Abstract
Pandemic outbreaks of influenza are caused by the emergence of a pathogenic and transmissible virus to which the human population is immunologically naïve. Recent outbreaks of highly pathogenic avian influenza (HPAI) of the H5N1 subtype are of particular concern because of the high mortality rate (60% case fatality rate) and novel subtype. In order to develop a vaccine that elicits broadly reactive antibody responses against emerging H5N1 isolates, we utilized a novel antigen design technique termed computationally optimized broadly reactive antigen (COBRA). The COBRA HA sequence was based upon HA amino acid sequences from clade 2 H5N1 human infections and the expressed protein retained the ability to bind the receptor, as well as mediate particle fusion. Non-infectious recombinant VLP vaccines using the COBRA HA were purified from a mammalian expression system. Mice and ferrets vaccinated with COBRA HA H5N1 VLPs had protective levels of HAI antibodies to a representative isolates from each subclade of clade 2. Furthermore, VLP vaccinated animals were completely protected from a lethal challenge of the clade 2.2 H5N1 virus A/Mongolia/Whooper Swan/244/2005. This is the first report describing the use of COBRA-based antigen design. The COBRA HA H5N1 VLP vaccine elicited broadly reactive antibodies and is an effective influenza vaccine against HPAI virus.
Introduction
The swine-origin H1N1 pandemic of 2009 reminded the worldwide community of the ever-present threat of pandemic influenza. Pandemic outbreaks of influenza are caused by the emergence and spread of a pathogenic and transmissible virus to which the human population is immunologically naïve [1]. Although predicting an emerging pandemic subtype of influenza is difficult, outbreaks of highly pathogenic avian influenza of the H5N1 subtype are of particular concern because of the high mortality rate (60% case fatality rate) and novel subtype [2]. To date, H5N1 influenza has not transmitted efficiently from person to person, but accumulation of mutations or reassortment with a human transmissible virus could result in a highly transmissible H5N1 virus [3]. H5N1 and contemporary H3N2 seasonal influenza viruses are able to generate stable reassortant viruses although the pathogenic potential of the reassortant viruses is less than that of the highly pathogenic H5N1 virus [4, 5]. Additionally, a recent report has demonstrated that not only can H1N1 pandemic viruses and highly pathogenic H5N1 viruses efficiently reassort, but these reassortants can replicate to higher titers than the source H5N1 virus [6]. The genetic compatibility between the influenza viruses, combined with the continued spread of both novel H1N1 in humans and highly pathogenic H5N1 in wild birds, highlights the potential for a new emerging H5N1 influenza infecting the human population and therefore, the need to develop effective vaccines against H5N1 isolates.
One of the challenges to developing effective H5N1 vaccines is the antigenic diversity within the subtype. H5N1 viruses are separated into distinct clades based upon phylogenetic distance among the hemagglutinin (HA) genes [7]. The clades are geographically diverse and are evolving under unique pressures specific to each respective location [8]. The majority of human infections were identified within the antigenically distinct clades 1 and 2, with clade 2 infections spanning over 60 countries and moving westward from Asia into Africa and the Middle East [9]. Genetic diversity within clade 2 has resulted in distinct subclades including 2.1, 2.2, 2.3, 2.4 and 2.5 with some subclades being further divided into additional sub-subclades [7]. Despite high levels of HA protein sequence homology between clades (>90%), there is little receptor blocking antibody cross-reactivity across clades and even within subclades [7]. Developing vaccines that are able to overcome the challenge of H5N1 antigenic diversity is a crucial step in pandemic preparedness.
The antigenic diversity of all subtypes of influenza is a challenge to influenza vaccine development in general. The current seasonal influenza vaccine uses a polyvalent formulation to address the issue of multiple subtypes simultaneously circulating in the human population. Even though a representative vaccine strain is selected and is expected to represent the most common strain for each subtype in a given season, vaccine escape occurs and yearly epidemics continue to happen. A recent report has demonstrated that a polyvalent H5N1 vaccine with components derived from various clades can elicit cross-clade antibody cross-reactivity and protective efficacy [10]. Alternative strategies that have been investigated for addressing the challenge of antigenic diversity include targeting conserved viral proteins, such as the M2 ion channel or nucleoprotein (NP) and targeting conserved domains of HA [11–14]. Additionally, engineering synthetic antigens that capture common immune epitopes from a population of primary viruses has the potential to overcome antigenic diversity.
Consensus-based H5N1 have been generated for several influenza proteins including HA, neuraminidase (NA) and matrix (M1) and each has elicited cross-reactive immune responses [15–18]. Consensus sequences are traditionally generated by aligning a population of sequences and selecting the most common residue at each position. These sequences are expected to effectively capture conserved linear epitopes and elicit cross-reactive cellular immune responses [15, 19–21]. Furthermore, consensus-based H5N1 HA immunogens expressed from DNA plasmids elicit broad antibody responses [16, 18]. However, consensus-based antigen design is inherently influenced by the input sequences used to generate the synthetic molecule and as such is subject to sampling bias. Using the NCBI Influenza Virus Resource database, the vast majority of HA sequences from both human and environmental isolates are from clades 1 and 2 [22]. Furthermore, within clade 2 the majority of human isolates are from infections in Indonesia and the subclade 2.1. The predominance of certain isolates, i.e. a majority of clade 2.1 isolates from Indonesia, can bias the output consensus sequence generated and may not accurately reflect the genetic diversity of H5N1 influenza viruses. In order to overcome these limitations, we report a new methodology of antigen design using multiple rounds of consensus generation termed computationally optimized broadly reactive antigen (COBRA). This method was designed to address the diversity specifically within clade 2 and utilized global surveillance efforts to generate a vaccine with the potential to elicit increased breadth of antibody responses within this antigenically diverse clade.
In this study, COBRA HA antigens are expressed on virus-like particles (VLPs) and purified as vaccine immunogens. VLPs are self-assembling, nonpathogenic, genomeless particles that are similar in size and morphology to intact virions [23]. VLPs can be produced in a variety of eukaryotic expression systems including yeast, insect and mammalian cells. Importantly, A VLP-based vaccine which protects against human papillomavirus has been approved for human use [24]. Influenza virus VLP vaccines are attractive alternatives to traditional split vaccines because they do not require the use of any live virus at any step of the vaccine production process [25]. Additionally, VLPs present surface antigens in their native oligomeric structures which are important for maintaining conformational epitopes..
We hypothesized that the COBRA HA delivered in the context of a VLP will elicit an antibody response that demonstrates increased breadth when compared to that of a VLP containing an HA molecule derived from a primary isolate. Here, we report the COBRA HA is a functional molecule, elicits broadly reactive antibody responses, and completely protects mice and ferrets from a heterologous, highly pathogenic H5N1 virus challenge.
Materials and Methods
Antigen construction and synthesis
Influenza A HA nucleotide sequences isolated from human H5N1 infections were downloaded from the NCBI Influenza Virus Resource database (see supporting materials for complete list of accession numbers and isolate descriptions) [22]. Nucleotide sequences were translated into protein sequences using the standard genetic code. Full length sequences from H5N1 clade 2 human infections from 2004 to 2006 were acquired and used for subsequent consensus generations. For each round of consensus generation, multiple alignment analysis was applied and the consensus sequence was generated using AlignX (Vector NTI). The final amino acid sequence, termed computationally optimized broadly reactive antigen (COBRA), was reverse translated and optimized for expression in mammalian cells, including codon usage and RNA optimization (GeneArt; Regensburg, Germany). This construct was then synthesized and inserted into the pTR600 expression vector.
In vitro expression
Human embryonic kidney (HEK) 293T cells (1×106) were transiently transfected with 3 µg DNA expressing COBRA. Cells were incubated for 72 h at 37°C, supernatants were removed, the cells were lysed with 1% Triton-X 100 and cell lysates were collected. Cell lysates were electrophoresed on a 10% SDS-PAGE gel and transferred to a PVDF membrane. The blot was probed with mouse polyclonal antisera pooled from mice infected with 6:2 reassortant H5N1 viruses with the surface glycoproteins derived from either A/Vietnam/1203/2004 or A/Whooper Swan/244/2005 and the HA-antibody complexes were detected using a goat anti-mouse IgG conjugated to horse radish peroxidase (HRP) (Southern Biotech; Birmingham, AL, USA). HRP was detected by chemiluminescent substrate (Pierce Biotechnology; Rockford IL, USA) and exposed to X-ray film (ThermoFisher; Pittsburgh, PA, USA).
Functional characterization
To determine receptor binding characteristics, virus-like particles (VLPs) containing COBRA HA proteins were purified from the supernatants of mammalian cell lines. HEK 293T cells were transiently transfected with plasmids expressing HIV Gag, COBRA HA and neuraminidase (NA, A/Thailand/1(KAN-1)/2004) and incubated for 72 h at 37°C. Supernatants were collected and VLPs were purified via ultracentrifugation (100,000 × g through 20% glycerol, weight per volume) for 4 h at 4°C. The pellets were subsequently resuspended in phosphate buffered saline PBS, pH 7.2 and stored at −80°C until use. Protein concentration was determined by Micro BCA™ Protein Assay Reagent Kit (Pierce Biotechnology, Rockford, IL, USA). COBRA HA VLPs were prepared in various amounts as measured by total protein and each individual preparation was two-fold serially diluted in v-bottom microtiter plates. An equal volume of either 1% turkey or 1% horse erythrocytes (RBC) (Lampire; Pipersville, PA, USA) in PBS was added to the diluted VLPs and incubated for 30–60 minutes at room temperature. The HA titer was determined by the reciprocal dilution of the last well which contained agglutinated RBC.
To determine endosomal fusion characteristics, COBRA-pseudotyped lentiviral vectors expressing a luciferase reporter gene were produced as described [26]. Briefly, 293T cells were co-transfected by using the following plasmids: 7µg of pCMVdeltaR8.2, 7µg of pHR CMV-Luc, 3µg pCMV/R N1(Kan-1) (all kindly provided by Dr. G. Nabel) and 3ug pTR600 COBRA. Cells were transiently transfected and incubated for 72h at 37°C. Supernatants were harvested, centrifuged to clear cell debris, filtered through a 0.22 µm syringe filter, aliquotted, and used immediately or frozen at −80°C. For fusion assays, 100 ul of pseudoviruses were added to confluent MDCK cells in 48-well plates (~30,000 cells per well). Plates were incubated at room temperature for 30 min, washed, and fresh medium added. Forty-eight hours after infection, cells were lysed in mammalian cell lysis buffer (Promega; Madison, WI, USA). A standard quantity of cell lysate was used in a luciferase assay with luciferase assay reagent (Promega; Madison, WI, USA) according to the manufacturer’s protocol.
Vaccine Preparation
HEK 293T cells were transiently transfected with plasmids expressing M1 (A/Puerto Rico/8/1934, optimized for expression in mammalian cells), NA (A/Thailand/1(KAN-1)/2004, optimized for expression in mammalian cells) and COBRA HA and incubated for 72h at 37°C. Supernatants were collected and cell debris removed by low speed centrifugation followed by vacuum filtration through a 0.22 µm sterile filter. VLPs were purified via ultracentrifugation (100,000 × g through 20% glycerol, weight per volume) for 4 h at 4C. The pellets were subsequently resuspended in PBS pH 7.2 and stored in single use aliquots at −80C until use. Total protein concentration was determined by Micro BCA™ Protein Assay Reagent Kit (Pierce Biotechnology, Rockford, IL, USA).
HA specific content was determined by western blot and densitometry. Purified recombinant COBRA HA and purified VLPs were prepared in standard total protein amounts and were electrophoresed on a 10% SDS-PAGE gel and transferred to a PVDF membrane. The blot was probed with mouse polyclonal antisera pooled from mice infected with 6:2 reassortant H5N1 viruses with the surface glycoproteins derived from either A/Vietnam/1203/2004 or A/Whooper Swan/244/2005 and the HA-antibody complexes were detected using a goat anti-mouse IgG conjugated to horse radish peroxidase (HRP) (Southern Biotech; Birmingham, AL, USA). HRP was detected by chemiluminescent substrate (Pierce Biotechnology; Rockford IL, USA) and exposed to X-ray film (ThermoFisher; Pittsburgh, PA, USA). Density of bands was determined using ImageJ software (NIH) [27]. Density of recombinant HA bands were used to calculate a standard curve and the density of the purified VLPs was interpolated using the results from the recombinant HA. Experiments were performed in triplicate and multiple exposure times were analyzed for all iterations.
Mouse studies
BALB/c mice (Mus musculis, females, 6–8 weeks) were purchased from Harlan Sprague Dawley, (Indianapolis, IN, USA) and housed in microisolator units and allowed free access to food and water and were cared for under USDA guidelines for laboratory animals. For dosing studies, mice (5 mice per group) were vaccinated with one of two doses of purified VLPs (1.5µg or 0.3µg), based upon HA content from the densitometry assay, via intramuscular injection at week 0 and then boosted with the same dose at week 3. For comparison studies, mice (20 mice per group) were vaccinated with purified VLPs (3µg), based upon HA content from the densitometry assay, via intramuscular injection at week 0 and then boosted with the same dose at week 3. Vaccines at each dose were formulated with Imject® alum adjuvant (Imject® Alum, Pierce Biotechnology; Rockford, IL, USA) according to the manufacturer’s protocol or vehicle alone. Fourteen to twenty-one days after each vaccination, blood was collected from anesthetized mice via the retro-orbital plexus and transferred to a microfuge tube. Tubes were centrifuged and sera was removed and frozen at −20 ± 5°C.
Three weeks after final vaccination, mice were challenged intranasally with 5×103 plaque forming units (PFU) of the highly pathogenic H5N1 virus A/Whooper Swan/Mongolia/244/2005 (Clade 2.2) in a volume of 50µl. The challenge dose represents approximately 50LD50 in mice. After infection, mice were monitored daily for weight loss, disease signs and death for 14 days after infection. Individual body weights, sickness scores and death were recorded for each group on each day after inoculation. Sickness score was determined by evaluating activity (0=normal, 1=reduced, 2=severely reduced), hunched back (0=absent, 1=present) and ruffled fur (0=absent, 1=present) [28]. Experimental endpoint was defined as >20% weight loss or display of neurological disease such as hind limb paralysis. All H5N1 influenza virus studies were performed under high-containment biosafety level 3 enhanced conditions (BSL3+). All procedures were in accordance with the NRC Guide for the Care and Use of Laboratory Animals, the Animal Welfare Act, and the CDC/NIH Biosafety in Microbiological and Biomedical Laboratories.
Ferret studies
Fitch ferrets (Mustela putorius furo, female, 6–12-months of age), influenza naïve and descented, were purchased from Marshall Farms (Sayre, PA, USA). Ferrets were pair housed in stainless steel cages (Shor-line, Kansas City, KS, USA) containing Sani-chips Laboratory Animal Bedding (P.J. Murphy Forest Products, Montville, NJ, USA). Ferrets were provided with Teklad Global Ferret Diet (Harlan Teklad, Madison, WI, USA) and fresh water ad libitum. The VLPs were diluted in PBS, pH 7.2 to achieve final concentration. Ferrets (n=9) were vaccinated with 15|µg of purified VLPs, based upon HA content as determined by densitometry assay, via intramuscular injection in the quadriceps muscle in a volume of 0.50 ml at week 0 and then boosted with the same dose at week 3. Vaccines were stored at −80°C prior to use and formulated with Imject® alum adjuvant (Imject® Alum; Pierce Biotechnology, Rockford, IL, USA) immediately prior to use according to manufacturer’s protocol. Animals were monitored for adverse events including weight loss, temperature, loss of activity, nasal discharge, sneezing and diarrhea weekly during the vaccination regimen. Prior to vaccination, animals were confirmed by HAI assay to be seronegative for circulating influenza A (H1N1 and H3N2) and influenza B viruses. Fourteen to twenty-one days after each vaccination, blood was collected from anesthetized ferrets via the anterior vena cava and transferred to a microfuge tube. Tubes were centrifuged and sera was removed and frozen at −20 ± 5°C.
Three weeks after final vaccination, ferrets were challenged intranasally with 1×106 plaque forming units (PFU) of the highly pathogenic H5N1 virus A/Whooper Swan/Mongolia/244/2005 (Clade 2.2) in a volume of 0.5 ml in each nostril for a total infection volume of 1 ml. After infection, ferrets were monitored daily for weight loss, disease signs and death for 14 days after infection. Individual body weights, sickness scores, and death were recorded for each group on each day after inoculation. Sickness score was determined by evaluating activity (0=normal, 1=alert and active with stimulation, 2=alert but not active after stimulation, 3=not alert or active after stimulation), nasal discharge (0=absent, 1=present), sneezing (0=absent, 1=present), decreased food intake (0=absent, 1=present), diarrhea (0=absent, 1=present), dyspnea (0=absent, 1=present) and neurological symptoms (0=absent, 1=present). Nasal washes were performed by instilling 3 ml of PBS into the nares of anesthetized ferrets each day for 14 days after inoculation. Washes were collected and stored at −80°C until use. Experimental endpoint was defined as >20% weight loss, development of neurological symptoms, or an activity score of 3 (not active or alert after stimulation). All H5N1 influenza virus studies were performed under high-containment biosafety level 3 enhanced conditions (BSL3+). All procedures were in accordance with the NRC Guide for the Care and Use of Laboratory Animals, the Animal Welfare Act, and the CDC/NIH Biosafety in Microbiological and Biomedical Laboratories.
ELISA assay
The ELISA assay was used to assess total antibody titer and IgG isotype titer to the HA. High binding, 96-well polystyrene plates (Costar; Lowell, MA, USA) were coated overnight with 50ng/well of recombinant HA. Coating antigens were derived from the following representative viral isolates: A/Vietnam/1203/2004 (clade 1), A/Indonesia/5/2005 (clade 2.1), A/Whooper Swan/244/2005 (clade 2.2) and A/Anhui/1/2005 (calde 2.3). Plates were blocked with 5% milk diluted in PBS with 0.05% Tween 20. Serum samples were diluted in blocking buffer and added to plates. Serum was two-fold serially diluted and allowed to incubate for 1 hour at room temperature. Plates were washed and species specific antibody against IgG, IgG1, IgG2a, IgG2b or IgG3 and linked to horseradish peroxidase (HRP) (Southern Biotech; Birmingham, AL, USA) were diluted in blocking buffer and added to plates. Plates were incubated for 1 hour at room temperature. Plates were washed and HRP was developed with TMB substrate (Sigma-Aldrich; St. Louis, MO, USA). Plates were incubated in the dark for 15 minutes and then the reaction was stopped with 2N H2SO4. Optical densities at a wavelength of 450nm (OD450) were read by a spectrophotometer (BioTek; Winooski, VT, USA) and end point dilution titers were determined. End point titers were determined as the reciprocal dilution of the last well which had an OD450 above the mean OD450 plus two standard deviations of naïve animal sera.
Hemagglutination inhibition (HAI) assay
The HAI assay was used to assess functional antibodies to the HA able to inhibit agglutination of horse erythrocytes. The protocol was adapted from the CDC laboratory-based influenza surveillance manual [29]. To inactivate non-specific inhibitors, sera were treated with receptor destroying enzyme (RDE; Denka Seiken, Co., Japan) prior to being tested [30–34]. Briefly, three parts RDE was added to one part sera and incubated overnight at 37°C. RDE was inactivated by incubation at 56°C for ~30 min. RDE treated sera was two-fold serially diluted in v-bottom microtiter plates. An equal volume of reassortant virus, adjusted to approximately 8 HAU/50µl, was added to each well. The reassortant viruses contained the internal genes from the mouse adapted strain A/Puerto Rico/8/1934 and the surface proteins HA and NA from the following representative viral isolates: A/Vietnam/1203/2004 (clade 1), A/Indonesia/5/2005 (clade 2.1), A/Whooper Swan/244/2005 (clade 2.2) and A/Anhui/1/2005 (clade 2.3). The plates were covered and incubated at room temperature for 20 min followed by the addition of 1% horse erythrocytes (HRBC) (Lampire Biologicals, Pipersville, PA, USA) in PBS. Red blood cells were stored at 4°C and used within 72 h of preparation. The plates were mixed by agitation, covered, and the RBCs were allowed to settle for 1 h at room temperature [35]. The HAI titer was determined by the reciprocal dilution of the last well which contained non-agglutinated RBC. Positive and negative serum controls were included for each plate. All mice were negative (HAI ≤1:10) for pre-existing antibodies to currently circulating human influenza viruses prior to vaccination.
Plaque Assay
Madin-Darby Canine Kidney (MDCK) cells were plated (5 × 105) in each well of a 6-well plate. Samples were diluted (final dilution factors of 100 to 10−6) and overlayed onto the cells in 100µl of DMEM supplemented with penicillin-streptomycin and incubated for 1hr. Samples were removed, cells were washed twice and media was replaced with 2 ml of L15 medium plus 0.8% agarose (Cambrex; East Rutherford, NJ, USA) and incubated for 72 h at 37°C with 5% CO2. Agarose was removed and discarded. Cells were fixed with 10% buffered formalin, and then stained with 1% crystal violet for 15 min. Following thorough washing in dH2O to remove excess crystal violet, plates were allowed to dry, plaques counted, and the plaque forming units (PFU)/ml were calculated.
Statistical Analysis
Statistical significance of the antibody data was determined using a two-way analysis of variance (ANOVA) with Bonferroni’s post-test to analyze differences between each vaccine group for the different test antigens (multiparametric). Differences in weight loss, sickness score, and viral titers were analyzed by two-way ANOVA, followed by Bonferroni's post test for each vaccine group at multiple time points. Significance was defined as p < 0.05. Statistical analyses were done using GraphPad Prism software.
Results
Computationally optimized broadly reactive antigen design
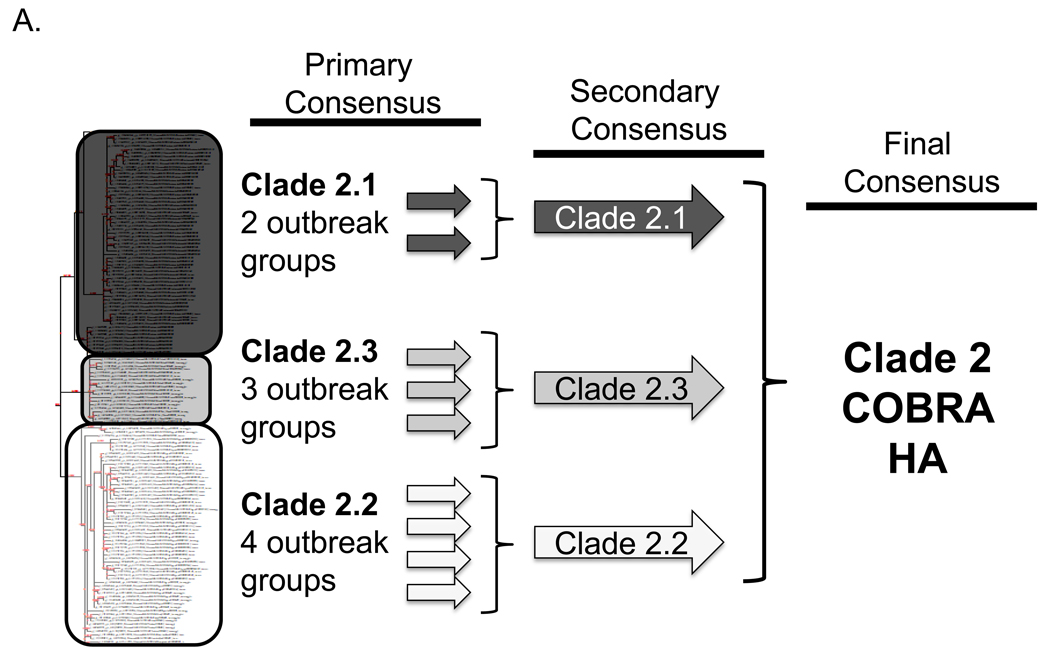
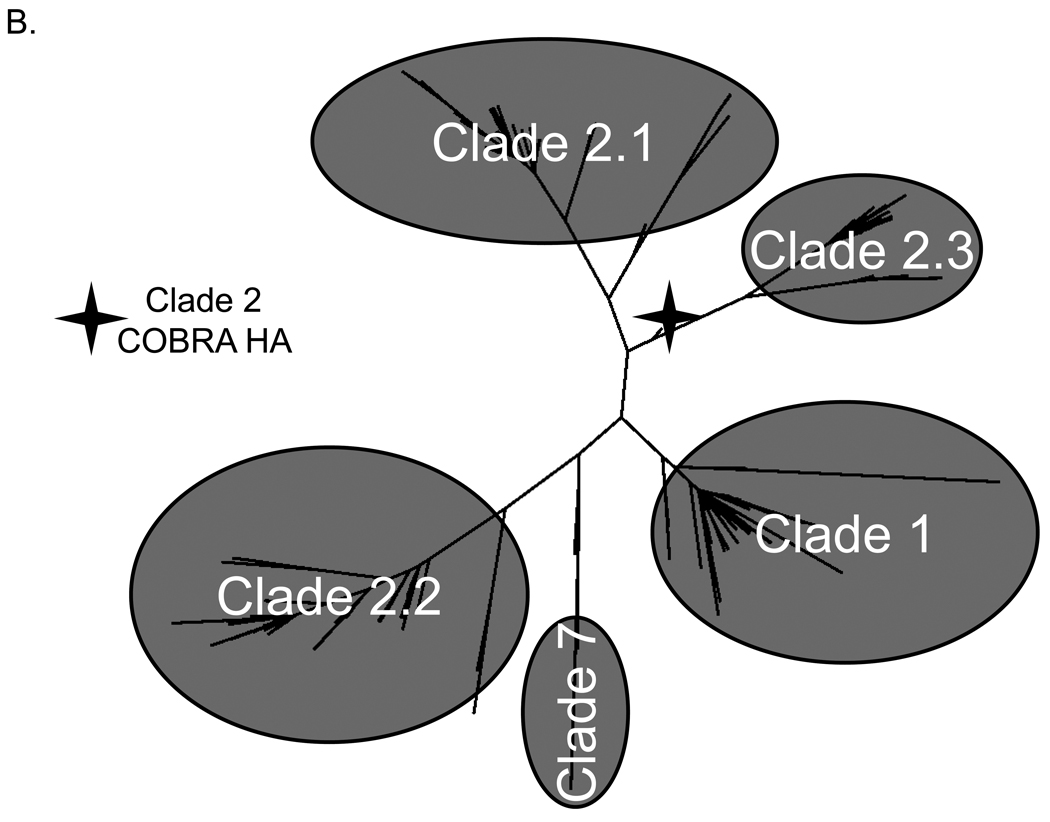
To address the challenge of antigenic diversity present in H5N1 influenza, we designed a computationally optimized broadly reactive antigen (COBRA). For the first step of antigen generation, 129 unique hemagglutinin (HA) sequences were downloaded from the NCBI Influenza Virus Resource (IVR) sequence database [22] representing clade 2 H5N1 viruses isolated from human infections between 2004 and 2006. The sequences were first grouped into phylogenetic subclades and then further divided into individual outbreak groups within each subclade based upon geographic location and time of isolation. HA amino acid sequences for each individual outbreak group were aligned and the most common amino acid at each position was determined resulting in primary consensus sequences representing each outbreak group within each subclade (FIGURE 1A). Primary consensus sequences within each subclade were then aligned and the most common amino acid was chosen resulting in secondary consensus sequences representing each subclade (FIGURE 1A). The secondary consensus sequences were aligned and the most common amino acid at each position was selected resulting in the final consensus sequence referred to as clade 2 COBRA HA (FIGURE 1A). Phylogenetic analysis of the COBRA HA with all human isolates of H5N1 HA proteins indicated that COBRA retained a clade 2-like sequence without being grouped specifically within any clade 2 subclade cluster (FIGURE 1B). Furthermore, a BLAST search using the COBRA HA sequence revealed that it is a unique sequence that has not been isolated from the environment (data not shown).
Figure 1. COBRA HA Design.
Schematic to describe how the COBRA HA molecule was designed (A). The phylogenetic tree was inferred from hemagglutinin amino acid sequences using the maximum likelihood method and clade/sub-clade groupings were identified. Primary consensus sequences were generated for each outbreak group. Secondary consensus sequences were then generated for each subclade using the primary sequences as input. The secondary consensus sequences were then aligned and the resulting consensus, designated COBRA, was generated. Phylogenetic analysis of the COBRA HA (B). The unrooted phylogenetic tree was inferred from hemagglutinin amino acid sequences from human H5N1 infections isolated from 2004 to 2009 and the clade/sub-clade groupings are indicated. The star represents the COBRA HA sequence relative to human H5N1 infections.
Characterization of COBRA
Since COBRA is a fully synthetic protein, the retention of natural hemagglutinin function was confirmed. Initially, COBRA expression was verified by transient transfection of mammalian cells. Analysis of the total cell lysate demonstrated that the COBRA HA migrates at its predicted molecular weight of approximately 73kDa (FIGURE 2A). Because the poly-basic cleavage site was retained in the COBRA HA sequence, both HA0 and the HA1 subunits were detected by immunoblot at similar molecular weights as recombinant HA and the HA on the H5N1 virion (FIGURE 2A). Virus-like particles (VLPs) with COBRA HA on the surface bound sialic acid in a dose-dependent manner and this binding was specific to COBRA, since empty lentiviral core alone did not bind to the receptor (FIGURE 2B).
Figure 2. COBRA HA Functional Characterization.
COBRA HA was translated in vitro and the cell culture lysates were analyzed by SDS-PAGE (A). Lane designations: 1) H5N1 recombinant HA; 2) COBRA HA; 3) Expression vector; 4) H5N1 reassortant virus. The COBRA HA (lane 2) migrates at its expected molecular weight confirming expression of the synthetic protein. COBRA HA VLPs were prepared in various amounts, serially diluted, and incubated with 1% erythrocytes to evaluate receptor binding (B). HA titer was determined as the last well in which the RBCs remained suspended in a lattice structure. COBRA HA and control lentiviral pseudoparticles packaging a CMV-Luc gene were generated in HEK 293T cells and used to infect MDCK cells with or without trypsin (C). Particle fusion was determined by luciferace production by infected cells.
To determine if the COBRA HA was functional, the protein was pseudotyped onto lentiviral Gagp24 to generate pseudoparticles [36, 37]. COBRA HA containing pseudoparticles mediated cell fusion as efficiently as H5N1 control pseudoparticles without the requirement for trypsin. In contrast, H1N1 pseudoparticles did require trypsin and pseudoparticles without surface HA produced luciferace at similar levels as the cell only controls (FIGURE 2C). Taken together, these results demonstrate that although the COBRA HA is a synthetic protein not found in nature, it retains all of the functions of a natural hemagglutinin protein.
Mouse Dosing Immunizations
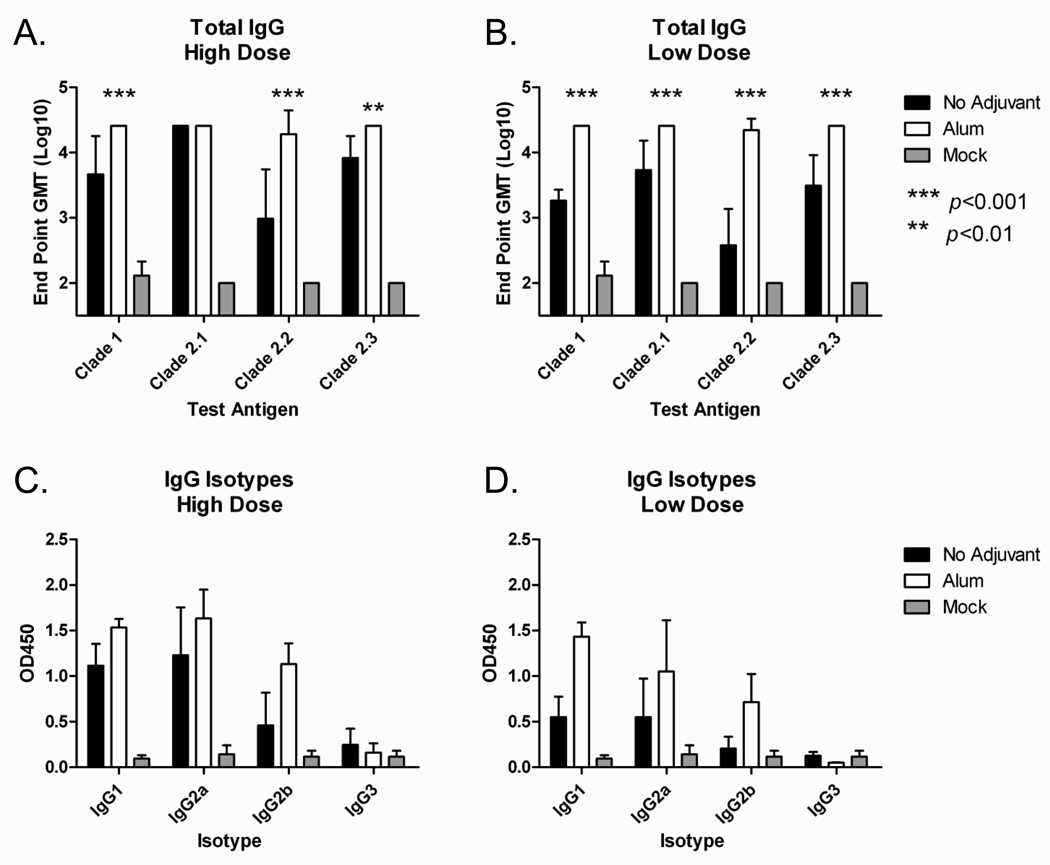
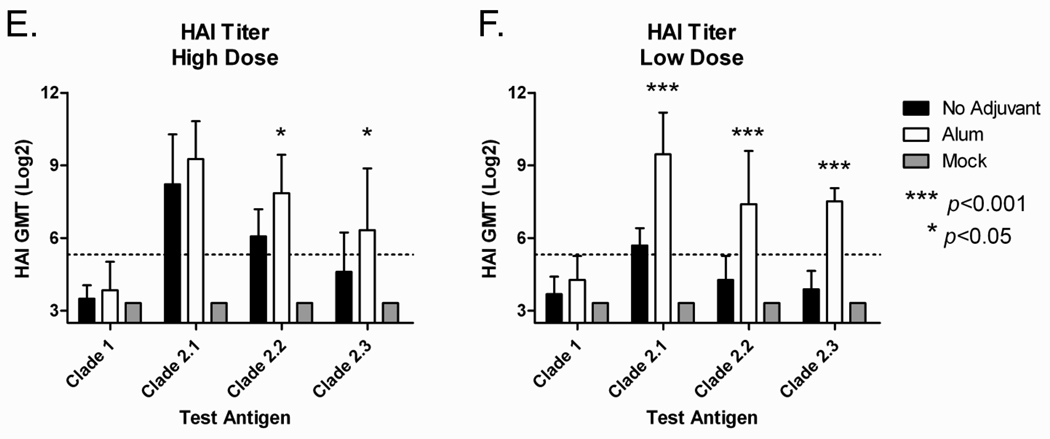
Mice (BALB/c; n=5) were vaccinated (week 0 and 3) via intramuscular injection with purified COBRA VLPs at either a high dose (1.5ug HA) or low dose (0.3ug HA) with and without Imject® alum adjuvant. At week 5, all COBRA VLP-vaccinated animals had anti-HA antibodies that recognized heterologous recombinant HA derived from both clade 1 and also subclades of clade 2 (FIGURE 3A and 3B). Imject® alum significantly increased anti-HA antibody titers in both low and high dose groups as compared to the non-adjuvanted groups (p<0.01). The IgG isotype subclasses elicited by the VLP vaccines against a clade 2.1 coating antigen consisted mainly of IgG1 and IgG2a, indicating a mixed T helper response (FIGURE 3C and 3D). Similar results were found for additional coating antigens representing clade 1, clade 2.2 and clade 2.3 (data not shown). In addition to recognizing HA, antibodies were also evaluated for the ability to block virus from binding its receptor via inhibition of viral-induced agglutination of horse erythrocytes (HAI). All mice administered Imject® alum adjuvanted vaccines, regardless of dose, had HAI titers ≥1:40 to viruses expressing HA from clades 2.1 and 2.2 and 90% of the mice had titers ≥1:40 to a clade 2.3 representative virus (FIGURE 3E and 3F). Non-adjuvanted vaccines elicited generally lower HAI antibody titers with 100% of high dose animals achieving titers ≥1:40 only against clade 2.1 viruses. Imject® alum adjuvanted vaccines elicited significantly higher HAI antibody titers to clade 2.2 and clade 2.3 viruses regardless of dose as compared to non-adjuvanted vaccines (p<0.05 for high dose and p<0.001 for low dose, respectively). None of the vaccines elicited high HAI titer antibodies to a clade 1 virus.
Figure 3. COBRA HA Mouse Dosing Immunogenicity.
BALB/c mice (n=5/group) were vaccinated at 0 and 3 weeks with blood collected at 14 to 21 days after each vaccination. Vaccines were formulated at high (1.5ug HA), and low (0.03ug HA) doses; with and without Imject® alum and delivered intramuscularly. Total IgG at week 5 was determined via ELISA for each vaccine group (A and B). Values represent the geometric mean titer (±95% confidence interval) of log10 transformed endpoint titers. IgG isotypes were evaluated via ELISA for each vaccine group (C and D). Values represent the mean OD450 of a 1:200 dilution of serum. Hemagglutination inhibition (HAI) serum antibody titer for each vaccine group was determined at week 5 using representative reassortant viruses (E and F). Values represent the geometric mean titer (±95% confidence interval) of log2 transformed titers. The dotted line represents the 1:40 titer. Significant differences were determined by two-way ANOVA with Bonferroni’s post-test to evaluate differences between the vaccine formulations for each test antigen. A p value of less than 0.05 was considered significant.
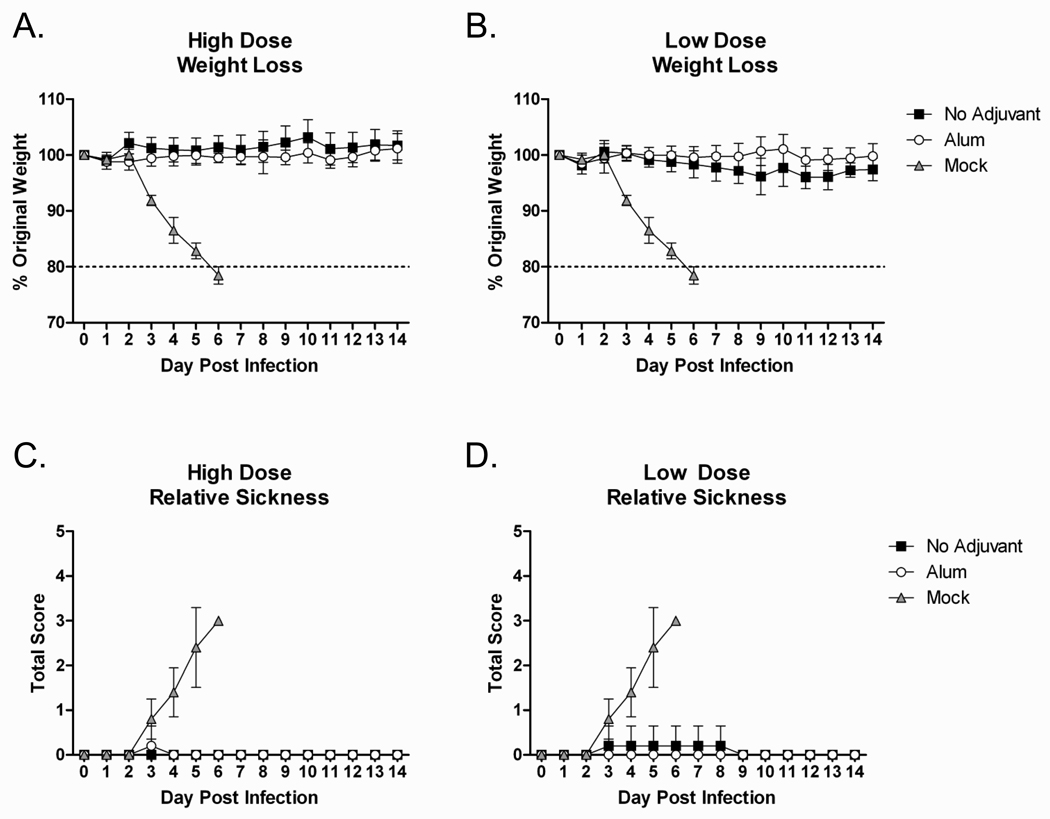
Mouse Dosing Challenge
Mice that received the COBRA VLP vaccines or mock vaccinated control mice were challenged intranasally with a lethal dose of clade 2.2 H5N1 highly pathogenic avian influenza (A/Mongolia/whooper swan/244/2005) to evaluate the protective efficacy of the different COBRA vaccine formulations. All COBRA vaccinated mice, regardless of dose or the presence of adjuvant, were protected from weight loss and death following lethal challenge, while all mock vaccinated animals rapidly lost weight and required euthanasia by day 6 post infection (FIGURE 4A and 4B). Additionally, COBRA VLP vaccinated mice had no signs of disease, while mock vaccinated animals developed such symptoms as ruffled fur, hunched back, and lethargy (FIGURE 4C and 4D).
Figure 4. COBRA HA Mouse Dosing Efficacy.
BALB/c mice (n=5/group) were vaccinated with COBRA HA VLPs with or without adjuvant. Mice were infected with 5×103 PFU of the highly pathogenic clade 2.2 H5N1 virus A/Whooper Swan/Mongolia/244/2005. Mice were followed to monitor weight loss (A and B) and sickness (C and D). Sickness score was determined by evaluating activity (0=normal, 1=reduced, 2=severely reduced), hunched back (0=absent, 1=present) and ruffled fur (0=absent, 1=present). All mock vaccinated mice reached the experimental endpoint and required euthanasia by 6 days post infection.
Mouse Comparison Immunizations
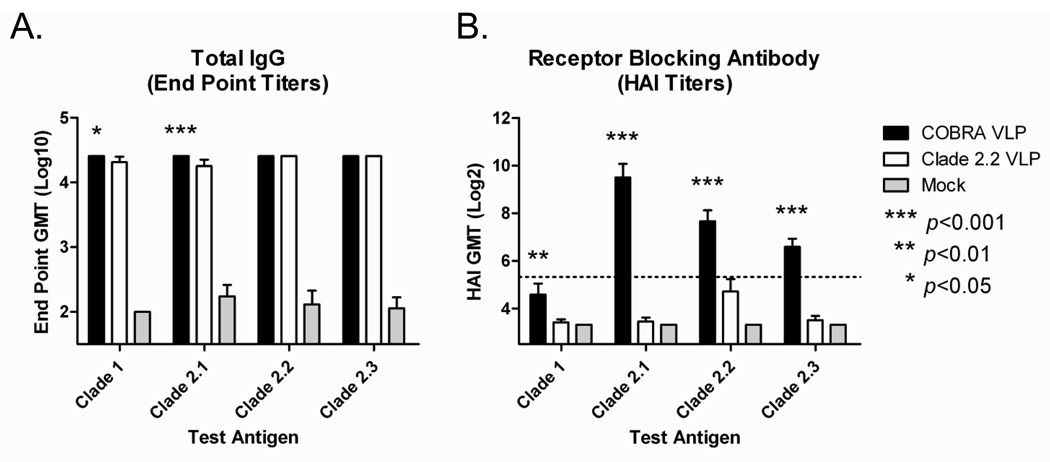
To determine if the COBRA HA vaccine elicits a broader antibody response compared to a vaccine derived from a primary isolate, an additional set of mice were vaccinated with either COBRA VLPs or clade 2.2 (A/Mongolia/whooper swan/244/2005) VLPs. Mice (BALB/c; n=20) were vaccinated (week 0 and 3) via intramuscular injection with either COBRA VLPs or clade 2.2 VLPs at a high dose (3ug HA) with Imject® alum adjuvant. At week 5, all COBRA VLP-vaccinated mice and all clade 2.2 VLP-vaccinated mice had anti-HA antibodies that recognized heterologous recombinant HA derived from both clade 1 and various subclades of clade 2 (FIGURE 5A). Although no significant differences were found in total IgG titers between vaccine groups, COBRA VLP-vaccinated animals had higher HAI antibody titers against all viruses tested as compared to clade 2.2 VLP-vaccinated animals (p<0.01; FIGURE 5B). Furthermore, COBRA VLP-vaccinated animals had an increased frequency of HAI titers of ≥1:40 compared to clade 2.2 VLP-vaccinated animals (TABLE 1).
Figure 5. Mouse Comparison Immunogenicity.
BALB/c mice (n=20/group) were vaccinated at 0 and 3 weeks with blood collected at 14 to 21 days after each vaccination. Vaccines were formulated at a high dose (3ug HA) with Imject® alum and delivered intramuscularly. Total IgG at week 5 was determined via ELISA for each vaccine group (A). Values represent the geometric mean titer (±95% confidence interval) of log10 transformed endpoint titers. Hemagglutination inhibition (HAI) serum antibody titer for each vaccine group was determined at week 5 using representative reassortant viruses (B). Values represent the geometric mean titer (±95% confidence interval) of log2 transformed titers. The dotted line represents the 1:40 titer. Significant differences were determined by two-way ANOVA with Bonferroni’s post-test to evaluate differences between the vaccine formulations for each test antigen. A p value of less than 0.05 was considered significant.
Table 1. Mouse seroconversion frequency.
Percentage of VLP-vaccinated animals achieving an HAI titer of ≥1:40 to each test antigen.
| Vaccine Antigen | Clade 1a | Clade 2.1b | Clade 2.2c | Clade 2.3d |
|---|---|---|---|---|
| COBRA | 45% (9/20) |
100% (20/20) |
100% (20/20) |
100% (20/20) |
| Clade 2.2c | 0% (0/20) |
0% (0/20) |
50% (10/20) |
0% (0/20) |
A/Vietnam/1203/2004
A/Indonesia/5/2005
A/Whooper Swan/Mongolia/244/2005
A/Anhui/1/2005
Mouse Comparison Challenge
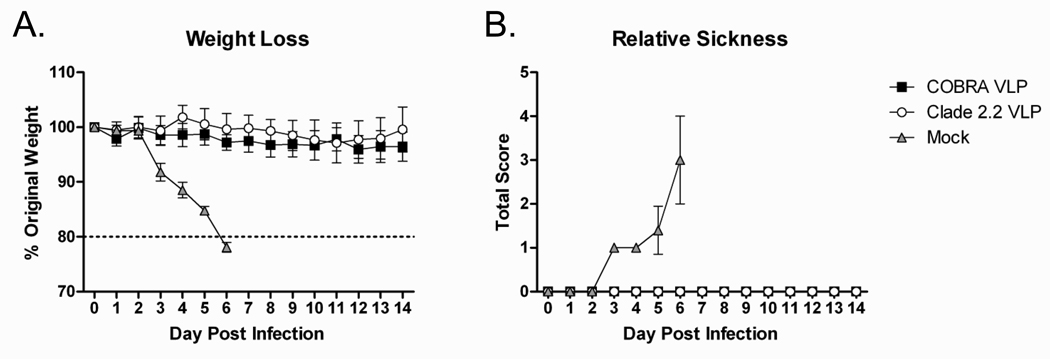
Mice that received the COBRA VLP vaccine, clade 2.2 VLP vaccine or mock vaccinated control mice were challenged intranasally with a lethal dose of clade 2.2 H5N1 highly pathogenic avian influenza (A/Mongolia/whooper swan/244/2005) to evaluate the protective efficacy of the VLP vaccines. All VLP-vaccinated mice were protected from weight loss and death following lethal challenge while all mock vaccinated animals rapidly lost weight and required euthanasia by day 6 post infection (FIGURE 6A). Additionally, VLP vaccinated mice did not show signs of disease, while mock vaccinated animals developed ruffled fur, hunched back, and lethargy (FIGURE 6B). Even though the clade 2.2 VLP was matched to the challenge virus, no significant differences were found between COBRA VLP and clade 2.2 VLP vaccinated mice in any of the parameters analyzed indicating that the COBRA VLP vaccine protected animals as efficiently as the homologous vaccine.
Figure 6. Mouse Comparison Efficacy.
BALB/c mice (n=20/group) were vaccinated with VLPs with adjuvant. Mice were infected with 5×103 PFU of the highly pathogenic clade 2.2 H5N1 virus A/Whooper Swan/Mongolia/244/2005. Mice were followed to monitor weight loss (A) and sickness (B). Sickness score was determined by evaluating activity (0=normal, 1=reduced, 2=severely reduced), hunched back (0=absent, 1=present) and ruffled fur (0=absent, 1=present). All mock (adjuvant-only) vaccinated mice reached the experimental endpoint and required euthanasia by 6 days post infection.
Ferret Comparison Immunizations
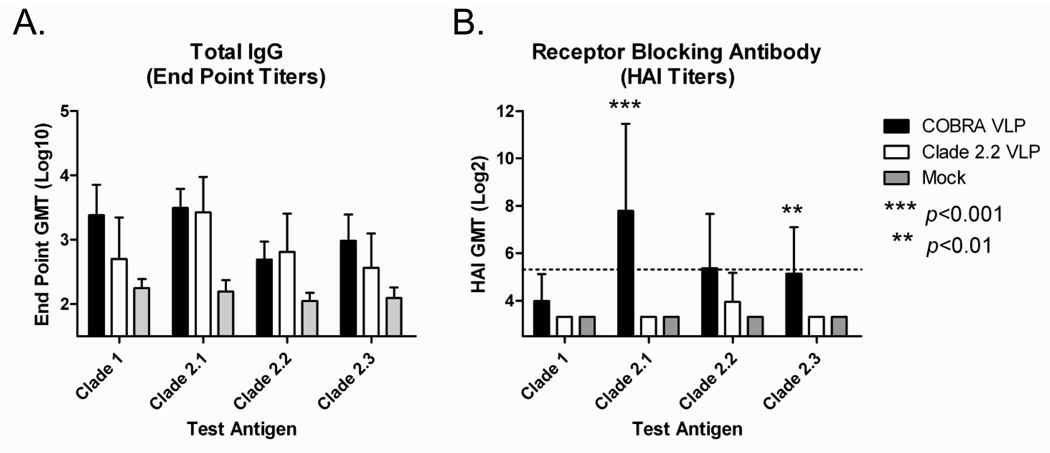
Ferrets are the most relevant model for influenza disease and as such the COBRA vaccine was tested in this more rigorous animal model. Ferrets (Fitch; n=9) were vaccinated (week 0 and 3) via intramuscular injection with COBRA VLPs or clade 2.2 VLPs at a high dose (15ug HA) with Imject® alum adjuvant. Serum was collected from ferrets at week 5 and antibody responses to the COBRA vaccines were evaluated. All vaccinated ferrets had anti-HA antibodies that recognized heterologous recombinant HA derived from both clade 1 and also subclades of clade 2 (FIGURE 7A). No significant difference in anti-HA antibody was found between the COBRA VLP vaccine and the clade 2.2 VLP vaccine for any of the antigens tested (p>0.05). In addition to recognizing HA, antibodies were also evaluated for HAI activity. COBRA VLP-vaccinated animals had higher HAI antibody titers against clade 2.1 and clade 2.3 viruses as compared to clade 2.2 VLP-vaccinated animals (p<0.01 FIGURE 7B). Similar to the mice, COBRA VLP-vaccinated ferrets displayed an increased rate of achieving HAI titers ≥1:40 when compared to clade 2.2 VLP-vaccinated ferrets (TABLE 2).
Figure 7. Ferret Immunogenicity.
Ferrets (n=9/group) were vaccinated with VLPs (15ug HA) with Imject® alum at weeks 0 and 3 and serum collected at week 5. Total IgG at week 5 was determined via ELISA for each vaccine group (A). Values represent the geometric mean titer (±95% confidence interval) of log10 transformed endpoint titers. Hemagglutination inhibition (HAI) serum antibody titer for each vaccine group was determined at week 5 using representative reassortant viruses (B). Values represent the geometric mean titer (±95% confidence interval) of log2 transformed titers. The dotted line represents the 1:40 titer. Significant differences were determined by two-way ANOVA with Bonferroni’s post-test to evaluate differences between the vaccine formulations for each test antigen. A p value of less than 0.05 was considered significant.
Table 2. Ferret seroconversion frequency.
Percentage of VLP-vaccinated animals achieving an HAI titer of ≥1:40 to each test antigen.
| Vaccine Antigen | Clade 1a | Clade 2.1b | Clade 2.2c | Clade 2.3d |
|---|---|---|---|---|
| COBRA | 0% (0/9) |
78% (7/9) |
56% (5/9) |
56% (5/9) |
| Clade 2.2c | 0% (0/9) |
0% (0/9) |
22% (2/9) |
0% (0/9) |
A/Vietnam/1203/2004
A/Indonesia/5/2005
A/Whooper Swan/Mongolia/244/2005
A/Anhui/1/2005
Ferret Comparison Challenge
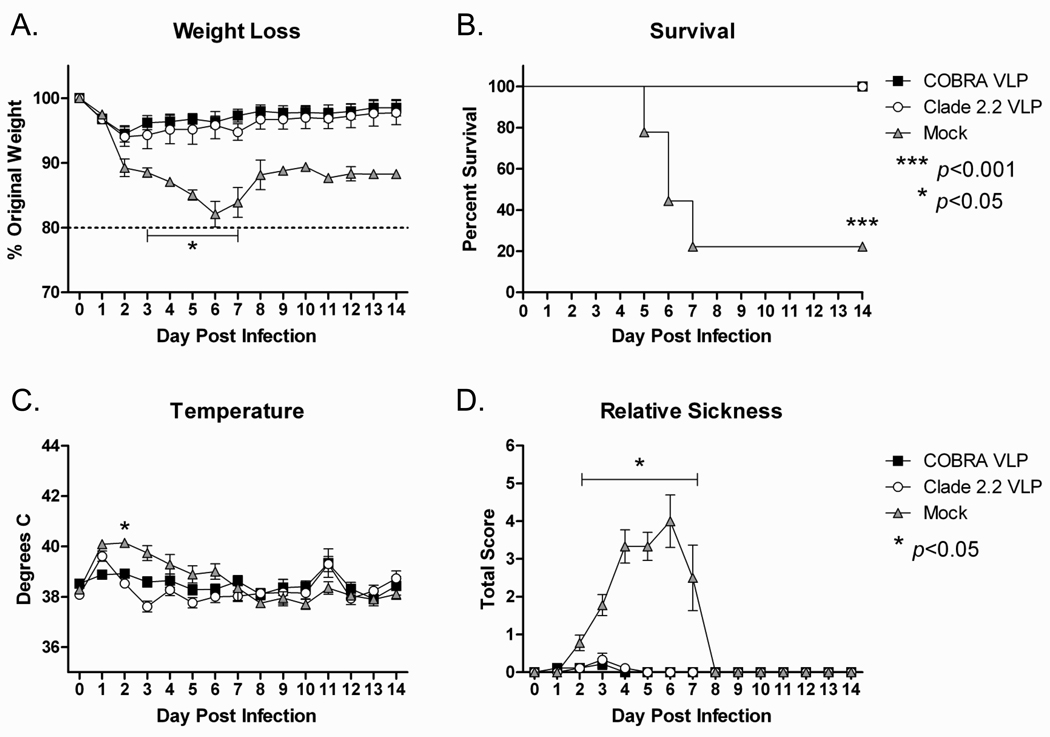
Ferrets that received the COBRA VLP vaccines, clade 2.2 VLP vaccines or mock vaccinated control animals were challenged intranasally with clade 2.2 H5N1 highly pathogenic avian influenza (A/Mongolia/whooper swan/244/2005) to evaluate the protective efficacy of the COBRA vaccine in the ferret model of influenza infection. All VLP vaccinated ferrets were protected from weight loss and death following viral challenge, while all mock vaccinated animals rapidly lost weight and 78% (7/9) of mock vaccinated animals required euthanasia by day 7 post-infection (FIGURE 8A and 8B). Additionally, both COBRA VLP-vaccinated and clade 2.2-vaccinated ferrets were protected from acute fever and failed to develop significant signs of disease while mock vaccinated animals had an elevated body temperature and developed such symptoms as lethargy, diarrhea and decreased food and water intake (FIGURE 8C and 8D). In addition to monitoring outward signs of disease progression, nasal washes were collected for determination of viral replication in the upper respiratory tract. Ferrets vaccinated with either COBRA VLPs or clade 2.2 VLPs did not have detectable virus at any point after infection, while mock vaccinated animals had high levels of viral replication for the first five days of the infection (FIGURE 8E). Importantly, no significant differences were found between COBRA VLP and clade 2.2 VLP vaccinated ferrets in any of the challenge parameters analyzed confirming the findings in mice that the COBRA VLP vaccine protected animals as efficiently as the homologous vaccine.
Figure 8. Ferret Efficacy.
Ferrets (n=9/group) were vaccinated with VLPs formulated with adjuvant. Ferrets were challenged with 1×106 PFU of the highly pathogenic clade 2.2 H5N1 virus A/Whooper Swan/Mongolia/244/2005. Animals were monitored daily for weight loss (A), survival (B), temperature (C) and clinical symptoms (D). Relative sickness scores were determined by measuring lethargy (0–3), runny nose (0–1), sneezing (0–1), loss of appetite (0–1) and diarrhea (0–1). Animals reaching experimental endpoint were euthanized according to institutional guidelines. Nasal washes were collected serially post infection and virus titers determined via plaque assay (E). Statistical significance was determined using a two-way ANOVA with Bonferroni’s post test. A p value of less than 0.05 was considered significant.
Discussion
In this study, we investigated the efficacy of a computationally optimized broadly reactive antigen (COBRA) approach designed for influenza HA as a H5N1 vaccine candidate. Centralized vaccines are a potential strategy for inducing broadly reactive immune responses and are comprised of synthetic antigens that represent a population of sequences. These vaccine antigens are generated by three different methods: center-of-the tree, ancestral, and consensus [38]. Center-of-the tree (COT) sequences are derived by constructing a phylogenetic tree and selecting the sequence that is equidistant from all points [39]. Ancestral sequences represent the most recent common ancestor for the population of sequences selected [40]. Consensus sequences encode the most common amino acid found at each position for the selected population [41]. Ancestral and COT sequences are expected to be an estimate of an actual sequence that existed in the past while consensus sequences are expected to be more relevant to the current epidemic [41]. A recent report has demonstrated the efficacy of using ancestral HA and NA sequences to generate infectious virus capable of eliciting broadly-reactive antibodies post infection in ferrets [42]. Consensus-based vaccines have been previously investigated as strategies for eliciting broadly reactive immune responses for multiple pathogens including HIV-1 [43–47] and influenza [11, 15–17]. Although the COBRA HA investigated in this report utilized a consensus-based approach, important design features differentiate COBRA from prior strategies. COBRA represents a novel method of sequence construction that features a layered building approach that is intended to capture the most common antigenic characteristics, while avoiding the complication of differential sequence availability that can bias a consensus sequence to the most prevalent antigenic cluster. Avoiding this sampling bias is essential to generating a centralized vaccine that accurately represents the population of input sequences. COBRA HA phylogenetic analysis in the context of H5N1 human infections demonstrates that using a layered consensus building approach can indeed overcome the challenges of sequence availability and results in a sequence that retains similarity to the input without localizing into any one antigenic cluster (FIGURE 1B).
Previously, consensus HA, NA and Matrix sequences developed for H5N1 DNA vaccination elicited robust cellular immune responses to each vaccine component, as well as antibodies to the HA [15–17]. This consensus HA consisted of 16 input HA sequences, primarily clade 1 viruses and derived from fatal human infections [15]. For this study, the COBRA antigen utilized over 100 input sequences from human clade 2 infections, which represents the more rapidly emerging clade of H5N1 influenza. Importantly, the work presented here confirms the earlier reports of the efficacy of a centralized H5N1 vaccine and extends those findings using DNA based vaccinations to a virus-like particle (VLP) platform. With any whole protein derived from a centralized sequence, proper folding and retention of function cannot be assumed. Although COBRA HA is a synthetic sequence, we demonstrated that the COBRA HA retained the binding and fusion functions of wild-type HA proteins (FIGURE 2).
Influenza vaccines delivered as VLPs represent a promising new generation of cell-based candidate vaccines [25, 48, 49]. The high virulence of H5N1 makes growing viral stocks for traditional inactivation and generation of split vaccines both difficult and dangerous [50]. VLPs are self assembling, nonpathogenic, genomeless particles that are similar in size and morphology to intact virions [23, 51]. The influenza VLPs described in these studies include the viral proteins HA, NA and matrix and are flexible enough to incorporate a synthetic HA without a decrease in stability or function. The COBRA HA vaccine was evaluated for immunogenicity in mice with and without adjuvant at multiple doses (FIGURE 3). The COBRA vaccine elicited anti-HA total IgG regardless of dose or adjuvant with an isotype profile indicative of a mixed T helper response. IgG1 and IgG2a have been shown to have non-redundant functions in influenza immunity with IgG1 associated with virus neutralization and IgG2a associated with viral clearance [52]. Inclusion of Imject® alum as an adjuvant did not impact the total IgG titers or isotype profile but resulted in increased HAI antibody responses at all doses tested. Aluminum salts have been used to enhance antibody responses to vaccines for almost 100 years [53]. Imject® alum was selected for use in these studies because it is an aluminum salt-based adjuvant and its commercial analogs are the most widely used adjuvants that have been used in Diphteria, Tetanus, Pertussis and Hepatitis A and B vaccines [54]. Aluminum salts enhance antigen uptake by antigen presenting cells and stimulate a pro-inflammatory response via activation of the NOD-like receptor protein 3 inflammasome pathway [55]. Inclusion of an adjuvant increased the breadth and intensity of HAI antibody responses in mice vaccinated with the COBRA HA compared to non-adjuvanted vaccine recipients. Additionally, the relative affinity of the anti-HA antibodies was increased in the adjuvanted groups compared to the non-adjuvanted groups (data not shown). Importantly, the increase in breadth of the HAI antibody response was not a non-specific effect of the adjuvant. An adjuvanted vaccine based on the HA of the clade 2.2 virus A/Whooper Swan/Mongolia/244/2005 failed to elicit significant HAI antibody titers to heterologous test antigens (FIGURES 5 and 7).
One of the challenges in developing a pre-pandemic vaccine for H5N1 is the diversity within the subtype. An important lesson from the novel H1N1 pandemic of 2009 is that predicting the origin of a pandemic virus is currently not impossible. If the next influenza pandemic is caused by H5N1, the clade or subclade of the pandemic virus may not be identified until the outbreak is well underway. An effective pre-pandemic vaccine should be immunogenic and broadly reactive so as to give the best chance of preventing disease caused by drift variants. Identifying vaccine candidate strains based on HA sequence may not effectively select an isolate that will elicit a broad antibody response. The COBRA HA sequence is similar to the clade 2.2 HA used as a vaccine in these studies (95% identity) and the two different HA molecules have similar identity profiles to the other test antigens investigated here (TABLE 3). Despite the high levels of identity between immunogens, structural modeling predicted the COBRA sequence to retain the most common structural features found in representative sequences while the clade 2.2 HA was divergent in potentially antigenic regions surrounding the receptor binding domain (data not shown). Consistent with these predictions, the COBRA based H5N1 vaccine elicited a broader antibody response than the clade 2.2 primary isolate based H5N1 vaccine. Although the breadth of this immunity was limited to clade 2, the antigen was designed specifically to address the wide diversity specifically within clade 2. Further work and antigen design is currently underway to include input sequences from more diverse clades to address the cross-clade diversity of H5N1. In addition to the broadened antibody responses, the COBRA vaccine protected experimentally infected animals as efficiently as a homologous vaccine.
Table 3. Percent Identity of Test Antigens.
HA amino acid sequences were aligned and percent identity across the entire protein was determined for the vaccine immunogens compared to the representative test antigens.
A/Vietnam/1203/2004
A/Indonesia/5/2005
A/Whooper Swan/Mongolia/244/2005
A/Anhui/1/2005
Although the HAI assay has historically been the most commonly used serological assay for monitoring influenza immunity, no correlation between HAI titer and protective efficacy against H5N1 infection has been reported [56–61]. For seasonal influenza, an HAI titer ≥1:40 against a seasonal virus protects ~50% of the vaccinated population [62]. Despite the lack of correlation of protection for H5N1 infection, the 1:40 HAI titer remains the standard for evaluating vaccine immunogenicity. Using the 1:40 titer as an indicator of positive responses, the adjuvanted COBRA vaccines consistently elicited a high rate of seroconversion to diverse clade 2 viruses (TABLES 1 and 2). Consistent with prior findings, even in the absence of positive HAI titers, vaccinated animals were protected from experimental infection (FIGURES 4, 6 and 8). Protection could be due to non-neutralizing anti-HA antibodies. All COBRA HA immunized animals had total IgG titers to a diverse number of H5N1 HA antigens. Additionally, both anti-NA antibodies and cellular responses elicited by these vaccines against influenza proteins could contribute to protection against H5N1 infection [63–66]. The COBRA HA sequence does not contain unique epitopes, as predicted by immune epitope software (www.immuneepitope.org), compared to representative primary clade 2 isolates [67]. However, VLPs elicit cellular immune responses [25] and while these responses are predicted to be equivalent between the COBRA and primary isolate vaccine groups, the protective capacity of the cell-mediated immune responses cannot be ruled out.
This is the first report of a consensus-based H5N1 vaccine delivered in the context of a virus-like particle. The COBRA HA represents a new generation of centralized vaccine design and elicited a broadly reactive antibody response that protected experimentally infected animals from a heterologous H5N1 challenge. Increasing the breadth of antibody responses is a challenge not only for H5N1, but all influenza viruses. Utilizing COBRA-based vaccine design may enable the development of influenza vaccines more resistant to antigenic drift variants and warrants further evaluation.
Supplementary Material
Acknowledgments
This work was supported by an NIH training grant award T32AI060525 to BMG, NIH/NIAID award U01AI077771 to TMR, the Center for Vaccine Research and a collaborative research grant from PATH Vaccine Solutions. This project is funded, in part, under a grant with the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions. The authors would like to thank Brianne Stein and Corey Crevar for technical assistance. In addition, we thank Ruben Donis and Richard Webby for providing the H5N1 reassortant viruses.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization. Avian influenza: assessing the pandemic threat. 2005 [cited; Available from http://www.who.int/csr/disease/influenza/WHO_CDS_2005_29/en/index.html.
- 2.World Health Organization. Cumulative Number of Confirmed Human Cases of Avian Influenza A/(H5N1) Reported to WHO. 2008 [cited; Available from: http://www.who.int/csr/disease/avian_influenza/country/cases_table_2008_02_26/en/index.html.
- 3.Li KS, Guan Y, Wang J, Smith GJD, Xu KM, Duan L, et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430(6996):209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 4.Chen L-M, Davis CT, Zhou H, Cox NJ, Donis RO. Genetic Compatibility and Virulence of Reassortants Derived from Contemporary Avian H5N1 and Human H3N2 Influenza A Viruses. PLoS Pathog. 2008;4(5):e1000072. doi: 10.1371/journal.ppat.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C, Hatta M, Nidom CA, Muramoto Y, Watanabe S, Neumann G, et al. Reassortment between avian H5N1 and human H3N2 influenza viruses creates hybrid viruses with substantial virulence. Proceedings of the National Academy of Sciences. 107(10):4687–4692. doi: 10.1073/pnas.0912807107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Octaviani CP, Ozawa M, Yamada S, Goto H, Kawaoka Y. High Level of Genetic Compatibility between Swine-Origin H1N1 and Highly Pathogenic Avian H5N1 Influenza Viruses. The Journal of Virology. 84(20):10918–10922. doi: 10.1128/JVI.01140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. [cited February 27, 2008];Antigenic and genetic characteristics of H5N1 viruses and candidate H5N1 vaccine viruses developed for potential use as human vaccines. 2008 February; 2008; Available from: http://www.who.int/csr/disease/avian_influenza/guidelines/H5VaccineVirusUpdate20080214.pdf.
- 8.Wang J, Vijaykrishna D, Duan L, Bahl J, Zhang JX, Webster RG, et al. Identification of the Progenitors of Indonesian and Vietnamese Avian Influenza A (H5N1) Viruses from Southern China. The Journal of Virology. 2008;82(7):3405–3414. doi: 10.1128/JVI.02468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webster RG, Govorkova EA. H5N1 Influenza -- Continuing Evolution and Spread. TheNew England Journal of Medicine. 2006;355(21):2174–2177. doi: 10.1056/NEJMp068205. [DOI] [PubMed] [Google Scholar]
- 10.Prabakaran M, He F, Meng T, Madhan S, Yunrui T, Jia Q, et al. Neutralizing Epitopes of Influenza Virus Hemagglutinin: Target for the Development of a Universal Vaccine against H5N1 Lineages. The Journal of Virology. 84(22):11822–11830. doi: 10.1128/JVI.00891-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tompkins SM, Zhao Z-S, Lo C-Y, Misplon JA, Liu T, Ye Z, et al. Matrix Protein 2 Vaccination and Protection against Influenza Viruses, Including Subtype H5N1. Emerging Infectious Diseases 2007 March. 2007;13(3):426–435. doi: 10.3201/eid1303.061125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kodihalli S, Kobasa DL, Webster RG. Strategies for inducing protection against avian influenza A virus subtypes with DNA vaccines. Vaccine. 2000;18(23):2592–2599. doi: 10.1016/s0264-410x(99)00485-5. [DOI] [PubMed] [Google Scholar]
- 13.Bianchi E, Liang X, Ingallinella P, Finotto M, Chastain MA, Fan J, et al. Universal Influenza B Vaccine Based on the Maturational Cleavage Site of the Hemagglutinin Precursor. The Journal of Virology. 2005;79(12):7380–7388. doi: 10.1128/JVI.79.12.7380-7388.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio. 2010;1(1):e00018–e00010. doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laddy DJ, Yan J, Corbitt N, Kobasa D, Kobinger GP, Weiner DB. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine. 2007;25(16):2984–2989. doi: 10.1016/j.vaccine.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laddy DJ, Yan J, Kutzler M, Kobasa D, Kobinger GP, Khan AS, et al. Heterosubtypic Protection against Pathogenic Human and Avian Influenza Viruses via <italic>In Vivo</italic> Electroporation of Synthetic Consensus DNA Antigens. PLoS ONE. 2008;3(6):e2517. doi: 10.1371/journal.pone.0002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laddy DJ, Yan J, Khan AS, Andersen H, Cohn A, Greenhouse J, et al. Electroporation of Synthetic DNA Antigens Offers Protection in Nonhuman Primates Challenged with Highly Pathogenic Avian Influenza Virus. The Journal of Virology. 2009;83(9):4624–4630. doi: 10.1128/JVI.02335-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen M-W, Cheng T-JR, Huang Y, Jan J-T, Ma S-H, Yu AL, et al. A consensus hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proceedings of the National Academy of Sciences. 2008;105(36):13538–13543. doi: 10.1073/pnas.0806901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBurney SP, Ross TM. Human immunodeficiency virus-like particles with consensus envelopes elicited broader cell-mediated peripheral and mucosal immune responses than polyvalent and monovalent Env vaccines. Vaccine. 2009;27(32):4337–4349. doi: 10.1016/j.vaccine.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver EA, Lu Z, Camacho ZT, Moukdar F, Liao H-X, Ma B-J, et al. Cross-Subtype T-Cell Immune Responses Induced by a Human Immunodeficiency Virus Type 1 Group M Consensus Env Immunogen. The Journal of Virology. 2006;80(14):6745–6756. doi: 10.1128/JVI.02484-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan J, Yoon H, Kumar S, Ramanathan MP, Corbitt N, Kutzler M, et al. Enhanced Cellular Immune Responses Elicited by an Engineered HIV-1 Subtype B Consensus-based Envelope DNA Vaccine. Mol Ther. 15(2):411–421. doi: 10.1038/sj.mt.6300036. [DOI] [PubMed] [Google Scholar]
- 22.Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, et al. The Influenza Virus Resource at the National Center for Biotechnology Information. The Journal of Virology. 2008;82(2):596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu C-Y, Yeh Y-C, Yang Y-C, Chou C, Liu M-T, Wu H-S, et al. Mammalian Expression of Virus-Like Particles for Advanced Mimicry of Authentic Influenza Virus. PLoS ONE. 5(3):e9784. doi: 10.1371/journal.pone.0009784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garland SM, Hernandez-Avila M, Wheeler CM, Perez G, Harper DM, Leodolter S, et al. Quadrivalent Vaccine against Human Papillomavirus to Prevent Anogenital Diseases. New England Journal of Medicine. 2007;356(19):1928–1943. doi: 10.1056/NEJMoa061760. [DOI] [PubMed] [Google Scholar]
- 25.Bright RA, Carter DM, Crevar CJ, Toapanta FR, Steckbeck JD, Cole KS, et al. Cross-Clade Protective Immune Responses to Influenza Viruses with H5N1 HA and NA Elicited by an Influenza Virus-Like Particle. PLoS ONE. 2008;3(1):e1501. doi: 10.1371/journal.pone.0001501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z-y, Chakrabarti BK, Xu L, Welcher B, Kong W-p, Leung K, et al. Selective Modification of Variable Loops Alters Tropism and Enhances Immunogenicity of Human Immunodeficiency Virus Type 1 Envelope. The Journal of Virology. 2004;78(8):4029–4036. doi: 10.1128/JVI.78.8.4029-4036.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- 28.Toapanta F, Ross T. Impaired immune responses in the lungs of aged mice following influenza infection. Respiratory Research. 2009;10(1):112. doi: 10.1186/1465-9921-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gillim-Ross L, Subbarao K. Emerging respiratory viruses: challenges and vaccine strategies. Clin Microbiol Rev 2006 Oct. 19(4):614–636. doi: 10.1128/CMR.00005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bright RA, Medina MJ, Xu X, Perez-Oronoz G, Wallis TR, Davis XM, et al. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 2005 Oct 1. 366(9492):1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- 31.Bright RA, Ross TM, Subbarao K, Robinson HL, Katz JM. Impact of glycosylation on the immunogenicity of a DNA-based influenza H5 HA vaccine. Virology 2003 Apr 10. 308(2):270–278. doi: 10.1016/s0042-6822(03)00008-4. [DOI] [PubMed] [Google Scholar]
- 32.Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. Jama 2006 Feb 22. 295(8):891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell JA, Green TD, Bright RA, Ross TM. Induction of heterosubtypic immunity to influenza A virus using a DNA vaccine expressing hemagglutinin-C3d fusion proteins. Vaccine 2003 Feb 14. 21(9–10):902–914. doi: 10.1016/s0264-410x(02)00539-x. [DOI] [PubMed] [Google Scholar]
- 34.Ross TM, Xu Y, Bright RA, Robinson HL. C3d enhancement of antibodies to hemagglutinin accelerates protection against influenza virus challenge. Nat Immunol. 2000;1(2):127–131. doi: 10.1038/77802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Askonas B, McMichael A, Webster R. The immune response to influenza viruses and the problem of protection against infection. In: Beare ASe., editor. Bascin and applied influenza research. CRC Press; 1982. pp. 159–188. [Google Scholar]
- 36.Nefkens, Garcia, Ling, Lagarde, Nicholls, Tang, et al. Hemagglutinin pseudotyped lentiviral particles: Characterization of a new method for avian H5N1 influenza sero-diagnosis. Journal of Clinical Virology. 2007;39(1):27–33. doi: 10.1016/j.jcv.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Haynes JR, Dokken L, Wiley JA, Cawthon AG, Bigger J, Harmsen AG, et al. Influenza-pseudotyped Gag virus-like particle vaccines provide broad protection against highly pathogenic avian influenza challenge. Vaccine. 2009;27(4):530–541. doi: 10.1016/j.vaccine.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 38.Kesturu GS, Colleton BA, Liu Y, Heath L, Shaikh OS, Rinaldo JCR, et al. Minimization of genetic distances by the consensus, ancestral, and center-of-tree (COT) sequences for HIV-1 variants within an infected individual and the design of reagents to test immune reactivity. Virology. 2006;348(2):437–448. doi: 10.1016/j.virol.2005.11.055. [DOI] [PubMed] [Google Scholar]
- 39.Nickle DC, Jensen MA, Gottlieb GS, Shriner D, Learn GH, Rodrigo AG, et al. Consensus and Ancestral State HIV Vaccines. Science. 2003;299(5612):1515c–1518c. doi: 10.1126/science.299.5612.1515c. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Molecular Biology and volution. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 41.Gao F, Liao H-X, Hahn BH, Letvin NL, Korber BT, Haynes BF. Centralized HIV-1 Envelope Immunogens and Neutralizing Antibodies. Current HIV Research. 2007;5(6):572–577. doi: 10.2174/157016207782418498. [DOI] [PubMed] [Google Scholar]
- 42.Ducatez MF, Bahl J, Griffin Y, Stigger-Rosser E, Franks J, Barman S, et al. Feasibility of reconstructed ancestral H5N1 influenza viruses for cross-clade protective vaccine development. Proceedings of the National Academy of Sciences. 108(1):349–354. doi: 10.1073/pnas.1012457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao F, Weaver EA, Lu Z, Li Y, Liao H-X, Ma B, et al. Antigenicity and Immunogenicity of a Synthetic Human Immunodeficiency Virus Type 1 Group M Consensus Envelope Glycoprotein. The Journal of Virology. 2005;79(2):1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamano T, Sawanpanyalert P, Yanai H, Piyaworawong S, Hara T, Sapsutthipas S, et al. Determination of HIV Type 1 CRF01_AE gag p17 and env-V3 Consensus Sequences for HIV/AIDS Vaccine Design. AIDS Research and Human Retroviruses. 2004;20(3):337–340. doi: 10.1089/088922204322996572. [DOI] [PubMed] [Google Scholar]
- 45.Krohn K, Stanescu I, Blazevic V, Vesikari T, Ranki A, Ustav M. A DNA HIV-1 vaccine based on a fusion gene expressing non-structural and structural genes of consensus sequence of the A–C subtypes and the ancestor sequence of the F–H subtypes. Preclinical and clinical studies. Microbes and Infection. 2005;7(14):1405–1413. doi: 10.1016/j.micinf.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 46.Mwau M, Cebere I, Sutton J, Chikoti P, Winstone N, Wee EGT, et al. A human immunodeficiency virus 1 (HIV-1) clade A vaccine in clinical trials: stimulation of HIV-specific T-cell responses by DNA and recombinant modified vaccinia virus Ankara (MVA) vaccines in humans. Journal of General Virology. 2004;85(4):911–919. doi: 10.1099/vir.0.19701-0. [DOI] [PubMed] [Google Scholar]
- 47.Malm M, Rollman E, Ustav M, Hinkula J, Krohn K, Wahren B, et al. Cross-Clade Protection Induced by Human Immunodeficiency Virus-1 DNA Immunogens Expressing Consensus Sequences of Multiple Genes and Epitopes From Subtypes A, B, C, and FGH. Viral Immunology. 2005;18(4):678–688. doi: 10.1089/vim.2005.18.678. [DOI] [PubMed] [Google Scholar]
- 48.Bright RA, Carter DM, Daniluk S, Toapanta FR, Ahmad A, Gavrilov V, et al. Influenza virus-like particles elicit broader immune responses than whole virion inactivated influenza virus or recombinant hemagglutinin. Vaccine. 2007;25(19):3871–3878. doi: 10.1016/j.vaccine.2007.01.106. [DOI] [PubMed] [Google Scholar]
- 49.Mahmood K, Bright R, Mytle N, Carter D, Crevar C, Achenbach J, et al. H5N1 VLP vaccine induced protection in ferrets against lethal challenge with highly pathogenic H5N1 influenza viruses. Vaccine. 2008;26:5393–5399. doi: 10.1016/j.vaccine.2008.07.084. [DOI] [PubMed] [Google Scholar]
- 50.Subbarao K, Joseph T. Scientific barriers to developing vaccines against avian influenza viruses. Nat Rev Immunol. 2007;7(4):267–278. doi: 10.1038/nri2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young KR, Ross TM. Particle-based Vaccines for HIV-1 Infection. Current Drug Targets - Infectious Disorders. 2003;3:151–169. doi: 10.2174/1568005033481213. [DOI] [PubMed] [Google Scholar]
- 52.Huber VC, McKeon RM, Brackin MN, Miller LA, Keating R, Brown SA, et al. Distinct Contributions of Vaccine-Induced Immunoglobulin G1 (IgG1) and IgG2a Antibodies to Protective Immunity against Influenza. Clinical and Vaccine Immunology. 2006;13(9):981–990. doi: 10.1128/CVI.00156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Janeway CA, Medzhitov R. INNATE IMMUNE RECOGNITION. Annual Review of Immunology. 2002;20(1):197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 54.Tritto E, Mosca F, De Gregorio E. Mechanism of action of licensed vaccine adjuvants. Vaccine. 2009;27(25–26):3331–3334. doi: 10.1016/j.vaccine.2009.01.084. [DOI] [PubMed] [Google Scholar]
- 55.Eisenbarth SC, Colegio OR, O/'Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453(7198):1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Govorkova E, Webby R, Humberd J, Seiler J, Webster R. Immunization with Reverse Genetics Produced H5N1 Influenza Vaccine Protects Ferrets against Homologous and Heterologous Challenge. The Journal of Infectious Diseases. 2006;194(2):159–167. doi: 10.1086/505225. [DOI] [PubMed] [Google Scholar]
- 57.Ninomiya A, Imai M, Tashiro M, Odagiri T. Inactivated influenza H5N1 whole-virus vaccine with aluminum adjuvant induces homologous and heterologous protective immunities against lethal challenge with highly pathogenic H5N1 avian influenza viruses in a mouse model. Vaccine. 2007;25(18):3554–3560. doi: 10.1016/j.vaccine.2007.01.083. [DOI] [PubMed] [Google Scholar]
- 58.Lipatov A, Hoffmann E, Salomon R, Yen H-L, Webster R. Cross-Protectiveness and Immunogenicity of Influenza A/Duck/Singapore/3/97(H5) Vaccines against Infection with A/Vietnam/1203/04(H5N1) Virus in Ferrets. The Journal of Infectious Diseases. 2006;194(8):1040–1043. doi: 10.1086/507709. [DOI] [PubMed] [Google Scholar]
- 59.Suguitan AL, McAuliffe J, Mills KL, Jin H, Duke G, Lu B, et al. Live, Attenuated Influenza A H5N1 Candidate Vaccines Provide Broad Cross-Protection in Mice and Ferrets. PLoS Medicine. 2006;3(9):e360. doi: 10.1371/journal.pmed.0030360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu X, Edwards LE, Desheva JA, Nguyen DC, Rekstin A, Stephenson I, et al. Cross- protective immunity in mice induced by live-attenuated or inactivated vaccines against highly pathogenic influenza A (H5N1) viruses. Vaccine. 2006;24(44–46):6588–6593. doi: 10.1016/j.vaccine.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 61.Hoelscher MA, Garg S, Bangari DS, Belser JA, Lu X, Stephenson I, et al. Development of adenoviral-vector-based pandemic influenza vaccine against antigenically distinct human H5N1 strains in mice. The Lancet. 2006;367(9509):475–481. doi: 10.1016/S0140-6736(06)68076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davenport F. Control of influenza. Medical Journal of Australia. 1973 Supplement:33–38. doi: 10.5694/j.1326-5377.1973.tb111174.x. [DOI] [PubMed] [Google Scholar]
- 63.Chen Z, Kadowaki S-e, Hagiwara Y, Yoshikawa T, Matsuo K, Kurata T, et al. Cross- protection against a lethal influenza virus infection by DNA vaccine to neuraminidase. Vaccine. 2000;18(28):3214–3222. doi: 10.1016/s0264-410x(00)00149-3. [DOI] [PubMed] [Google Scholar]
- 64.Sandbulte MR, Jimenez GS, Boon ACM, Smith LR, Treanor JJ, Webby RJ. Cross- Reactive Neuraminidase Antibodies Afford Partial Protection against H5N1 in Mice and Are Present in Unexposed Humans. PLoS Medicine. 2007;4(2):e59. doi: 10.1371/journal.pmed.0040059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kreijtz JHCM, Bodewes R, van Amerongen G, Kuiken T, Fouchier RAM, Osterhaus ADME, et al. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine. 2007;25(4):612–620. doi: 10.1016/j.vaccine.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 66.Furuya Y, Chan J, Regner M, Lobigs M, Koskinen A, Kok T, et al. Cytotoxic T Cells Are the Predominant Players Providing Cross-Protective Immunity Induced by {gamma}-Irradiated Influenza A Viruses. The Journal of Virology. 84(9):4212–4221. doi: 10.1128/JVI.02508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, et al. The Immune Epitope Database 2.0. Nucleic Acids Research. 38 suppl 1:D854–D862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.