pluripotent factors comparing OSKM versus miR302/367 during the first eight days after viral transduction. (E) FACS analysis of miR302/367 reprogrammed Oct4-GFP MEFs compared to OSKM reprogrammed MEFs at six and eight days post-viral transduction. Scale bars=50 μm.
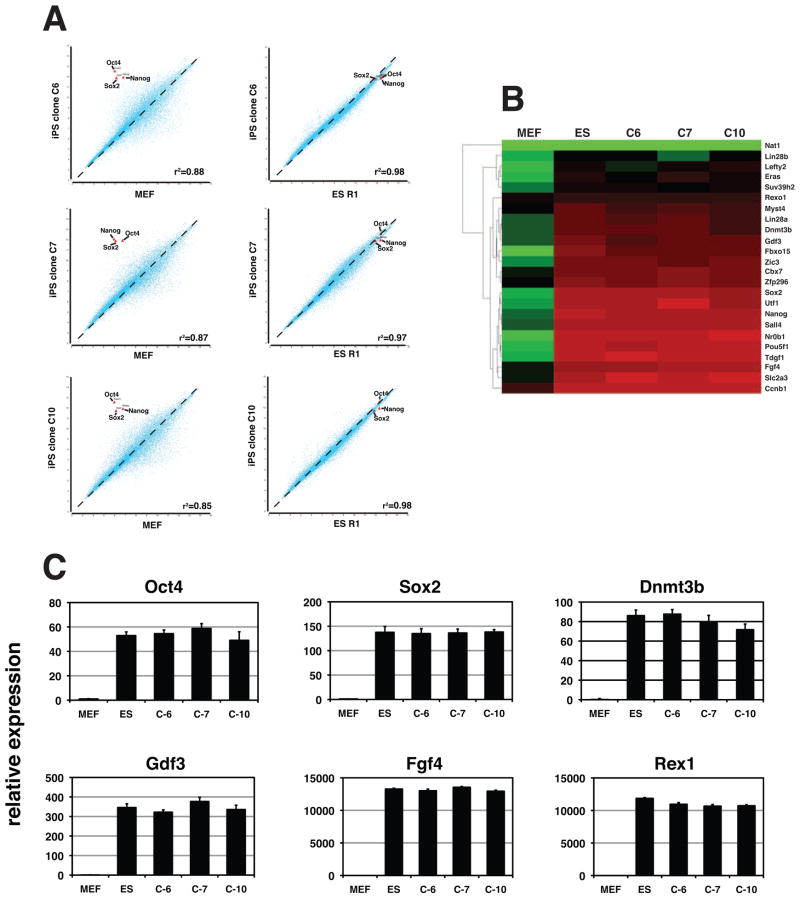
We further characterized the miR302/367 generated iPS clones by microarray analysis for their similarity at the global gene expression level to the mouse ES cell line R1. We used clones at passage 15 for these analyses. These data show a very high degree of correlation with global gene expression in the R1 ES cell line (Fig. 2A and B). These clones lacked integration of any of the OSKM factors that we use as controls but did contain viral integration of the miR302/367 lentivirus into the genome (Supplemental Figure 1). miR302/367 iPS clones that have been passaged serially maintain their ES like morphology and Q-PCR shows that they exhibit identical expression of pluripotent genes as mouse ES cells (Fig. 2C and data not shown). Moreover, the miR302/367 lentivirus is silenced at later passages (Supplemental Figure 2). These results suggest that expression of miR302/367 in addition to VPA was able to reprogram mouse MEFs to an iPS cell state without expression of other previously described pluripotent factors.
Figure 2. miR302/367 iPSC clones have a similar expression profile as mouse ES cells.
(A) Microarray experiments were used to show the similarity between miR302/367 iPS cell clones C6, C7, and C10 at passage 15 and the mouse ES cell line R1. (B) Heatmap of pluripotent gene expression of mouse ES cell line R1 and miR302/367 iPS cell clones C6, C7, and C10 from experiment in A. (C) Q-PCR of pluripotent gene expression of miR302/367 iPS cell clones C6, C7, and C10 at and mouse ES cell line R1. See also Figures S1 and S2.
miR302/367 reprogramming is more efficient than OSKM reprogramming
The rapid appearance of miR302/367 reprogrammed iPS cells suggested that expression of these miRNAs improved the temporal kinetics of reprogramming. To test this hypothesis, we expressed in parallel miR302/367 and the OSKM genes using an identical number of starting MEFs and viral titer. VPA was included in both OSKM as well as miR302/367 reprogramming experiments. Previous studies have demonstrated that using the OSKM factors, an average colony forming reprogramming efficiency of 0.2–0.8% is observed (Huangfu et al., 2008a). Using miR302/367, we consistently observe Oct4-GFP positive clones seven days after starting viral transduction, which is sooner than cells transduced in parallel with the OSKM factors (Fig. 3A). By counting the number of clones with ES like morphology at eight and ten days after starting viral transduction, we show that expression of miR302/367 produces two orders of magnitude more iPS clones than when the OSKM factors are used (Fig 3B). At day 10, 79.8% of miR302/367 iPS clones exhibited robust expression of Oct4-GFP which is greater than clones expressing the OSKM factors, of which only approximately 50% express Oct4-GFP (Fig. 3C).
To better quantify this increase in iPS reprogramming efficiency, we performed quantitative real time PCR (Q-PCR) for pluripotent marker genes during the first eight days of the reprogramming process on primary induction plates. The experiment used the same number of starting MEFs and viral titer for infection. These data indicate that while cells transduced with the OSKM factors expressed only very low levels of pluripotent marker genes during this time period, miR302/367 transduced cells expressed all of the genes examined at robust levels by day 8 (Fig. 3D). The numbers of clones were such that after 8–10 days, the plates containing the miR302/367 iPS clones became overcrowded resulting in decreased cell viability unless they were isolated and expanded. We also assessed the efficiency of reprogramming by miR302/367 using fluorescent activated cell sorting (FACS) for expression of GFP from the Oct4 locus in Oct4-GFP MEFs (Lengner et al., 2007). OSKM reprogrammed MEFs do show Oct4-GFP expression at both six and eight days of the reprogramming process with up to 17% of cells expressing GFP by day eight which is in the same range as previously reported (Fig. 3E and (Huangfu et al., 2008a)). However, miR302/367 is able to activate Oct4-GFP expression in up to 80% of MEFs after eight days of reprogramming (Fig. 3E). These data support the conclusion that miR302/367 is able to reprogram fibroblasts to a pluripotent state up to two-orders of magnitude more efficiently than OSKM factors.
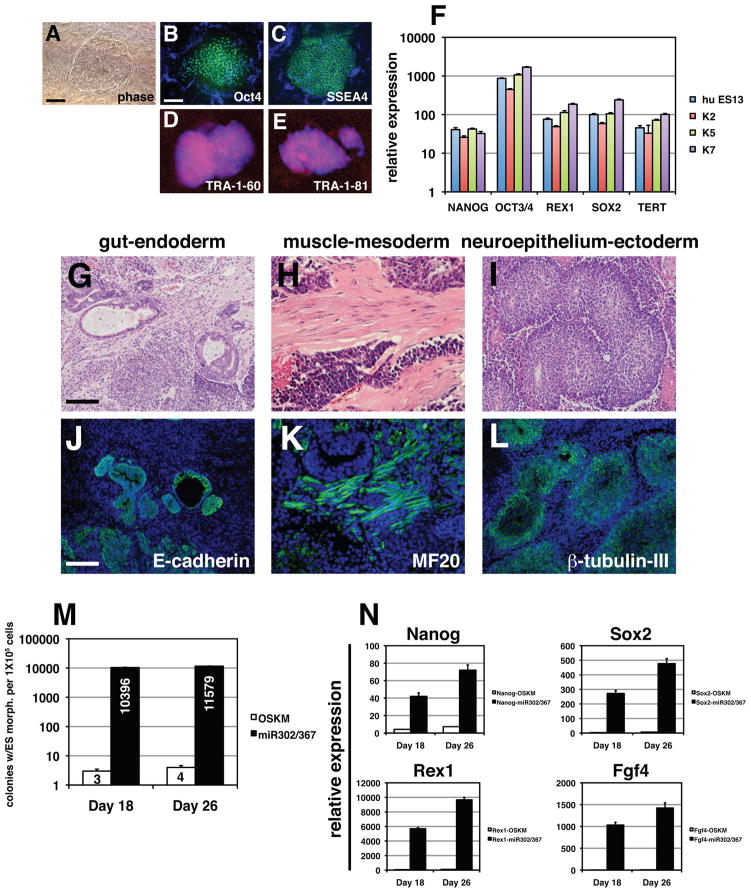
miR302/367 iPS cells can generate derivatives of mesoderm, endoderm, and ectoderm in teratomas, generate adult chimeras, and contribute to the mouse germline
To more fully characterize the pluripotent characteristics of miR302/367 iPS cells, we generated teratomas in immune deficient mice with multiple miR302/367 iPS clones. miR302/367 iPS derived teratomas formed readily and exhibited tissues representing all three germ layers as noted by structures resembling muscle fibers, keratinized epidermal cells, and luminal structures lined with gut-like epithelium (Fig. 4A). Supporting these morphological findings, neural epithelial-like structures were positive for βIII-tubulin expression, muscle-like structures were positive for myosin heavy chain expression, and gut-like epithelium was positive for E-cadherin expression (Fig. 4B). A more stringent assay for pluripotency is determining whether miR302/367 iPS cells can generate tissues within the developing embryo using chimeric embryo analysis. Therefore, we generated miR302/367 iPS clones from MEFs made from the Rosa26lacZ mouse line which expresses β-galactosidase ubiquitously (Friedrich and Soriano, 1991). Injection of these miR302/367 iPS clones generated high percentage chimeras in more than 50% of the injected embryos (Fig. 4C and data not shown). Most of these chimeras exhibited 80–95% contribution from miR302/367 iPS cells to all tissues examined (Fig. 4C and Supplemental Figure. 3).
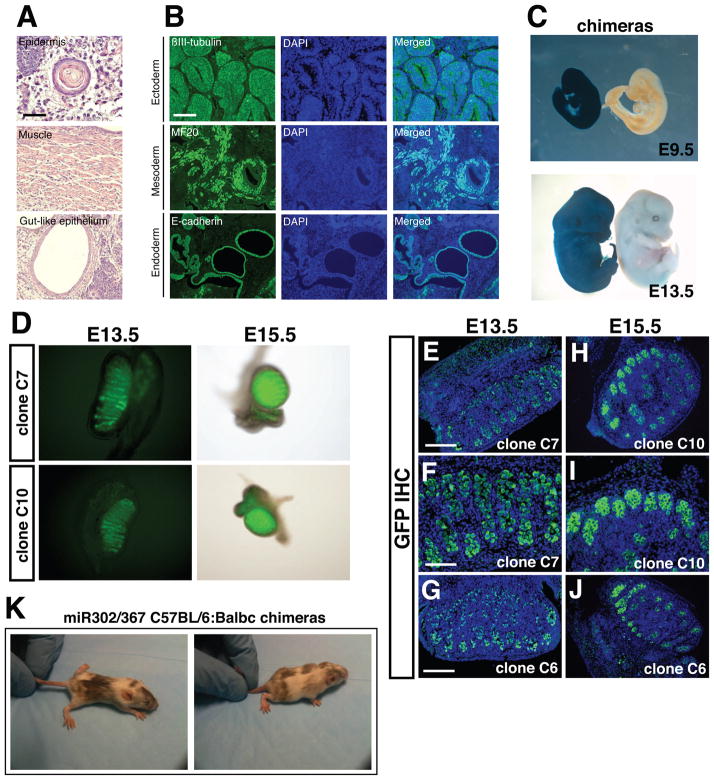
Figure 4. miR302/367 iPS cells can generate derivatives of mesoderm, endoderm, and ectoderm and contributue to the germline of mice.
(A) Hematoxylin and eosin staining of teratomas derived from miR302/367 iPS cell clones showing skin epidermal-like structures, muscle, and gut-like epithelium. These data are representative of five different miR302/367 iPS cell clones which were injected and all produced teratomas. (B) Immunostaining of miR302/367 iPS derived teratoma tissues showing expressing of βIII-tubulin positive neural epithelium, MF20 positive striated muscle, and E-cadherin positive endodermal cells. (C) miR302/367 iPS clones can generate all tissues within the developing embryo as shown by lacZ histochemical staining of high percentage chimeric embryos derived from Rosa26-miR302/367 iPS clones at both E9.5 and E13.5. (D) Both whole mount fluorescence (D) and immunostaining for Oct4-GFP protein expression (E–J) show high-level contribution of miR302/367 iPS cell clones to the germline within the gonads of recipient mice. The data are representative of three clones (C6, C7, C10) which were injected into blastocysts and all three contributed to the germline. (K) miR302/367 iPSCs generated from C57BL/6 MEFs generate high percentage postnatal chimeras as noted by coat color. See also Figure S3. Scale bars: A=100 μm and B, D, G, H, J=150 μm, F and I=100 μm.
To test whether miR302/367 iPS cells could contribute to the germline of mice, we injected three different mouse miR302/367 iPS clones derived from Oct4-GFP MEFs. Mouse gonads were collected at E13.5 and E15.5 and visualized both by whole mount fluorescence and then fixed and sectioned for immunostaining for GFP expression. All three clones contributed efficiently to germ cells in the gonads of chimeric mice (Fig. 4D–J). Moreover, miR302/367 iPS clones generated from C57BL/6 MEFs can generate high percentage postnatal chimeras, although germline transmission has not yet been examined (Fig. 4K). Thus, miR302/367 iPS clones are pluripotent, are competent to generate all three germ layers, and contribute efficiently to the germline of mice. A summary of mouse clones tested for pluripotency is found in Supplemental Table 1.
miR302/367 can reprogram human fibroblasts to a pluripotent state more efficiently than OSKM factors
To assess whether miR302/367 can reprogram human fibroblasts, we transduced human foreskin and dermal fibroblasts with the miR302/367 lentivirus. Within 12–14 days, we observed clones with the classic human ES cell morphology (Fig. 5A). Immunostaining of these clones showed they expressed OCT4, SSEA4, TRA-1-60, and TRA-1-81 (Fig. 5B–E). Q-PCR using three different miR302/367 hiPS cell clones shows that they all express pluripotent markers at levels equivalent to the hES cell line HUES13 (Fig. 5F). We reprogrammed the human foreskin fibroblast cell line BJ and performed DNA fingerprinting to show that clones from miR302/367 reprogramming are derived from the original parental BJ line (Supplemental Figure 4). Moreover, these human clones did not contain any integrants of the OSKM viruses and the miR302/367 virus was silenced in later passages (Supplemental Figures 1 and 2). Interestingly, VPA was not required for reprogramming human fibroblasts and its addition did not affect the efficiency of reprogramming (see below and data not shown). Teratomas were generated from seven different miR302/367 hiPS clones and all exhibited formation of mesoderm, endoderm, and ectoderm (Fig. 5G–L). A summary of human clones tested for pluripotency is found in Supplemental Table 1.
Figure 5. miR302/367 reprograms human fibroblasts to a pluripotent state more efficiently than OSKM factors.
(A–E) Colony morphology and OCT4, SSEA4, TRA-1-60, and TRA-1-81 immunostaining of miR302/367 reprogrammed human fibroblasts. (F) Q-PCR of pluripotent stem cell marker genes in three different miR302/367 reprogrammed human fibroblast lines as compared to the human ES line HUES13. (G–I) Hematoxylin and eosin staining of teratomas derived from miR302/367 human iPS cell clones showing endoderm (gut), mesoderm (muscle), and ectoderm (neural epithelium) like structures. These data represent the results from seven human miR302/367 iPS cell clones. (J–L) Immunostaining of miR302/367 human iPS cell derived teratoma tissues showing expressing of E-cadherin positive endodermal cells, MF20 positive striated muscle, and βIII-tubulin positive neural epithelium. (M) Efficiency of miR302/367 reprogramming in human foreskin fibroblasts by colony counts of clones with human ES like morphology at 18 and 26 days post-viral transduction. Data are the average of three assays ± S.E.M. (N) Q-PCR of pluripotent gene expression in miR302/367 reprogrammed human foreskin fibroblasts at 18 and 26 days post-viral transduction. Data are the average of three assays ± S.E.M. See also Figures S1, S2, and S4. Scale bars: A–E= 50 μm, G–L=150 μm.
We next assessed whether there was an increase in human reprogramming efficiency similar to what we observed in MEFs. Starting with the same number of human foreskin fibroblasts and OSKM and miR302/367 viral titers, the number of colonies with ES like morphology formed at 18 and 26 days after starting viral transduction is two-orders of magnitude greater for miR302/367 than when using OSKM expression (Fig. 5M). Based on the cell counts, approximately 10% of human fibroblasts used for viral transduction produce iPS cell clones (Fig. 5K). Q-PCR from primary induction plates also reveals a dramatic increase in pluripotent gene expression in miR302/367 expressing versus OSKM expressing human foreskin fibroblasts (Fig. 5N). These data indicate that miR302/367 can reprogram human as well as mouse fibroblasts to an iPS cell state with greatly increased efficiency.
miR367 expression is required for miR302/367 iPS reprogramming
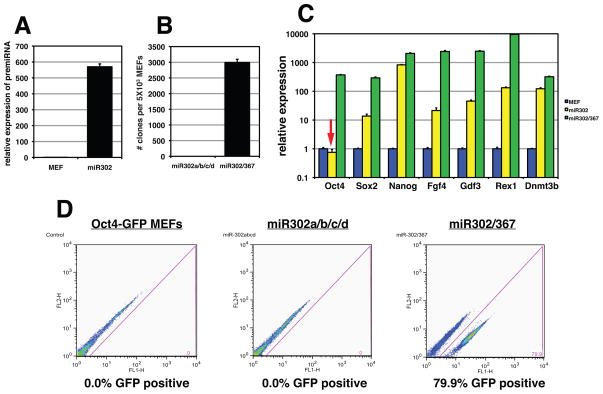
The miR302/367 cluster contains five different miRNAs, miR302a/b/c/d and miR367. All are expressed from a common promoter located in intron 8 of the Larp7 gene (Card et al., 2008). miR302a/b/c/d all share a common seed sequence suggesting that they target a similar set of mRNAs and thus may act redundantly (Fig. 1A). However, miR367 has a different seed sequence and thus may target a different set of mRNAs (Fig. 1A). Therefore, we tested whether miR367 expression is required for miR302/367 iPS cell reprogramming. Using a lentivirus lacking the miR367 sequence, we infected Oct4-GFP MEFs alongside the miR302/367 lentivirus and assessed pluripotent reprogramming by colony counts, Q-PCR and FACS analysis. The miR302a/b/c/d virus lacking miR367 is expressed at high levels in MEFs (Fig. 6A). However, miR302a/b/c/d did not generate any iPS cell colonies when expressed in MEFs at day 10 of reprogramming (Fig. 6B). Continued culture for up to three weeks did not result in formation of any iPS cell colonies from miR302a/b/c/d transduced MEFs (data not shown). Moreover, expression of miR367 alone did not reprogram fibroblasts (data not shown). Q-PCR of primary induction plates eight days after viral transduction shows that several important pluripotent genes were expressed at lower levels in miR302a/b/c/d transduced MEFs versus miR302/367 transduced MEFs (Fig. 6C). Importantly, Oct4 expression is not observed at detectable levels in response to miR302a/b/c/d expression (Fig. 6C, arrow). Using FACS analysis and Oct4-GFP MEFs, we show that there is no induction of Oct4 gene expression when expressing miR302a/b/c/d without miR367 while miR302/367 expression induces robust Oct4-GFP expression by day eight (Fig. 6D). These data show that without miR367 expression, miR302a/b/c/d expression was unable to reprogram mouse MEFs and that this correlated with a lack of induction of Oct4 gene expression. Thus, the coordinated action of the miR302a/b/c/d family along with miR367 is required for iPS cell reprogramming.
Figure 6. miR367 expression is required for miR302/367 iPS cell reprogramming.
(A) The miR302a/b/c/d pre-miRNA is expressed at high levels in transduced MEFs. (B) Number of colonies generated after 10 days of miR302a/b/c/d or miR302/367 expression. Data are the average of four assays ± S.E.M. (C) Pluripotent gene expression from primary induction plates eights days after viral induction of miR302a/b/c/d or miR302/367 viruses. Note lack of Oct4 gene expression in miR302a/b/c/d expressing cells (red arrow). Data are the average of three assays ± S.E.M. (D) FACS analysis of Oct4-GFP MEFs eight days after transduction with either miR302a/b/c/d or miR302/367 viruses.
Low levels of Hdac2 permit miR302/367 reprogramming
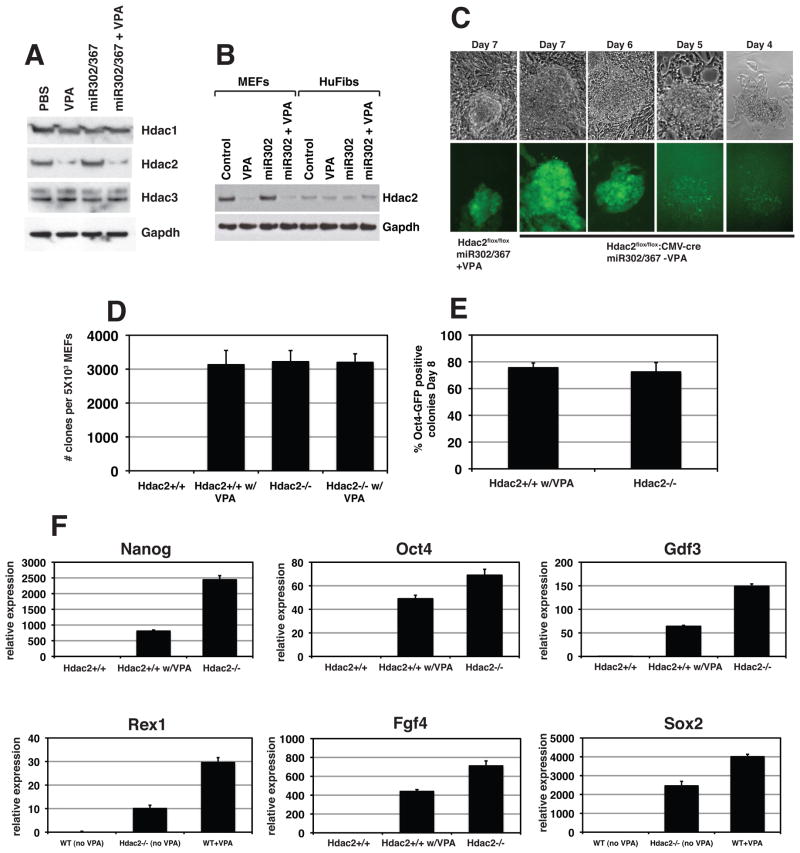
Recent evidence has pointed to an important role for chromatin remodeling factors in regulating the ES cell pluripotent state (Lagarkova et al., 2010; Mali et al., 2010). Previous data has shown that VPA, a known Hdac inhibitor, enhances OSKM reprogramming suggesting an important role for Hdac mediated chromatin remodeling in iPS reprogramming (Huangfu et al., 2008a). We initially found that in the absence of VPA, miR302/367 was unable to efficiently reprogram MEFs to iPS cells and of the few clones that did develop, none survived clonal replating (Fig. 7D and F and data not shown). Interestingly, VPA was not necessary for reprogramming of human foreskin or dermal fibroblasts (Fig. 5). VPA has been reported to specifically degrade Hdac2 protein (Kramer et al., 2003). Therefore, we assessed whether expression of class I Hdacs was altered by miR302/367 or VPA treatment by performing Western blots for Hdac1, 2, and 3 expression during miR302/367 mediated reprogramming. While Hdac1 and Hdac3 expression levels were unchanged in all conditions, VPA caused degradation of Hdac2 protein in MEFs (Fig. 7A). Expression of miR302/367 did not affect the levels of Hdac1, 2, or 3 in the presence or absence of VPA in MEFs (Fig. 7A). In contrast, human foreskin fibroblasts expressed much lower levels of Hdac2 protein and the protein levels of Hdac2 were not affected by VPA in these cells (Fig. 7B). These data suggest that low levels of Hdac2 may significantly enhance or even be required for miR302/367 reprogramming and that human fibroblasts express much lower levels of Hdac2 than MEFs.
Figure 7. VPA specifically degrades Hdac2 protein and suppression of Hdac2 is required for iPS reprogramming by miR302/367.
(A) VPA specifically degrades Hdac2 but not Hdac1 or Hdac3 proteins. Expression of miR302/367 alone did not have any affect on Hdac1, −2, or −3 protein levels. (B) Human foreskin fibroblasts express much lower levels of Hdac2 than MEFs. (C) Hdac2−/− MEFs start to reprogram between six and seven days post viral transduction which is similar to wild-type MEFs treated with VPA. (D) Number of clones generated with Hdac2−/− MEFs in the absence of VPA is similar to Hdac2+/+ MEFs with VPA at eight days post-viral transduction. Hdac2+/+ MEFs without VPA treatment did not generate any viable clones and VPA addition to Hdac2−/− MEFs did not increase the number of clones generated. (E) Percentage of Oct4-GFP positive clones is similar for Hdac2+/+ MEFs with VPA treatment and Hdac2−/− MEFs without VPA treatment at eight days post-viral transduction. (F) Q-PCR for pluripotent stem cell marker genes shows enhanced expression of pluripotency markers at day eight of reprogramming by miR302/367 in wild-type and Hdac2−/− MEFs versus Hdac2+/+ MEFs without VPA treatment. Data are the average of three assays ± S.E.M. See also Figures S5 and S6.
To test whether suppression of Hdac2 is specifically required for efficient reprogramming by miR302/367, we generated Hdac2−/− MEFs from Hdac2flox/flox mice using adenoviral mediated cre excision of Hdac2 and determined whether loss of Hdac2 altered the efficiency of miR302/367 reprogramming of MEFs in the absence of VPA (Supplemental Figure 5). We found that in Hdac2−/− MEFs transduced with the miR302/367 virus, Oct4-GFP positive clones were observed as early as six days post-viral infection (Fig. 7C). Eight days after viral transduction, Hdac2−/− MEFs had formed significant numbers of iPS cell clones in the absence of VPA whereas wild-type MEFs in the absence of VPA did not generate any viable clones (Fig. 7D). VPA addition to Hdac2−/− MEFs did not change the number of iPS cell clones obtained (Fig. 7D). The number of iPS cell clones generated and the percentage of clones that were Oct4-GFP positive with miR302/367 transduced wild-type MEFs plus VPA and miR302/367 transduced Hdac2−/− MEFs lacking VPA were similar (Fig. 7D and E). Loss of Hdac2 expression or VPA addition did not affect proliferation rates in MEFs (Supplemental Figure 6). Q-PCR to assess expression of pluripotency related genes also shows increased reprogramming by miR302/367 in Hdac2−/− MEFs compared to wild-type MEFs without VPA (Fig. 7F). Thus, low levels of Hdac2 or suppression of Hdac2 is required for efficient pluripotent stem cell reprogramming by miR302/367.
DISCUSSION
Current strategies for generating iPS cells rely upon expression of multiple pluripotent stem cell associated transcription factors. We show that a single miRNA cluster, miR302/367, can reprogram fibroblasts more efficiently than the standard OSKM method. With ongoing advances in miRNA biology, these findings may lead to a non-viral, non-transcription factor mediated procedure for generating iPS cells for use not only in basic stem cell biology studies but also for high throughput generation of human iPS clones from large patient populations.
Previous studies have demonstrated the usefulness of iPS cells not only in the study of basic stem cell biology but also in the ability to generate patient specific iPS clones which can then be further differentiated into the cell lineage of choice including hematopoietic, cardiomyocyte, and hepatocyte cell lineages (Moretti et al., 2010a; Moretti et al., 2010b; Raya et al., 2010; Si-Tayeb et al., 2010). However, at this point the low efficiency of iPS reprogramming is an impediment to adapting the process to high throughput approaches. Such approaches would allow for the generation of iPS clones from large patient populations obtained from genome wide association studies for use in characterizing the identified genomic differences at the cell biological level. Our finding that reprogramming by miR302/367 is up to two orders of magnitude more efficient than the OSKM factors suggests that this method may prove to be amenable for use in large scale iPS cell generation. Several other reports have demonstrated that using techniques including Sendai viral expression as well as direct transfection of synthesized mRNAs for the OSKM factors can improve upon the efficiency of iPS reprogramming (Seki et al., 2010; Warren et al., 2010). Based on our data, we obtain efficiencies that are greater than either of these techniques and using human fibroblasts the percent of cells that generate iPS cell clones approaches 10%. Thus, miR302/367 iPS cell reprogramming is more efficient than previously described methods including transfection of synthetic mRNAs for OSKM factors (Warren et al., 2010).
The mechanism underlying the increased efficiency of miR302/367 iPS reprogramming is likely to revolve in part around the nature of miRNA biology. First, miRNA expression does not require protein translation and thus leads to a fast response in protein expression based on inhibition of mRNA translation and stability. Second, miRNAs generally target scores or hundreds of mRNAs that coordinate expression of many different proteins which can rapidly impose a dominant phenotypic change in cell identity. This ability to target many different mRNAs simultaneously also increases the complexity underlying the mechanism of miR302/367 function. miR302/367 collectively targets hundreds of different mRNA targets including those that regulate chromatin remodeling and cell proliferation based on bioinformatic prediction algorithms (Betel et al., 2008; Grimson et al., 2007; Krek et al., 2005). Our data indicate that miR367 expression is essential for iPS cell reprogramming by the miR302/367 cluster. As miR367 has a different seed sequence suggesting a different set of mRNA targets, analysis of the combinatorial regulation of miR302a/b/c/d and miR367 targets may provide important information regarding both the pluripotent gene network and also factors whose expression is required to be suppressed for efficient iPS cell reprogramming.
Our studies underscore the role of Hdac2 in iPS cell reprogramming. The specific degradation of Hdac2 protein by VPA is likely the reason that this small molecule has been found to be more efficacious than other Hdac enzymatic inhibitors in enhancing iPS reprogramming (Huangfu et al., 2008a). Several recent studies have demonstrated the importance of other chromatin remodeling processes in iPS cell reprogramming (Bhutani et al., 2010; Lagarkova et al., 2010; Mali et al., 2010). Hdac2 has also been found to be part of an extended regulatory network for pluripotency in ES cells by interacting with both Oct4 and Myc (Kim et al., 2008). Since iPS cell reprogramming involves the resetting of the epigenetic state of a differentiated cell to a pluripotent “ground state”, additional studies into the necessity of chromatin remodeling will likely lead to better insight into cell lineage trans-differentiation events. Our finding that human cells, which express much lower levels of Hdac2 protein, do not require VPA for miR302/367 mediated reprogramming suggests that differing levels of Hdac2 may account, at least in part, for the different iPS cell reprogramming efficiencies exhibited by different cell lineages. Moreover, Hdac2 expression may decline during development such that adult cells have little Hdac2 protein resulting in an absence of an affect by VPA. Future studies into whether these correlations exist more broadly in other cell lineages may be beneficial for optimizing reprogramming by other methods including the OSKM factors.
Our studies show that miRNAs can be powerful tools for mediating iPS cell reprogramming without the need for pluripotent factors including the OSKM factors. The current focus on developing miRNAs for therapeutic use could lead to a non-viral mediated method of altering miR302/367 expression, which could in turn allow for a rapid miRNA/small molecule approach for iPS cell reprogramming.
MATERIALS AND METHODS
Lentiviral Vector Construction
A mouse genomic DNA fragment comprising of miR302/367 or miR302a/b/c/d family of miRNA was amplified by PCR using primers listed in Supplementary Table 1. The amplified fragment was cloned into Acc65I and XhoI restriction enzyme sites of pENTR1A entry vector (Invitrogen) and verified by sequencing. The fragment was excised from the entry vector and ligated into BsrGI site of pLOVE destination vector (Blelloch et al., 2007) resulting in pLOVE-miR302/367 vector. The pLOVE-miR302a/b/c/d vector was generated in the same fashion but using a different 3′ primer that excluded the miR367 sequence.
Cell Culture, Viral Production and Induction of Pluripotent Stem Cells
Mouse fibroblasts were isolated from Oct4-GFP, Rosa26-LacZ and Hdac2flox/flox embryos at E13.5 and cultured in fibroblast medium as described (Takahashi et al., 2007). Hdac2 was excised by infection of Hdac2flox/flox MEFs with adeno-cre virus. Human dermal fibroblast were cultured in DMEM/F12, 15% FBS, penicillin/streptomycin and L-glutamine. Viral particles were generated by transfection of plated 293T cells with pLOVE vectors encoding miR302/367, Oct4, Sox2, Klf4, or N-myc along with pMD.G and psPAX2 vectors as described (Blelloch et al., 2007). Supernatant from the transfected cells were collected every 24 hr for 48 hrs and titered. The titered viral suspension was mixed with 0.5μl of 10μg/mL polybrene (American Bioanalytical, MA) per milliliter of viral suspension and used to infect fibroblasts. After viral infection, mouse fibroblast were cultured in mouse ES medium supplemented with or without valproic acid at a final concentration of 2mM for the indicated length of time. Infected human fibroblast were culture in human ES medium as described (Huangfu et al., 2008a; Takahashi et al., 2007).
Immunostaining
Clones were washed twice in PBS (with Mg2+ and Ca2+) and fixed in 3.7% formaldehyde. Cells were permeabilized in 0.2% Nonidet P40 (Roche) and blocked in 10% goat serum. Cells were incubated in the following primary antibodies at 4°C overnight: Oct3/4 (Santa Cruz Biotechnology), Sox2 (R &D Systems), Nanog (Abcam), SSEA1 and SSEA4 (Developmental Studies Hybridoma Bank), TRA-1-60 and TRA-1-81 (Millipore, Inc.), and GFP (Clontech). Secondary antibodies are Alexa Fluor 488 and 568 (Invitrogen). The mounting medium used was Vectorshield with DAPI (Vector Laboratories). Alkaline phosphatase histochemical staining was performed using SIGMAFAST Fast Red TR/Naphtol AS-MX tablets following manufacturer’s instructions (Sigma-Aldrich).
RNA isolation, quantitative RT-PCR, and microarray experiments
Total RNA was isolated using Trizol (Invitrogen). Two micrograms of RNA was used to synthesize cDNA using Superscript First Stand Synthesis Kit (Invitrogen). Real time PCR was performed using SYBR Green (Applied Biosystems) by 7900HT Fast Real Time PCR System (Applied Biosystems). Real time primer sequences are listed in Supplemental Table 1. For microarray experiments, the Affymetrix Mouse Gene 1.0 ST arrays were used. Microarray data were analyzed using Robust Multichip Analysis (RMA) and Principal Component Analysis (PCA) and the Partek Genomics Suite v6.5.
Teratoma Formation and DNA fingerprinting analysis
miR302/367 iPS cells were passaged twice on 0.1% gelatin coated plates for an hour to remove feeders. 5 × 105 cells were mixed with Matrigel and injected into each flank of NOD-SCID mice. Tumors were harvested at 4 weeks post-injection, fixed in 4% paraformaldehyde and embedded in paraffin. Sectioned tumors were stained for hematoxylin and eosin. For immunofluoresence staining, the primary antibodies were β-III tubulin (Abcam), MF-20 (Developmental Studies Hybridoma Bank) and E-cadherin (Cell Signaling). Genomic DNA from human miR302/367 iPS cell clones was used for DNA fingerprinting analysis (Cell Line Genetics, LLC, Madison, Wisc.).
Generation of Mouse Chimeras with miR302/367 iPS cell clones
miR302/367 iPS cells were generated using Rosa26-LacZ mouse embryonic fibroblasts (Friedrich and Soriano, 1991). The cells were passaged twice on 0.1% gelatin coated plates for an hour to remove feeders and injected into E3.5 C57BL/6 blastocysts. Embryos were harvested at E9.5 and E13.5 and stained for LacZ activity using previously described methods (Shu et al., 2002). For germline contribution experiments, miR302/367 iPS cell clones C6, C7, and C10, which were generated from Oct4-GFP MEFs, were used for blastocyst injection. Gonads were harvested from E13.5 and E15.5 embryos, visualized by fluorescence microscopy and then fixed and sectioned for GFP immunostaining. Embryos and tissues were embedded in paraffin and sectioned as described (Cohen et al., 2009; Shu et al., 2002). All three clones contributed to the germline.
Western Blots
Total cell lysates were prepared for Western blotting as previously described (Trivedi et al., 2008). Equal amounts of protein were resolved by SDS-PAGE and transferred to polyvinylidenedifluoride membranes. Membranes were incubated with Hdac1 antibody (1:1000 dilution, Cell Signaling), Hdac2 antibody (1:1000 dilution, Invitrogen) or Hdac3 antibody (1:1000 dilution, Sigma). Primary antibody binding was visualized by HRP-conjugated secondary antibody and detected by enhanced chemiluminescence (LumiGlo, Cell Signaling). For loading control, membranes were reprobed with primary antibody against GAPDH (1:2500 dilution, Abcam).
Proliferation assays
Proliferation assays for MEFs were performed using the CellTiter 96 Aqueous One Solution Cell Proliferation kit (Promega, Inc.). 20 μl of CellTiter Reagent, which functions by being incorporated by viable cells into a colorimetric product that can be measured at 490nm, was added to 100 μL of culture medium, incubated at 37°C for and absorbance was measured at 490nm at 1.5 hours, 2.5 hours, and 4.5 hours.
Generation of conditional Hdac2flox/flox mice
The Hdac2flox/flox allele was generated by flanking exon 2 with loxP recombination sites using the targeting vector depicted in Supplemental Figure 5A. Upon cre-mediated recombination, exon 2 is deleted and the resulting mRNA is out of frame with multiple early stop codons producing premature termination and loss of Hdac2 protein. This construct was electroporated into R1 ES cells; correctly targeted ES clones were identified using Southern blot analysis (Supplemental Figure 5B) and used to generate high percentage chimeras and germline transmission of the Hdac2flox//+ allele. Ubiquitous CMV-Cre transgenic mice were used to delete Hdac2 and to demonstrate the resulting loss of Hdac2 protein by Western blot analysis (Supplemental Figure 4C). Hdac2flox/flox mice were crossed with Oct4-GFP knock-in mice (Lengner et al., 2007) to generate Hdac2flox/flox:Oct4-GFP mouse embryonic fibroblasts which were treated with adenovirus expressing cre recombinase to delete Hdac2 for reprogramming experiments.
Supplementary Material
Supplemental Figure 1. The miR302/367 virus is integrated into the genome and miR302/367 iPS cell clones do not contain Oct4, Sox2, Klf4, or Myc expressing viruses. PCR was used on genomic DNA from miR302/367 mouse clones C6, C7, and C10 (A) and K1, K2, and K5 human clones (B) to show presence of the miR302/367 lentivirus and absence of any other reprogramming viruses. Primers used are listed in Supplemental Table 1. Relates to Figures 1, 2, and 5.
Supplemental Figure 2. Expression of the miR302/367 pre-miRNA is extinguished in late passage miR302/367 iPS cell clones. Q-PCR for the pre-miRNA for miR302/367 virally expressed sequences in MEFs, mouse ES line R1, and miR302/367 clones C6, C7, and C10 at passage 3 and 15 (A) and the same in human clones K2, K5, and K7 at passages 3 and 12 (B). Primers used are listed in Supplemental Table 1. Relates to Figures 1, 2, and 5.
Supplemental Figure 3. Contribution of miR302/367 iPS cells to mouse tissues. miR302/367 iPS clones were injected into blastocysts and embryos were harvested at either E9.5 (A) or E13.5 (B) and histochemically stained for β-galactosidase expression. Expression is observed throughout the developing embryo at E9.5 (A). Representative lacZ staining in brain (B), gut (C), liver (D), whisker follicles (E), and eye (F) at E13.5. Relates to Figure 4. Scale bars=150 μm.
Supplemental Figure 4. DNA fingerprinting of human miR302/367 iPS cell clones generated from the human foreskin cell line BJ. Fingerprinting was performed by Cell Line Genetics LLC. Relates to Figure 5.
Supplementary Figure 5. Generation of conditional Hdac2flox/flox allele. (A) Targeting construct to generate conditional Hdac2flox/flox. LoxP sites were introduced upstream and downstream of exon 2. (B) Southern blot confirming proper targeting in 3 ES clones for the floxed Hdac2 allele. (C) Western blot analysis demonstrating complete deletion of Hdac2 in P0 Hdac2flox/flox:CMV-cre hearts. Alpha-tubulin expression was used as a loading control. Relates to Figure 7.
Supplemental Figure 6. Cell proliferation of MEFs is not affected by loss of Hdac2 expression or VPA. Cell titer proliferation assays were performed on wild-type (Oct4-GFP) and Hdac2−/− MEFs in the presence or absence of VPA. Cell proliferation changes were measured as noted in the Methods section. Relates to Figure 7.
Supplemental Table 1. Summary of clones tested for pluripotency
K clones are derived from human BJ foreskin fibroblasts. D clones are derived from dermal fibroblasts.
Acknowledgments
These studies were supported by funding from the NIH to E.E.M. (HL064632, HL087825, and HL100405), J.A.E. (HL071546 and HL100405), C.M.T. (HL098366), and an American Heart Association Jon Holden DeHaan Myogenesis Center Award to E.E.M. and J.A.E.. The authors thank Ken Zaret for the Rosa26-lacZ MEFs and gratefully acknowledge the support of the Penn Institute for Regenerative Medicine during these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R, Venere M, Yen J, Ramalho-Santos M. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell. 2007;1:245–247. doi: 10.1016/j.stem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen ED, Ihida-Stansbury K, Lu MM, Panettieri RA, Jones PL, Morrisey EE. Wnt signaling regulates smooth muscle precursor development in the mouse lung via a tenascin C/PDGFR pathway. J Clin Invest. 2009;119:2538–2549. doi: 10.1172/JCI38079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008a;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008b;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, Brill B, Groner B, Bach I, Heinzel T, et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lagarkova MA, Shutova MV, Bogomazova AN, Vassina EM, Glazov EA, Zhang P, Rizvanov AA, Chestkov IV, Kiselev SL. Induction of pluripotency in human endothelial cells resets epigenetic profile on genome scale. Cell Cycle. 2010;9:937–946. doi: 10.4161/cc.9.5.10869. [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, Scholer HR, Tomilin A, Jaenisch R. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A, Bellin M, Jung CB, Thies TM, Takashima Y, Bernshausen A, Schiemann M, Fischer S, Moosmang S, Smith AG, et al. Mouse and human induced pluripotent stem cells as a source for multipotent Isl1+ cardiovascular progenitors. FASEB J. 2010a;24:700–711. doi: 10.1096/fj.09-139477. [DOI] [PubMed] [Google Scholar]
- Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, Dorn T, Goedel A, Hohnke C, Hofmann F, et al. Patient-Specific Induced Pluripotent Stem-Cell Models for Long-QT Syndrome. N Engl J Med. 2010b doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- Raya A, Rodriguez-Piza I, Navarro S, Richaud-Patin Y, Guenechea G, Sanchez-Danes A, Consiglio A, Bueren J, Izpisua Belmonte JC. A protocol describing the genetic correction of somatic human cells and subsequent generation of iPS cells. Nat Protoc. 2010;5:647–660. doi: 10.1038/nprot.2010.9. [DOI] [PubMed] [Google Scholar]
- Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Seki T, Yuasa S, Oda M, Egashira T, Yae K, Kusumoto D, Nakata H, Tohyama S, Hashimoto H, Kodaira M, et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7:11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Shu W, Jiang YQ, Lu MM, Morrisey EE. Wnt7b regulates mesenchymal proliferation and vascular development in the lung. Development. 2002;129:4831–4842. doi: 10.1242/dev.129.20.4831. [DOI] [PubMed] [Google Scholar]
- Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, Duncan SA. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Trivedi CM, Lu MM, Wang Q, Epstein JA. Transgenic overexpression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J Biol Chem. 2008;283:26484–26489. doi: 10.1074/jbc.M803686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Blelloch R. Cell cycle regulation by MicroRNAs in embryonic stem cells. Cancer Res. 2009;69:4093–4096. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. The miR302/367 virus is integrated into the genome and miR302/367 iPS cell clones do not contain Oct4, Sox2, Klf4, or Myc expressing viruses. PCR was used on genomic DNA from miR302/367 mouse clones C6, C7, and C10 (A) and K1, K2, and K5 human clones (B) to show presence of the miR302/367 lentivirus and absence of any other reprogramming viruses. Primers used are listed in Supplemental Table 1. Relates to Figures 1, 2, and 5.
Supplemental Figure 2. Expression of the miR302/367 pre-miRNA is extinguished in late passage miR302/367 iPS cell clones. Q-PCR for the pre-miRNA for miR302/367 virally expressed sequences in MEFs, mouse ES line R1, and miR302/367 clones C6, C7, and C10 at passage 3 and 15 (A) and the same in human clones K2, K5, and K7 at passages 3 and 12 (B). Primers used are listed in Supplemental Table 1. Relates to Figures 1, 2, and 5.
Supplemental Figure 3. Contribution of miR302/367 iPS cells to mouse tissues. miR302/367 iPS clones were injected into blastocysts and embryos were harvested at either E9.5 (A) or E13.5 (B) and histochemically stained for β-galactosidase expression. Expression is observed throughout the developing embryo at E9.5 (A). Representative lacZ staining in brain (B), gut (C), liver (D), whisker follicles (E), and eye (F) at E13.5. Relates to Figure 4. Scale bars=150 μm.
Supplemental Figure 4. DNA fingerprinting of human miR302/367 iPS cell clones generated from the human foreskin cell line BJ. Fingerprinting was performed by Cell Line Genetics LLC. Relates to Figure 5.
Supplementary Figure 5. Generation of conditional Hdac2flox/flox allele. (A) Targeting construct to generate conditional Hdac2flox/flox. LoxP sites were introduced upstream and downstream of exon 2. (B) Southern blot confirming proper targeting in 3 ES clones for the floxed Hdac2 allele. (C) Western blot analysis demonstrating complete deletion of Hdac2 in P0 Hdac2flox/flox:CMV-cre hearts. Alpha-tubulin expression was used as a loading control. Relates to Figure 7.
Supplemental Figure 6. Cell proliferation of MEFs is not affected by loss of Hdac2 expression or VPA. Cell titer proliferation assays were performed on wild-type (Oct4-GFP) and Hdac2−/− MEFs in the presence or absence of VPA. Cell proliferation changes were measured as noted in the Methods section. Relates to Figure 7.
Supplemental Table 1. Summary of clones tested for pluripotency
K clones are derived from human BJ foreskin fibroblasts. D clones are derived from dermal fibroblasts.