Abstract
We investigated the overlap shared between the immunoglobulin (Ig) proteome of the cerebrospinal fluid (CSF) and the B cell Ig-transcriptome of CSF and the central nervous system (CNS) tissue of three patients with multiple sclerosis. We determined the IgG-proteomes of CSF by mass spectrometry, and compared them to the IgG-transcriptomes from CSF and brain lesions, which were analyzed by cDNA cloning. Characteristic peptides that were identified in the CSF-proteome could also be detected in the transcriptomes of both, brain lesions and CSF, providing evidence for a strong overlap of the IgG repertoires in brain lesions and in the CSF.
Keywords: multiple sclerosis, B cells, cerebrospinal fluid, central nervous system, oligoclonal bands
1. Introduction
The role of B cells and antibodies in the pathogenesis of multiple sclerosis (MS) has become more appreciated (Hafler et al., 2005; Meinl et al., 2006). B cells and their products, antibodies, are detected in all compartments of the central nervous system (CNS): in the parenchyma (Ozawa et al., 1994), in the meninges, where they may form germinal-center-like structures (Serafini et al., 2004), but also in the cerebrospinal fluid (CSF), where oligoclonal B cell expansions and expanded antibody populations, so-called "oligoclonal bands" (OCBs), are observed (Baranzini et al., 1999; Colombo et al., 2000; Monson et al., 2005; Obermeier et al., 2008; Owens et al., 1998, 2009; Qin et al., 1998). We recently showed that the repertoires of IgG-proteins and of B cells in the CSF overlap to a great extent. This provided evidence that the CSF-resident B cells may contribute to the production of OCB-antibodies (Obermeier et al., 2008). However, it is not clear if CSF B cells are the exclusive source of OCB antibodies as we recently learned that the MS CNS harbors a B cell network of clonally related B cells that populate both the CSF and distinct regions of the CNS (Lovato et al., in press).
We here asked the question to what extent the CSF IgG-protein repertoire, i.e. the CSF-proteome, is derived from the B cell receptor repertoires, i.e. the transcriptomes, of CSF and of parenchymal brain lesions. The starting point of this analysis therefore was the analysis of the IgG-protein repertoire from CSF by mass spectrometry. We then compared this repertoire to the IgG-transcriptome repertoires from CSF and CNS tissue, which we determined in parallel experiments by cDNA cloning. These data demonstrate that B cell clones are often shared between the CSF and CNS tissue and that these clones produce immunoglobulins present in the CSF.
2. Materials and Methods
2.1 Clinical samples
CNS tissue was collected at autopsy from two patients with clinically defined MS (MS-4 and MS-B2A) at the Department of Pathology at Brigham and Women’s Hospital. White matter lesions (plaques) were macroscopically identified, dissected, and immediately snap-frozen. Because of initial diagnostic uncertainty a brain biopsy was performed on patient L-296 revealing an inflammatory demyelinating process consistent with MS. The CSF of MS-4 and MS-B2A was removed post-mortem and centrifuged. The CSF of case L-296 was taken during routine diagnostic work-up of the patient. The IgG quotients IgGq=[IgG]CSF×103/[IgG]serum were MS-4: 19.3, MS-B2A: 11.9, and L-296: 8.1. All isolated cell pellets, supernatants, and brain tissue specimens were stored at −80 °C. See Supplementary Methods for clinical details. The study was approved by the human research internal review boards of the Ludwig-Maximilians-University, Munich, the Georg-August-University, Göttingen, and Partner´s Healthcare.
2.2 Analysis of the CSF IgG proteome
IgG antibodies from CSF supernatant were purified as described (Obermeier et al., 2008) with minor modifications: IgG-molecules were deglycosylated after purification by Protein G, we used the OFFGEL Fractionator (Agilent, Böblingen, Germany) for isoelectric focusing, and we further separated IgG-chain by non-reducing SDS-PAGE to separate Heavy and Light chain complexes and free chains. See Supplementary Methods for details. Mass spectrometric analysis of brain-resident IgG was not possible due to insufficient amount of autopsy material. Patient-specific IgG-transcriptomes obtained by cDNA cloning served as databases for the identification of peptide masses using the program MASCOT (Matrix Science, London, UK) (Obermeier et al., 2008).
2.3 Analysis of the IgG-transcriptomes from CSF and brain lesions by cDNA cloning
For patients MS-4 and MS-B2A, B cell IgG variable region libraries were built from tissue sections prepared on a cryostat. RNA was extracted from tissue sections 8- to 15-µm thick using the Absolutely RNA Nanoprep Kit (Stratagene, Cedar Creek, TX) according to the manufacturer’s instructions. From the total RNA, cDNA was synthesized and human IgG variable region genes were amplified as described (Willis et al., 2009). A fraction of the heavy chain data from patient MS-4 is also included in the study of Lovato et al. (Lovato et al., in press). For patient L-296 we isolated total RNA as described (Chomczynski and Sacchi, 1987) and generated IgG-H and -L chain transcriptomes (Obermeier et al., 2008). In addition we used the IgG2-specific primer HG-CH2-aa15-21-rev-in (5’-AGGGCGGCTGTGCTCTCG) in the second round of nested PCR.
2.4 PCR analysis of formaldehyde fixed brain tissue samples
We deparaffinated 80 sections of 10 µm thickness (1.5 mm diameter) using the High Pure FFPE RNA Micro Kit (Roche, Mannheim, Germany). We transcribed RNA into cDNA (Obermeier et al., 2008) and amplified the IgG-chains by two subsequent rounds of semi-nested PCR steps for 40 cycles each of 30 s at 94 °C, 30 s at 53 °C (1st PCR) and 55 °C (2nd PCR) and 30 s at 72 °C. Primer sequences are listed in Supplementary Table 1.
3. Results
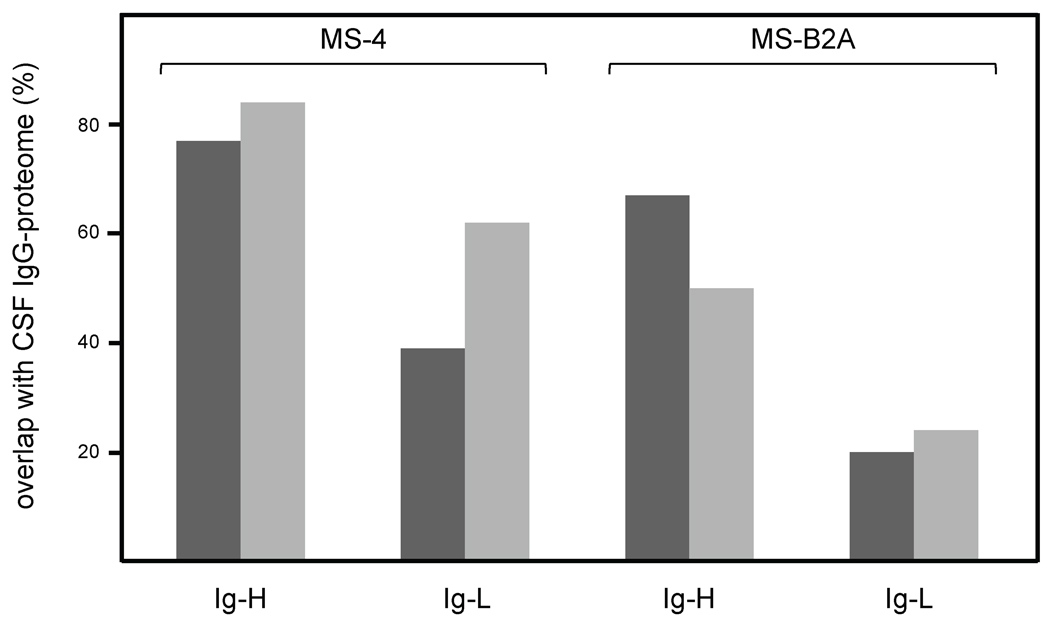
To analyze the IgG-proteome from CSF, we separated OCBs from CSF of three MS patients by non-reducing two dimensional gel-electrophoresis, isolated Heavy and Light IgG-chains (IgG-H and IgG-L) from the gels (Supplementary Fig. 1) and subjected them to MALDI and ESI mass spectrometry. For more than 50% of the peptides we could verify the sequences by tandem mass spectrometry. We then aligned the peptides to patient-specific IgG-transcript databases that were obtained independently from CSF and from CNS lesions by PCR-amplification, cloning, and sequencing of Ig-H-, κ-, and λ-chain transcripts (Willis et al., 2009). All transcripts from patients MS-4, MS-B2A, and L-296 are listed in Supplementary figs. 2, 3, 4. In these lists we highlighted the peptides, which we identified by mass spectrometry. We distinguish two fundamentally different types of peptides: peptides that contain only germline sequences, and "characteristic" peptides, i.e. peptides that contain amino acids, which were introduced by somatic hypermutation or comprise the CDR3 region. The characteristic peptides are therefore unique to each individual patient (Obermeier et al., 2008). Accordingly, we further considered only characteristic peptides. To determine the degree of overlap between the CSF IgG-proteome and the IgG-transcriptomes from CSF and CNS of patients MS-4 and MS-B2A, we counted the CSF- and CNS-transcripts that contained matching characteristic peptides (Fig. 1). We found that CSF IgG-proteomes of both patients covered to high percentages the transcriptomes from both, CSF and CNS. The overlaps were better for the H-chains than for the L-chains. The CSF IgG-proteome of patient MS-4 (Fig. 1, left panel) covered 84% of all identified CSF IgG-H chains and 77% of the CNS IgG-H chains. Of note, we identified 3 different H-chain sequences that were shared by 8 transcripts from both the CSF and distinct CNS lesions, where the entire transcript sequences were completely identical, and where we in addition could identify characteristic peptides. They belong to the V-H chains 1-02/b (two lesions and CSF), 2-05/b (one lesion and CSF), and 3-48/c (two lesions and CSF) (Supplementary Fig. 2A). The analysis for the IgG-L-chains from patient MS-4, and the IgG-H-chains from patient MS-B2A (Fig. 1, right panel), also showed considerable overlap of proteomes and transcriptomes between 39% and 67%. The IgG-L-chain overlap from patient MS-B2A was lower, because most identified peptides contained germline sequences (Supplementary Fig. 3B,C). The overlaps were decreased in patient MS-B2A compared to MS-4, presumably due to its lower IgG-quotient.
Figure 1.
Overlaps of CSF-proteome with the transcriptomes from CNS and CSF of patients MS-4 and MS-B2A. IgG-transcriptomes from CNS lesions and CSF were obtained by cDNA cloning. Proteome data were obtained by analyzing purified IgG antibodies from CSF by mass spectrometry. Repertoires were considered as overlapping when at least one characteristic peptide matched to the corresponding transcript. Characteristic peptides carry somatic hypermutated amino acids, or amino acids introduced by VDJ-recombination. Transcripts fulfilling this condition were attributed to the overlapping populations of CNS lesions (dark bars) and CSF (light bars). Their numbers relative to all detected characteristic peptides are given in percent. Transcripts that were found at different morphologically distinct brain lesions were counted independently. The left panel shows the overlaps of Ig-H and Ig-L chain repertoires from patient MS-4. The right panel shows the overlaps of Ig-H and Ig-L chain repertoires from patient MS-B2A. Here we identified less characteristic peptides, because most of the peptides identified by mass spectrometry derived from germline coded sequences.
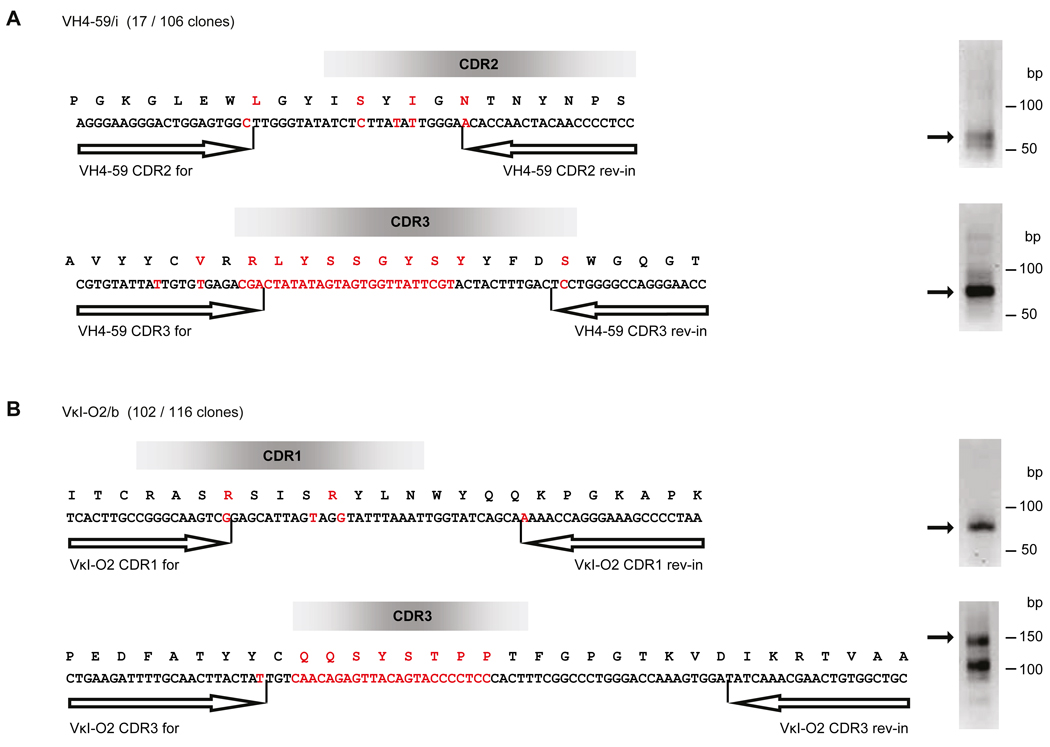
To analyse the CNS sample of patient L-296 we had to modify the protocol, because the biopsy sample was formaldehyde-fixed, which only allows amplification of short PCR-products. Therefore, we aligned the CSF-proteome only to the CSF-transcriptome, and then searched for relevant transcripts in the CNS-specimen by clone-specific PCR. To this end we modified the protocol for CSF-transcript analysis so that we could identify expanded B cell populations (Obermeier et al., 2008). We generated a large database comprising 383 independent IgG-H and IgG-L clones from CSF B cells. Three H-chains (VH4-39/b, VH4-59/i, VH3-74/b), one κ- (VκI-O2/b) and one λ-chain (Vλ3-h/e) were overrepresented (Supplementary Fig. 4). For these chains we designed clone-specific primers to amplify short, characteristic sequence fragments directly from the formaldehyde-fixed brain specimens. For two chains, VH4-59/i and VκI-O2/b, we obtained two different independent PCR products that contained the correct sequences (Fig. 2). This shows that these transcripts were present in both compartments, CSF and CNS lesion.
Figure 2.
Detection of overlapping transcriptome of CSF and brain lesion by clone-specific PCR using a formaldehyde-fixed biopsy sample from patient L-296. We determined clonally expanded B cells from CSF by cDNA cloning and then searched for identical clones in the lesion using clone-specific primers. The putative positions of the CDRs are shown as bars. Nucleotides and the respective amino acids that were exchanged by somatic hypermutation are displayed in red letters. We found identical sequences in CSF and the brain lesion for Heavy chain VH4-59/i (A) and Kappa chain VκI-O2/b (B). In brackets we indicate the degree of clonal expansion: the chain VH4-59/i was identified 17 times out of 106 analyzed IgG1 clones and VκI-O2/b was found 102 times out of 116 analyzed κ-chain clones. Arrows indicate the primer positions of the inner PCR primers. For each chain we amplified two independent sequences at different areas in the chains. The areas were chosen so that the primers ended with nucleotides introduced by somatic hypermutation, and that the amplified sequences contained additional somatically mutated nucleotides. PCR products were separated by gel electrophoresis (right panel), excised from the gels and analyzed by sequencing. All indicated VH4-59/i and VκI-O2/b sequences could be confirmed.
4. Discussion
We found strong overlaps of the CSF IgG-H and Ig-L-chain proteomes with the Ig-transcriptomes from CSF and from brain lesions. This means that the antibody repertoire present in the CSF reflects Ig-transcripts of B cells populating both compartments. Our data therefore confirm our previous study where we showed a strong overlap between CSF-proteome and CSF-transcriptome (Obermeier et al., 2008), providing evidence that CSF-resident B cells produce CSF-resident antibodies. Here we extend this finding, showing that CNS-resident B cells that are shared between the CSF and CNS synthesize antibodies that can be detected in the CSF. Collectively, at least for the three patients studied here, these data provide direct evidence that B cells and IgGs in the CSF to a large extent accurately represent those components present at the site of tissue damage, the CNS lesion. Although we do not have direct experimental evidence for obvious reasons, we speculate that a considerable fraction of the antibodies and B cells found in CSF may have been flushed out or migrated from the brain parenchyma or the meninges.
We identified several clonal variants, which differed only at positions that may be regarded as irrelevant for antigen recognition (i.e. outside the CDR regions). Such clones may therefore be functionally identical (Supplementary Figs. 2 and 3). Such functionally pervasive clones, which may be detected in different brain regions, have recently been demonstrated for T cell populations in multiple sclerosis brains (Junker et al., 2007), and most recently also for B cells (Lovato et al., in press). This indicates that the repertoires of T cells and B cells in different compartments, including the CSF, are structured in a similar manner.
As expected (Baranzini et al., 1999; Colombo et al., 2000; Monson et al., 2005; Obermeier et al., 2008; Owens et al., 1998, 2009; Qin et al., 1998), all transcripts and most peptides show features of antigen-driven responses, because they carry extensive somatic hypermutation. A number of antigens recognized by CSF samples have been described (Kanter et al., 2006; Robinson et al., 2003), but the specific antigens of recombinant antibodies that represented matching H- and L-chains from single cells have so far not been identified (Owens et al., 2009; von Büdingen et al., 2008). Further, specific antibodies directed against CNS antigens (O’Connor et al., 2005; Warren and Catz, 1991) were isolated directly from MS brain tissue. Our study shows that the CSF proteome mirrors to a large extent the IgG-transcriptome of inflamed CNS tissue, at least in the patients studied here. Therefore, analysis of the antigens of CSF derived antibodies provides a direct link to site of tissue pathology in the inflamed brain. Many of the antibodies detected in CSF belong to the IgG family, are activating complement, and contain somatic hypermutations. All these features indicate sustained exposure to specific antigens. CSF may therefore a valuable source for detecting brain-antigens that are directly involved in MS-pathogenesis in near future.
Supplementary Material
Acknowledgments
We thank Ingrid Eiglmeier for expert technical assistance and Edgar Meinl and Gurumoorthy Krishnamoorthy for comments on the manuscript. This work was supported by grants from Deutsche Forschungsgemeinschaft (SFB 571-A1, KD and RH), the German Ministry for Education and Research (BMBF, “German Competence Network Multiple Sclerosis” (KKNMS), RH), NeuroproMiSe (LSHM-CT-2005-018637-WPM5, KD, HW and WB). L.L. was supported by a training research fellowship FISM – Fondazione Italiana Sclerosi Multipla - Cod. 2008/B/3. S.N.W. is supported by a C. J. Martin Postdoctoral Fellowship from the National Health and Medical Research Council of Australia. K.C.O. was supported by a Career Transition Fellowship from the National Multiple Sclerosis Society. This work was also supported by a Jacob Javits Neuroscience Investigator Merit Award (R37 NS024247) to D.A.H.; grants from the US National Institutes of Health (D.A.H., P01AI39671); from the National Multiple Sclerosis Society (D.A.H.; RG2172C9 and RG3308A10)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baranzini SE, Jeong MC, Butunoi C, Murray RS, Bernard CCA, Oksenberg JR. B cell repertoire diversity and clonal expansion in multiple sclerosis brain lesions. Journal of Immunology. 1999;163:5133–5144. [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Analytical Biochemistry. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Colombo M, Dono M, Gazzola P, Roncella S, Valetto A, Chiorazzi N, Mancardi GL, Ferrarini M. Accumulation of clonally related B lymphocytes in the cerebrospinal fluid of multiple sclerosis patients. Journal of Immunology. 2000;164:2782–2789. doi: 10.4049/jimmunol.164.5.2782. [DOI] [PubMed] [Google Scholar]
- Hafler DA, Slavik JM, Anderson DE, O'Connor KC, De Jager P, Baecher-Allan C. Multiple sclerosis. Immunological Reviews. 2005;204:208–231. doi: 10.1111/j.0105-2896.2005.00240.x. [DOI] [PubMed] [Google Scholar]
- Junker A, Ivanidze J, Malotka J, Eiglmeier I, Lassmann H, Wekerle H, Meinl E, Hohlfeld R, Dornmair K. Multiple sclerosis: T-cell receptor expression in distinct brain regions. Brain. 2007;130(11):2789–2799. doi: 10.1093/brain/awm214. [DOI] [PubMed] [Google Scholar]
- Kanter JL, Narayana S, Ho PP, Catz I, Warren KG, Sobel RA, Steinman L, Robinson WH. Lipid microarrays identify key mediators of autoimmune brain inflammation. Nature Medicine. 2006;12(1):138–143. doi: 10.1038/nm1344. [DOI] [PubMed] [Google Scholar]
- Lovato L, Willis SN, Rodig SJ, Caron T, Almendinger SE, Howell O, Reynolds R, O’Connor KC, Hafler DA. Related B cell clones populate the Meninges and Parenchyma of Patients with Multiple Sclerosis. Brain. 2010 doi: 10.1093/brain/awq350. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinl E, Krumbholz M, Hohlfeld R. B lineage cells in the inflammatory CNS environment: Migration, maintenance, local antibody production and therapeutic modulation. Annals of Neurology. 2006;59(6):880–892. doi: 10.1002/ana.20890. [DOI] [PubMed] [Google Scholar]
- Monson NL, Brezinschek H-P, Brezinschek R, Mobley A, Vaughan GK, Frohman EM, Racke MK, Lipsky PE. Receptor revision and atypical mutational characteristics in clonally expanded B cells from the cerebrospinal fluid of recently diagnosed multiple sclerosis patients. Journal of Neuroimmunology. 2005;158:170–181. doi: 10.1016/j.jneuroim.2004.04.022. [DOI] [PubMed] [Google Scholar]
- O'Connor KC, Appel H, Bregoli L, Call ME, Catz I, Chan JA, Moore NH, Warren KG, Wong SJ, Hafler DA, Wucherpfennig KW. Antibodies from inflamed central nervous system tissue recognize myelin oligodendrocyte glycoprotein. Journal of Immunology. 2005;175(3):1974–1982. doi: 10.4049/jimmunol.175.3.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermeier B, Mentele R, Malotka J, Kellermann J, Wekerle H, Lottspeich F, Hohlfeld R, Dornmair K. Matching of oligoclonal Ig transcriptomes and proteomes of cerebrospinal fluid in multiple sclerosis. Nature Medicine. 2008;14(6):688–693. doi: 10.1038/nm1714. [DOI] [PubMed] [Google Scholar]
- Owens GP, Kraus H, Burgoon MP, Smith-Jensen T, Devlin ME, Gilden DH. Restricted use of VH4 germline segments in an acute multiple sclerosis brain. Annals of Neurology. 1998;43(2):236–243. doi: 10.1002/ana.410430214. [DOI] [PubMed] [Google Scholar]
- Owens GP, Bennett JL, Lassmann H, O'Connor KC, Ritchie AM, Shearer A, Lam C, Yu XL, Birlea M, DuPree C, Williamson RA, Hafler DA, Burgoon MP, Gilden D. Antibodies produced by clonally expanded plasma cells in multiple sclerosis CSF. Annals of Neurology. 2009;65(6):639–649. doi: 10.1002/ana.21641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa K, Suchanek G, Breitschopf H, Brück W, Budka H, Jellinger K, Lassmann H. Patterns of oligodendroglia pathology in multiple sclerosis. Brain. 1994;117:1311–1322. doi: 10.1093/brain/117.6.1311. [DOI] [PubMed] [Google Scholar]
- Qin Y, Duquette P, Zhang Y, Poole R, Antel JP. Clonal expansion and somatic hypermutation of VH genes of B cells from cerebrospinal fluid in multiple sclerosis. Journal of Clinical Investigation. 1998;102(5):1045–1050. doi: 10.1172/JCI3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WH, Fontoura P, Lee BJ, De Vegvar HEN, Tom J, Pedotti R, DiGennaro CD, Mitchell DJ, Fong D, Ho PPK, Ruiz PJ, Maverakis E, Stevens DB, Bernard CCA, Martin R, Kuchroo VK, Van Noort JM, Genain CP, Amor S, Olsson T, Utz PJ, Garren H, Steinman L. Protein microarrays guide tolerizing DNA vaccine treatment of autoimmune encephalomyelitis. Nature Biotechnology. 2003;21(9):1033–1039. doi: 10.1038/nbt859. [DOI] [PubMed] [Google Scholar]
- Serafini B, Rosicarelli B, Magliozzi R, Stigliano E, Aloisi F. Detection of ectopic B-cell follicles with germinal centers in the meninges of patients with secondary progressive multiple sclerosis. Brain Pathology. 2004;14(2):164–144. doi: 10.1111/j.1750-3639.2004.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Büdingen H-C, Harrer MD, Kuenzle S, Meier M, Goebels N. Clonally expanded plasma cells in the cerebrospinal fluid of MS patients produce myelin-specific antibodies. European Journal of Immunology. 2008;38(7):2014–2023. doi: 10.1002/eji.200737784. [DOI] [PubMed] [Google Scholar]
- Warren KG, Catz I. Purification of autoantibodies to myelin basic protein by antigen specific affinity chromatography from cerebrospinal fluid IgG of multiple sclerosis patients. Immunoreactivity studies with human myelin basic protein. Journal of Neurological Sciences. 1991;103:90–96. doi: 10.1016/0022-510x(91)90289-j. [DOI] [PubMed] [Google Scholar]
- Willis SN, Stadelmann C, Rodig SJ, Caron T, Gattenlöhner S, Mallozzi SS, Roughan JE, Allmendinger SE, Blewett MM, Brück W, Hafler DA, O'Connor KC. Epstein-Barr virus infection is not a characteristic feature of multiple sclerosis brain. Brain. 2009;132(12):3318–3328. doi: 10.1093/brain/awp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.