Abstract
In recent years, the number of known peptide natural products that are synthesized via the ribosomal pathway has rapidly grown. Taking advantage of sequence homology among genes encoding precursor peptides or biosynthetic proteins, in silico mining of genomes combined with molecular biology approaches has guided the discovery of a large number of new ribosomal natural products, including lantipeptides, cyanobactins, linear thiazole/oxazole-containing peptides, microviridins, lasso peptides, amatoxins, cyclotides, and conopeptides. In this review, we describe the strategies used for the identification of these ribosomally-synthesized and posttranslationally modified peptides (RiPPs) and the structures of newly identified compounds. The increasing number of chemical entities and their remarkable structural and functional diversity may lead to novel pharmaceutical applications.
1. Introduction
The power of peptides to recognize biological targets is well-appreciated from studies using various display techniques and synthetic libraries. Linear peptides, however, are often poor drug candidates due to rapid degradation and poor biodistribution. Cyclic peptides, sometimes containing non-proteinogenic structures, provide protection against proteolysis and greatly decrease the number of possible conformations, thereby favoring specific and tight interactions with the target. Recent strategies to increase peptide stability and improve productivity, together with alternative routes of administration, have resulted in an increasing number of peptide-based drugs and drug candidates [1], with currently more than 60 therapeutic peptides used commercially and more than 150 in clinical trials [1,2].
Among natural products, the therapeutic potential of cyclic peptides of non-ribosomal origin is well-recognized, as illustrated by the cyclic antimicrobial compounds vancomycin or daptomycin, the immunosuppressant cyclosporin A, or the antitumor agent bleomycin A. In addition, recent years have seen the discovery of a growing number of cyclic peptides that are biosynthesized by a ribosomal pathway, followed by extensive posttranslational modifications to produce the mature compounds. These pathways confer several clear advantages for bioengineering because of the direct link between gene sequence and natural product and because of their relatively short biosynthetic pathways. Furthermore, many of the biosynthetic enzymes have demonstrated substrate promiscuity. The number of families of RiPPs has expanded considerably in recent years with the discovery of the biosynthetic origin for cyanobactins, thiopeptides, microviridins, and amatoxins. These findings have shown that the ribosomal world is much more diverse than originally anticipated, and have revealed an enormous structural diversity that is just being appreciated [3,4]. This diversity is increasingly exploited by application of in silico analysis of genome data and by the use of the polymerase chain reaction (PCR) with degenerate primers to identify precursor peptides and to predict the structure of the mature compounds from the genomic context and from previously characterized molecules. This review focuses on RiPPs discovered using this strategy between 2007 and 2010.
2. Genome Mining for Lantipeptides
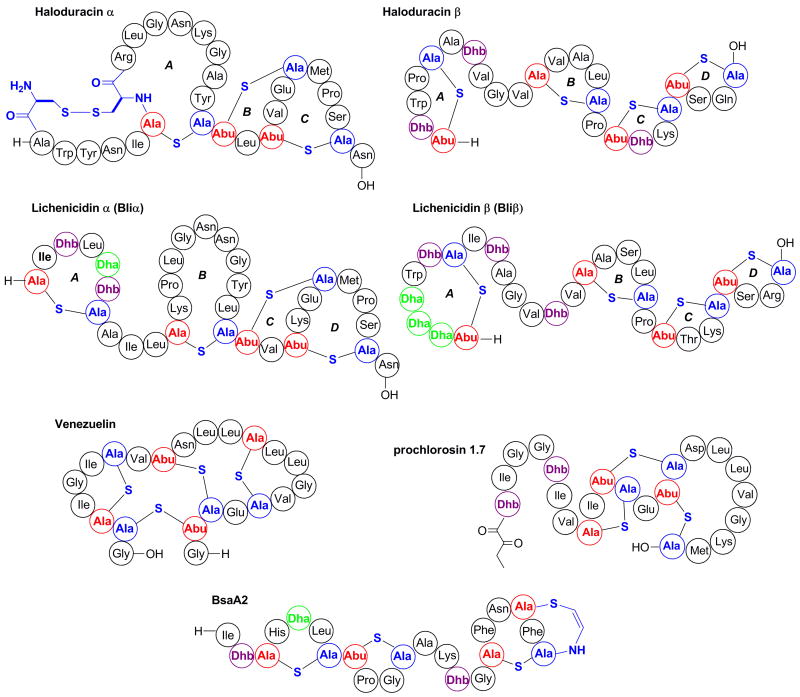
Lantipeptides are defined by their characteristic lanthionine and/or methyllanthionine thioether crosslinks and they often contain the unsaturated amino acids dehydroalanine and dehydrobutyrine [5]; lantipeptides with confirmed antimicrobial activity are called lantibiotics. They can be categorized in four classes according to the enzymes responsible for ring formation. In class I lantipeptides, the enzymes LanB and LanC catalyze the dehydration of serine and threonine residues and the subsequent intramolecular addition of cysteine residues, respectively, resulting in the formation of the thioether crosslinks. For the other classes, bifunctional enzymes are responsible for both dehydration and cyclization steps (LanM in class II, RamC-like for class III, and LanL in class IV) [5-7]. Genome mining for lanthionine synthetase homologs has recently guided the discovery of several lantipeptides. In the first such example, a bioinformatic analysis of the genome of Bacillus halodurans C-125 revealed the presence of two genes encoding for precursor peptides clustered with two additional open reading frames (ORF) encoding for LanM-type synthetases. Analysis of cell-free supernatants by mass spectrometry (MS), in vitro reconstitution of the LanM enzymes, antimicrobial assays, and mutagenesis experiments, allowed the identification and structural characterization of the two-component lantibiotic haloduracin (Figure 1) [8-10]. Using similar bioinformatic approaches or PCR amplification of conserved DNA sequences, several lanM-containing biosynthetic gene clusters were discovered subsequently, including the cluster encoding for lichenicidin in the genome of Bacillus licheniformis ATCC14580 (or DSM13) and VK21 [8,11-13]. In follow-up studies, this two-component lantibiotic was detected in bacterial cultures by antimicrobial assays and MS analysis (Figure 1) [11-13]. In a similar study, several variants of the lantibiotic epidermin, designated Bsa, and produced by methicillin-resistant Staphylococcus aureus strains were identified (Figure 1), suggesting that these bacteriocins may confer a competitive ecological advantage on community acquired infections [14].
Figure 1. Lantipeptides recently discovered by genome mining.
The two-component lantibiotics haloduracin from B. halodurans C-125 and lichenicidin from B. licheniformis ATCC 14580, DSM13, and VK21 were discovered after genome mining for LanM lanthionine synthetases. Venezuelin, a lantipeptide predicted from he genome sequence of S. venezuelae ATCC 10712, was produced in vitro after reconstitution of a novel LanL lanthionine synthetase. Prochlorosin 1.7 is one of 29 lantipeptides produced by Prochlorococcus MIT9313. BsaA2 was discovered after genome sequence analysis of several S. aureus strains. Dha: dehydroalanine, Dhb: dehydrobutyrine, Abu: α-aminobutyric acid. For the thioether crosslinks, residues derived from Ser/Thr are shown in red and residues originating from Cys are shown in blue.
A new route to lantipeptides was discovered by analysis of the draft genome sequence of S. venezuelae ATCC10712 revealing a lantibiotic-like gene cluster with an ORF encoding for a novel class of putative lanthionine synthetase (LanL) [5]. In vitro reconstitution of the enzyme activity with the predicted precursor peptide resulted in the production of venezuelin, the first class IV lantipeptide (Figure 1). LanL homologs were also identified in other species of actinobacteria and firmicutes [5]. Another recent genome database analysis revealed that several strains of marine Prochlorococcus and Synechococcus contain multiple ORFs encoding a wide diversity of lantipeptide precursor peptides but only a single gene encoding a LanM-like synthetase [15]. The precursor peptides have highly homologous leader peptides but display great diversity in the core peptide, the region of the precursor peptide that is modified to the mature natural product (note: throughout this review we use the term core peptide because the often-used term propeptide has an opposite meaning for different classes of RiPPs. For a discussion, see the Supporting Information of reference [4]). The enzymatic activity of the predicted LanM from Prochlorococcus MIT9313 was reconstituted in vitro and 17 selected precursor peptides (out of the 29 encoded in the genome) were converted to the corresponding lantipeptides (prochlorosins), providing an example of natural combinatorial biosynthesis. Analysis of the spent media of MIT9313 by MS showed in vivo production of at least three of these compounds (e.g. Figure 1), confirming that lantipeptide production is not restricted to Gram-positive or soil bacteria as long believed [15]. A search of sequences in the Global Oceanic Survey uncovered more than 150 prochlorosin precursor genes from many different locations suggesting prochlorosin production is widespread.
In another study, PSI-BLAST searches of the NCBI database for LanM homologs revealed more than 60 lanthionine synthetase genes, including examples in phyla (chloroflexi and proteobacteria) not previously identified as lantipeptide producers [11]. Similarly, a computerized discovery strategy using the genome-mining software BAGEL2 revealed approximately 150 putative lantipeptide gene clusters based on conserved biosynthetic, transport, and immunity machinery [16]. By using BAGEL, the biosynthetic gene cluster of pneumococcin A1 and A2 was identified in the genome sequence of S. pneumonia R6 [17]. Interestingly, when the predicted pneumococcin core peptides (belonging to class II lantipeptides) were modified in vivo using the nisin synthetase machinery (a class I system), lanthionine containing-peptides with antimicrobial activity were produced [17].
3. Genome mining for cyanobactins
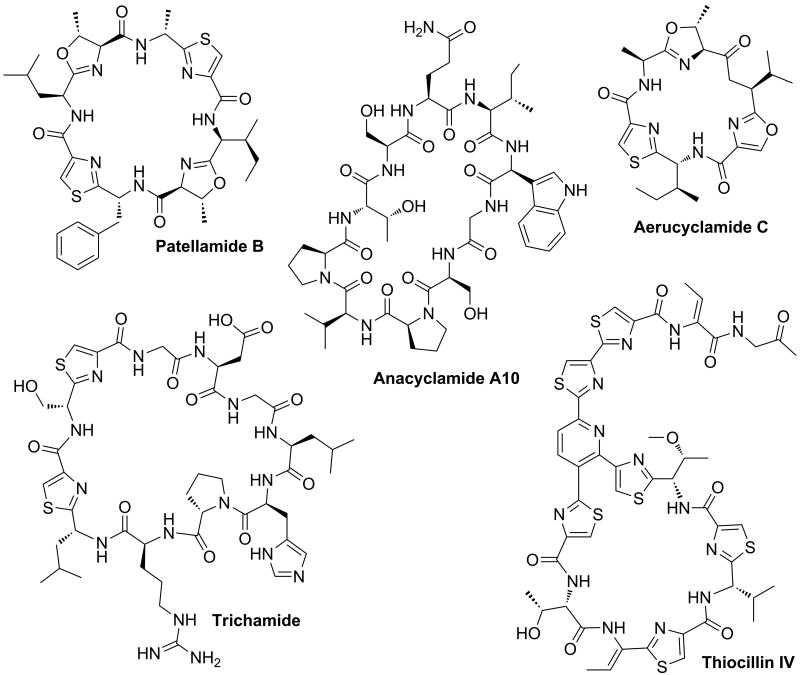
Cyanobactins are small cyclic peptides produced by a wide variety of cyanobacteria that typically, but not always, contain oxazoline and thiazoline structures, or their oxidized derivatives oxazole and thiazole. The inclusion of cyanobactins amongst the RiPPs was first revealed by the identification of the biosynthetic gene cluster encoding for patellamides in Prochloron didemni and heterologous expression in Escherichia coli [18,19]. This discovery has lead to the identification of several other related compounds. For instance, analysis of metagenome samples from 46 Prochloron-containing ascidians using PCR primers targeting patE-like genes (the gene encoding for the precursor peptide in patellamide biosynthesis) predicted 29 patellamide variants including the novel patellamide B (Figure 2). This study showed that hypervariable cassettes encoding for the core peptides within a conserved genetic background leads to the production of cyanobactin libraries with readily predictable chemical structures [20]. In addition to PatE, all cyanobactin gene clusters encode two proteases (PatA and PatG) that cleave at the N-and C-terminal boundaries of the core peptide and catalyze its head-to-tail cyclization. These proteins are highly conserved at the DNA sequence level across different strains, aiding detection of novel biosynthetic clusters in genomes. For example, when a collection of cyanobacteria was screened by PCR using degenerate primers specific for patA-like genes, about one third of the tested strains spanning different genera (Microcystis, Anabaena, Nodularia, Nostoc, Planktothrix, among others) were confirmed to contain cyanobactin biosynthetic gene clusters, suggesting the widespread occurrence of this family of metabolites [21]. In some cases, identification of the canonical protease cleavage sites in the precursor peptide allowed accurate prediction of the chemical structure of the mature compounds, facilitating detection and purification of the compounds from bacterial cultures. For instance, the trichamide biosynthetic gene cluster from Trichodesmium spp. was discovered after BLAST searches of GenBank, and subsequently the predicted trichamide structure was confirmed by MS (Figure 2) [22]. In a similar study, analyses of genomic data of the cyanobacterial strain Microcystis aeruginosa PCC7806 resulted in the discovery of the hexapeptides microcyclamide 7806A and 7806B, which were later shown to be the hydrolysis products of aerucyclamide C (Figure 2) [23,24]. A patellamide-like cluster was also discovered in the genome sequence of Lyngbya aestuarii CCY9616 and two novel cyanobactins were predicted [25]. More recently, annotation of the Anabaena sp. 90 genome led to the identification of a gene cluster with patA and patG homologs [26]. Screening of a collection of Anabaena strains using degenerated PCR primers specific for the genes encoding for one of the proteases as well as the precursor peptide revealed several strains with the genetic potential to biosynthesize cyanobactins [26]. Subsequent liquid chromatography (LC)-MS analysis together with isotope labeling experiments guided the isolation and identification of a novel family of 18 cyclic peptides termed anacyclamides (Figure 2). The peptides, composed of 7 to 20 amino acids, exhibit considerable sequence variation with only a conserved terminal proline. Interestingly, the isolated anacyclamides lack heterocyclized residues, as was predicted from the gene clusters [26]. Similar bioinformatic analyses of the genome of phylogenetically distant bacteria revealed several biosynthetic gene clusters that potentially encode for other unknown cyclic peptides [22].
Figure 2. Cyanobactins and thiopeptides discovered by genome mining approaches.
Patellamide B produced by P. didemmni was discovered by sequencing of metagenomic samples. Trichamide was isolated from Trichodesmium sp. and microcyclamide 7806A and 7806B from M. aeurignosa PCC7806 were found after identification of orthologue patellamide biosynthetic gene clusters. Anacyclamide A10 was isolated after PCR detection of its biosynthetic gene cluster in Anabaena sp90. The thiopeptide thiocillin IV was discovered in the culture supernatant of B. cereus ATCC 14579 after identification of its biosynthetic gene cluster through genome analysis.
4. Genome mining for thiazolyl peptides
Thiazolyl peptides or thiopeptides are a family of highly modified antibacterial compounds that harbor a central pyridine, hydropyridine, or dehydropiperidine ring decorated with thiazole substituents and at least one macrocycle (Figure 2). Their ribosomal origin was recently demonstrated by four independent research groups that reported the biosynthetic gene clusters for several members of this family (micrococcin P1-P2, thiostrepton, thiocillin I, siomycin A, thiomuracin A-I, nosiheptide, and GE2270A) in nearly simultaneous publications [27-31]. All thiazolyl peptide biosynthetic gene clusters contain, in addition to the gene encoding for the precursor peptide (TsrA for thiostrepton using the nomenclature in [28]), a set of at least five genes that encode enzymes likely mediating heterocyclization (TsrFH), dehydration (TsrCD), and a hypothetical protein (TsrE) that potentially catalyzes the formation of the central 6-membered nitrogen heterocycle. The characterization of thiopeptide gene clusters has facilitated the discovery of novel thiopeptides by bioinformatic analysis of genome sequences from other bacteria. For instance, several new clusters potentially encoding for thiopeptides were identified from Bacillus, Salinospora, Streptomyces, Frankia, Propionibacterium, and Nocardiopsis [27,29,32]. In the case of Bacillus cereus ATCC14579, eight compounds were detected in the culture supernatant, including the novel thiocillin IV (Figure 2) [27].
5. Genome mining for linear thiazole/oxazole-modified microcins
Microcin B17 (MccB17) and streptolysin S (SLS) are produced by E. coli and Streptococcus pyogenes, respectively. The precursor peptide (SagA in the case of SLS) is modified by a synthetase protein complex (SagBCD) that introduces several thiazole and oxazole heterocycles [33,34]. Gene clusters encoding homologs have been identified in the genomes of archaea as well as bacteria spanning six phyla, including cyanobacteria, actinobacteria, and proteobacteria [34]. Interestingly, in vitro incubation of a predicted precursor peptide (ClosA) identified in the genome of the pathogenic bacteria Clostridium botulinum ATCC3502 and C. sporogenes ATCC15579 with the SagBCD complex (or with SagB/ClosCD) resulted in a previously unknown peptide with hemolytic phenotype. Although the native toxin, named clostridiolysin S, has thus far not been isolated from the producer organisms, the high similarity of the sag and clos gene clusters suggests that the compound produced in vitro may resemble the peptide encoded by the native host [34,35]. Subsequent searches for SagB homologs in genome databases identified a gene cluster in Listeria monocytogenes F2365 and H7858 that encodes a novel peptide designated listeriolysin S. Gene deletions in the native host confirmed that the cluster encodes for a virulence peptide displaying hemolytic activity [36]. A similar analysis predicted the production of staphylosin S by S. aureus ET3-1, although the functionality of the cluster has not yet been confirmed [36]. Interestingly, a recent search for SagBCD homologs using a combination of informatics tools, uncovered three large families of precursor peptides with highly conserved and long leader regions in phylogenetically diverse bacteria [37,38]. Some of the mature peptides are predicted to contain thiazole/oxazole heterocycles. However, the characteristic sagBCD homologs are absent from other genomes encoding these precursor peptides, suggesting that additional biosynthetic pathways and structurally diverse RiPPs remain to be discovered [38].
6. Genome mining for microviridins
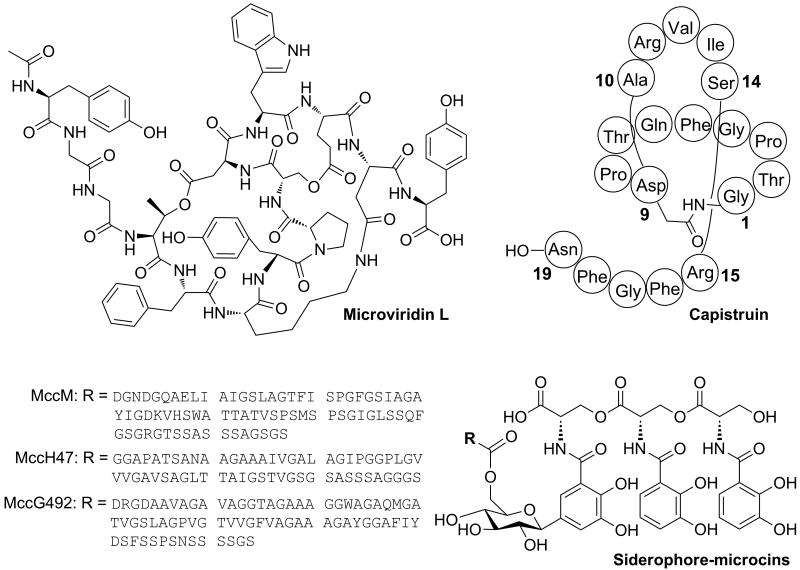
Microviridins are a family of N-acetylated tricyclic depsipeptides produced from a ribosomal precursor by cyanobacteria such as Microcystis aeruginosa and Planktothrix agardhii [39,40]. The precursor peptides are modified by ATP grasp ligases (MdnB, MdnC) that introduce ω-ester and ω-amide bonds (Figure 3) prior to cleavage of the leader peptide by a bifunctional transporter-peptidase. Analyses of genome data showed that microviridin-related biosynthetic gene clusters are present in several cyanobacterial genera including Anabaena, Nostoc, and Nodularia [39,40]. Recently, a collection of Microcystis strains, isolated from bodies of water around the world, was screened by PCR using degenerate primers for mdnB/C homologs [41]. Microviridin-like gene clusters were found in all tested strains, suggesting their global occurrence. Amplification of some of the precursor genes by PCR, followed by sequencing, uncovered seven novel microviridin precursors, and analysis of metagenomic DNA from field samples revealed eight additional precursor variants. To determine the functionality of the clusters, the Microcystis aeruginosa NIES843 gene cluster was heterologously expressed in E. coli allowing isolation of the novel peptide microviridin L and confirmation of the predicted chemical structure (Figure 3) [41].
Figure 3. Ribosomal peptides discovered by genome mining containing lactones and lactams or C-terminal siderophores.
The microviridin L biosynthetic gene cluster was detected in the genome of M. aeruginosa NIES843 after PCR screening and genomic DNA library construction. The capistruin gene cluster was identified in B. thailandensis E624 following orthologue neighborhood analysis of the gene cluster of the related peptide microcin J25 from E. coli. The siderophore-microcins MccM, MccH47, and MccG492 were discovered after genome sequence analysis of different Enterobacteriaceae.
7. Genome mining for lasso peptides
Lasso peptides are characterized by an N-terminal macrolactam ring that irreversibly traps a C-terminal tail, producing a complex and very stable three-dimensional lasso structure (e.g. Figure 3). The biosynthetic route to microcin J25 produced by E. coli AY25 was recently elucidated [42,43]. Two enzymes (McjB and McjC) catalyze the intramolecular cyclization of the ribosomally synthesized precursor molecule (McjA) and concomitant steric trapping of the C-terminal tail [42]. Ortholog neighborhood analysis of mcjB-like genes revealed several gene clusters from Bacillus, Burkholderia, Caulobacter, and Sphingopyxis encoding hypothetical lasso peptides with predictable structures [42,44]. In the case of Burkholderia thailandensis E264, screening for the molecular mass of the predicted cyclic peptide by LC-MS resulted in the detection and isolation of the novel lasso peptide capistruin (Figure 3) [44]. The structure of the peptide was determined by NMR spectroscopy and the compound was heterologously produced in E. coli confirming the involvement of the cluster in its biosynthesis [44].
8. Genome mining for siderophore-microcins
Siderophore-microcins are ribosomally synthesized peptides that contain a C-terminal glycosylated siderophore generated by the nonribosomal pathway. The gene cluster and biosynthetic pathway of MccE492 from Klebsiella pneumonia RYC492 have been characterized and four proteins (MceCDIJ) are known to be essential for the acquisition of the siderophore moiety [45-47]. Several bioinformatic studies suggested the existence of novel siderophore-microcins produced by members of the Enterobacteriaceae family [48,49]. In a recent study, two of the previously predicted microcins, MccM and MccH47, were isolated and characterized, demonstrating that both peptides contain the C-terminal salmochelin-like siderophore found in MccE492 (Figure 3) [50]. Additionally, further analysis of the MccE492 gene cluster suggested the existence of an additional siderophore-peptide named MccG492 (Figure 3) [50].
9. Genome mining for linaridins
Linaridins are linear peptides that contain dehydrated amino acids and additional posttranslational modifications such as N-terminal methylations or a C-terminal S-[(Z)-2-aminovinyl]-d-cysteine ring (AviCys). The biosynthetic gene cluster of cypemycin, the only structurally characterized member of the family, was recently identified [51], demonstrating that the characteristic dehydrobutyrine residues are potentially introduced by CypH/CypL, a pair of enzymes without homology to the corresponding dehydratases in lantipeptide biosynthesis. Searches for cypH/cypL homologs allowed the identification of 10 additional gene clusters present in different phyla of bacteria and archaea, establishing a novel family of ribosomal peptides [51].
10. Genome mining for amatoxins and phallotoxins from mushrooms
Amatoxins and phallotoxins are bicyclic octa- and heptapeptides, respectively, containing a Trp-Cys crossbridge (tryptathionine). They are produced by poisonous mushrooms of the genera Amanita, Galerina, Lepiota, and Conocybe [52]. Searches for homologs of the precursor genes of α-amanitin and phallacidin in the A. bisporigera draft genome revealed more than 20 ORFs having upstream and downstream conserved sequences and a hypervariable core region potentially encoding for amatoxin-like peptides [52]. Using degenerate primers that encode the conserved regions in the precursor peptides, the A. phalloides and A. ocreata genomes were screened by PCR, uncovering five additional genes likely encoding similar peptides, including one previously known compound [52]. Although the predicted peptides have not been isolated, the finding suggests that RiPP libraries of bioactive compounds are produced by several mushroom families.
11. Genome mining for cyclotides from plants
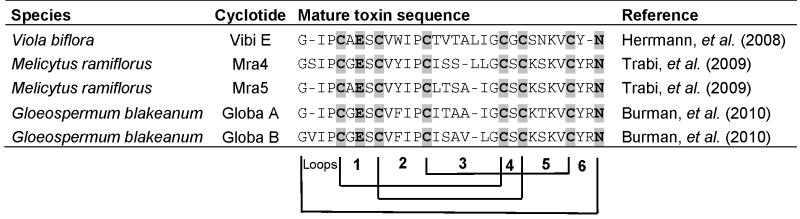
Cyclotides are head-to-tail cyclized peptides with a knotted arrangement of disulfide bonds produced by the Violaceae (violet), Rubiaceae (coffee), Cucurbitaceae, and Apocynaceae plant families (e.g. Table 1). They are biosynthesized as precursor peptides comprising an N-terminal endoplasmic reticulum (ER) signal, a leader peptide, and one to three core regions encoding the backbone(s) of the mature cyclotide(s) separated by small recognition sequences [53]. Screening programs suggest that individual plants contain genes encoding for up to 100 cyclotides, but only a few precursor genes have been identified. Cyclotide precursor sequences are only moderately conserved, with sequence identities of approximately 60%, which is challenging for development of a robust PCR strategy for the detection of novel compounds as described in section 12 for conopeptides. In a recent experiment, mRNA from Viola biflora was isolated and cDNA clones encoding cyclotides were generated by using 3′ rapid amplification of cDNA ends (RACE) and primers encoding a relatively well-conserved amino acid sequence in the ER signal peptide. Four novel cyclotide sequences were discovered using this strategy including one structure that was confirmed by LC-MS (Table 1) [54]. Similar studies in Melicytus ramiflorus, Gloeospermum blakeanum, G. pauciflorum, and Viola baoshanensis resulted in the identification of more than 40 novel cyclotide precursor genes, but only four molecules have been detected thus far by LC-MS (Table 1) [55-57]. An alternative strategy, based on the sequence analysis of an expression sequence tag (EST) library of Oldenlandia affinis (the producer of kalata B1, the first cyclotide discovered), revealed a new precursor gene putatively encoding the peptide kalata B19 [58], which has not been detected in O. affinis extracts. An extensive bioinformatics and expression analysis, combining BLAST searches for peptide sequences with the characteristic Cys spacing pattern, detection of N-terminal signal peptides, and analysis of EST databases uncovered several genes encoding cyclotide-like peptides from Poaceae species (including crops such as maize, wheat, and rice) [59]. Although none of these compounds has been isolated, and they may not be head-to-tail cyclized based on core peptide sequence, the finding suggests that ribosomal peptide natural products are considerably more widespread in plants than previously known.
Table 1.
Recently discovered cyclotides from genome mining approaches and LC-MS analysis. Six Cys residues involved in the Cys knot and two additional residues within the loops (a Glu in loop1 and an Asn/Asp in loop 6) are strictly conserved.
|
12. Genome mining for conopeptides from snails
The predatory marine cone snails produce a repertoire of small peptide-based neurotoxins named conopeptides with therapeutic potential [60]. Most conopeptides are highly crosslinked by disulfide bridges that generate a well-defined three-dimensional conformation, and many are decorated with other posttranslational modifications. The precursor peptides contain an N-terminal signal peptide, a leader region, and a hypervariable core peptide, and are posttranslationally modified in epithelial cells of the venom ducts. Some estimations suggest that the 500-700 living Conus species have evolved 50,000 to 140,000 different conopeptides, providing a large diversity of peptide-based neurotoxins, but less than 2% of these peptides have been isolated [60,61]. Recently, the discovery of new chemical entities has been dramatically accelerated by prediction of the mature peptide sequence from genomic DNA or from cDNA sequences, derived from mRNA extracted from the venom duct [62]. PCR with primers designed to recognize the conserved nucleotide sequence for the signal peptide and the 3′ untranslated region (3′-UTR) was used to sequence precursor genes of previously uncharacterized peptides from several Conus species [63-68] (Table 2). Direct sequencing of venom duct cDNA libraries from C. striatus and C. litteratus, together with bioinformatic analysis, has also provided more than 200 gene sequences encoding conopeptides [69,70]. Predicting the posttranslational modifications remains challenging, but structural homology with characterized peptide families can provide useful insights [68].
Table 2.
Examples of predicted mature conopeptides from cDNA sequencing.
| Species | Conotoxin | Mature peptide sequence | Molecular target | Reference |
|---|---|---|---|---|
| C. regius | α-RgIA | GCCSDPRCRYRCR | Nicotinic acetylcholine receptor, subunits α9 and α10 | Ellison, et al. (2006) |
| C. consors | CnlllA | GRCCDVPNACSGRWCRDHAQCC* | Tetrodotoxin-resistant sodium channels | Zhang, et al. (2006) |
| C. consors | CnlllB | ZGCCGEPNLCFTRWCRNNARCCRQQ | Tetrodotoxin-resistant sodium channels | Zhang, et al. (2006) |
| C. magnus | MIIA | ZGCCNVPNGCSGRWCRDHAQCC* | Tetrodotoxin-resistant sodium channels | Zhang, et al. (2006) |
| C. catus | CIIIA | GRCCEGPNGCSSRWCKDHARCC* | Tetrodotoxin-resistant sodium channels | Zhang, et al. (2006) |
| C. tulipa, C. striatus | TIIIA | RHGCCKGOKGCSSRECROQHCC* | Voltage gated sodium channel, subtypes Nav1.2 and Nav1.4 | Lewis, et al. (2007) |
| C. purpurascens | Con-P | GEγγHSKYQγCLRγlRVNKVQQγC* | N-methyl-d-aspartate receptor, subunits NR2B and NR2A | Gowd, et al. (2008) |
| C. ermine us | Con-E | GEγγHSKYQγCLRγlRVNNVQQγC* | N-methyl-d-aspartate receptor, subunits NR2B and NR2A | Gowd, et al. (2008) |
| C. pulicarius | PuSG1.1 | DDCCPDPACRQNHPELCSTR | Nicotinic acetylcholine receptors (undetermined) | Biggs, et al. (2008) |
| C. pulicarius | PuSG1.2 | DDCCPDPACRQNHPELCSSR | Nicotinic acetylcholine receptors (undetermined) | Biggs, et al. (2008) |
| C. bullatus | BulllA | VTDRCCKGKRECGRWCRDHSRCC* | Voltage gated sodium channel subtype Nav1.4 | Holford, et al. (2009) |
| C. bullatus | BulllB | VGERCCKNGKRGCGRWCRDHSRCC* | Voltage gated sodium channel subtype Nav1.4 | Holford, et al. (2009) |
| C. bullatus | BulllC | IVDRCCNKGNGKRGCSRWCRDHSRCC* | Voltage gated sodium channel subtype Nav1.4 | Holford, et al. (2009) |
z: pyroglutamate,
γ: γ-carboxyglutamate,
C-terminal amidation.
All cysteine residues are involved in define disulfide bridges (omitted).
Conus are not the only organisms producing RiPP toxins. For instance, novel molecules were recently discovered from venom mollusks from the turrid group that comprise approximately 10,000 species and that are likely to produce over a million pharmacologically active venom components [71]. Similarly, scorpions are expected to produce over 100,000 venom molecules, while millions of neurotoxic peptides are probably synthesized by 38,000 species of spiders [72]. Thus, the pool of biologically active peptides is enormous and advanced genome and transcriptome techniques will facilitate their discovery.
Summary and Outlook
The ever-increasing amount of DNA sequence data and the recent knowledge about the biosynthetic routes of RiPPs have lead to the discovery of novel gene clusters encoding previously unknown compounds. Upon identification of the clusters, prediction of the chemical structures of the mature peptides based on the genomic context of the precursor genes, in combination with several chemical and biological strategies, has aided bioassay-independent discovery and isolation of novel natural products. It is now clear that many species, from bacteria to eukarya, have taken advantage of combinatorial libraries of ribosomal peptides to efficiently evolve bioactive molecules that can mediate intra- and inter-species interactions. The recently discovered structural diversity introduced by posttranslational modifications has generated an exceptional pool of chemical entities that may aid in the development of novel peptide-based drug therapies.
Acknowledgments
Our work on RiPPs is supported by the National Institutes of Health (GM58822 to WAV). JEV was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award T32 GM070421.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review (2007-2010), have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Vlieghe P, Lisowski V, Martinez J, Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug Discov Today. 2010;15:40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Danho W, Swistok J, Khan W, Chu XJ, Cheung A, Fry D, Sun H, Kurylko G, Rumennik L, Cefalu J, et al. Opportunities and challenges of developing peptide drugs in the pharmaceutical industry. Adv Exp Med Biol. 2009;611:467–469. doi: 10.1007/978-0-387-73657-0_201. [DOI] [PubMed] [Google Scholar]
- 3.McIntosh JA, Donia MS, Schmidt EW. Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Nat Prod Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goto Y, Li B, Claesen J, Shi Y, Bibb MJ, van der Donk WA. Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol. 2010;8:e1000339. doi: 10.1371/journal.pbio.1000339. •This paper reports a new route for lantipeptide biosynthesis, discovered by mining of genome databases. The cryptic lantipeptide venezuelin was produced in vitro, after reconstitution of a novel lanthionine synthetase that defines a new class (class IV) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willey JM, van der Donk WA. Lantibiotics: Peptides of Diverse Structure and Function. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 7.Müller WM, Schmiederer T, Ensle P, Süssmuth RD. In vitro biosynthesis of the prepeptide of type-III lantibiotic labyrinthopeptin A2 including formation of a C-C bond as a post-translational modification. Angew Chem Int Ed Engl. 2010;49:2436–2440. doi: 10.1002/anie.200905909. •The authors demonstrate the first in vitro activity of a class III lanthionine synthetase, which contains a kinase domain for phosphorylation of Ser/Thr prior to elimination to generate dehydro amino acids. The enzyme also installs a novel crosslink (a labionine) that connects two Ala residues with a carbon-carbon bond. [DOI] [PubMed] [Google Scholar]
- 8.McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. Discovery and in vitro biosynthesis of haloduracin, a new two-component lantibiotic. Proc Natl Acad Sci USA. 2006;103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawton EM, Cotter PD, Hill C, Ross RP. Identification of a novel two-peptide lantibiotic, Haloduracin, produced by the alkaliphile Bacillus halodurans C-125. FEMS Microbiol Lett. 2007;267:64–71. doi: 10.1111/j.1574-6968.2006.00539.x. [DOI] [PubMed] [Google Scholar]
- 10.Cooper LE, McClerren AL, Chary A, van der Donk WA. Structure-Activity Relationship Studies of the Two-Component Lantibiotic Haloduracin. Chem Biol. 2008;15:1035–1045. doi: 10.1016/j.chembiol.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Begley M, Cotter PD, Hill C, Ross RP. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl Environ Microbiol. 2009;75:5451–5460. doi: 10.1128/AEM.00730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dischinger J, Josten M, Szekat C, Sahl HG, Bierbaum G. Production of the novel two-peptide lantibiotic lichenicidin by Bacillus licheniformis DSM 13. Plos One. 2009;4:e6788. doi: 10.1371/journal.pone.0006788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shenkarev ZO, Finkina EI, Nurmukhamedova EK, Balandin SV, Mineev KS, Nadezhdin KD, Yakimenko ZA, Tagaev AA, Temirov YV, Arseniev AS, et al. Isolation, structure elucidation, and synergistic antibacterial activity of a novel two-component lantibiotic lichenicidin from Bacillus licheniformis VK21. Biochemistry. 2010;49:6462–6472. doi: 10.1021/bi100871b. [DOI] [PubMed] [Google Scholar]
- 14.Daly KM, Upton M, Sandiford SK, Draper LA, Wescombe PA, Jack RW, O'Connor PM, Rossney A, Götz F, Hill C, et al. Production of the Bsa lantibiotic by community-acquired Staphylococcus aureus strains. J Bacteriol. 2010;192:1131–1142. doi: 10.1128/JB.01375-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li B, Sher D, Kelly L, Shi Y, Huang K, Knerr PJ, Joewono I, Rusch D, Chisholm SW, van der Donk WA. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc Natl Acad Sci U S A. 2010;107:10430–10435. doi: 10.1073/pnas.0913677107. •• A genome mining strategy was used to find examples of natural combinatorial biosynthesis. Single strains of the marine cyanobacteria Prochlorococcus produce a library of lantipeptides using a single lanthionine synthetase. The findings also indicate that lantipeptide biosynthesis is more widespread than appreciated previously. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Jong A, van Heel AJ, Kok J, Kuipers OP. BAGEL2: mining for bacteriocins in genomic data. Nucleic Acids Res. 2010;38(Suppl):W647–651. doi: 10.1093/nar/gkq365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majchrzykiewicz JA, Lubelski J, Moll GN, Kuipers A, Bijlsma JJ, Kuipers OP, Rink R. Production of a class II two-component lantibiotic of Streptococcus pneumoniae using the class I nisin synthetic machinery and leader sequence. Antimicrob Agents Chemother. 2010;54:1498–1505. doi: 10.1128/AAC.00883-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long PF, Dunlap WC, Battershill CN, Jaspars M. Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. ChemBioChem. 2005;6:1760–1765. doi: 10.1002/cbic.200500210. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 21.Leikoski N, Fewer DP, Sivonen K. Widespread occurrence and lateral transfer of the cyanobactin biosynthesis gene cluster in cyanobacteria. Appl Environ Microbiol. 2009;75:853–857. doi: 10.1128/AEM.02134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sudek S, Haygood MG, Youssef DT, Schmidt EW. Structure of trichamide, a cyclic peptide from the bloom-forming cyanobacterium Trichodesmium erythraeum, predicted from the genome sequence. Appl Environ Microbiol. 2006;72:4382–4387. doi: 10.1128/AEM.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziemert N, Ishida K, Quillardet P, Bouchier C, Hertweck C, de Marsac NT, Dittmann E. Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: from structure to genes and vice versa. Appl Environ Microbiol. 2008;74:1791–1797. doi: 10.1128/AEM.02392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Portmann C, Blom JF, Kaiser M, Brun R, Juttner F, Gademann K. Isolation of aerucyclamides C and D and structure revision of microcyclamide 7806A: Heterocyclic ribosomal peptides from Microcystis aeruginosa PCC 7806 and their antiparasite evaluation. J Nat Prod. 2008;71:1891–1896. doi: 10.1021/np800409z. [DOI] [PubMed] [Google Scholar]
- 25.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat Chem Biol. 2008;4:341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leikoski N, Fewer DP, Jokela J, Wahlsten M, Rouhiainen L, Sivonen K. Highly diverse cyanobactins in strains of the genus Anabaena. Appl Environ Microbiol. 2010;76:701–709. doi: 10.1128/AEM.01061-09. •• In this paper, a strategy is outlined for the discovery of cyanobactins from Anabena spp. based on genome data, bioinformatic analysis, isotope labeling, and MS. Several novel predicted compounds were detected in bacterial crude supernatants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wieland-Brown LCW, Acker MG, Clardy J, Walsh CT, Fischbach MA. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc Natl Acad Sci U S A. 2009;106:2549–2553. doi: 10.1073/pnas.0900008106. • One of four papers revealing the ribosomal origin of the thiopeptides. The authors used genome mining for the discovery of the biosynthetic gene cluster for thiocillin in Bacillus cereus. They also report the production of a previously unknown thiopeptide. Bioinformatic analysis indicates that similar clusters are widespread among bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J Am Chem Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. • One of four papers revealing the ribosomal origin of the thiopeptides. The authors report the biosynthetic gene cluster for thiostrepton. [DOI] [PubMed] [Google Scholar]
- 29.Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem Biol. 2009;16:141–147. doi: 10.1016/j.chembiol.2009.01.007. • One of four papers revealing the ribosomal origin of the thiopeptides. The authors report the biosynthetic gene cluster for thiostrepton and siomycin A, and demonstrate the production of thiocillins by B. cereus that was predicted by genome mining. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, Lamarche MJ, Parker CN, Burrer N, Esterow S, et al. Ribosomally Synthesized Thiopeptide Antibiotics Targeting Elongation Factor Tu. J Am Chem Soc. 2009:5946–5955. doi: 10.1021/ja900488a. • One of four papers revealing the ribosomal origin of the thiopeptides. The authors report the biosynthetic gene clusters for thiomuracin and GE2270A. [DOI] [PubMed] [Google Scholar]
- 31.Yu Y, Duan L, Zhang Q, Liao R, Ding Y, Pan H, Wendt-Pienkowski E, Tang G, Shen B, Liu W. Nosiheptide Biosynthesis Featuring a Unique Indole Side Ring Formation on the Characteristic Thiopeptide Framework. ACS Chem Biol. 2009;4:855–864. doi: 10.1021/cb900133x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engelhardt K, Degnes KF, Kemmler M, Bredholt H, Fjaervik E, Klinkenberg G, Sletta H, Ellingsen TE, Zotchev SB. Production of a new thiopeptide antibiotic, TP-1161, by a marine-derived Nocardiopsis species. Appl Environ Microbiol. 2010;76:4969–4976. doi: 10.1128/AEM.00741-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science. 1996;274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 34.Lee SW, Mitchell DA, Markley AL, Hensler ME, Gonzalez D, Wohlrab A, Dorrestein PC, Nizet V, Dixon JE. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc Natl Acad Sci USA. 2008;105:5879–5884. doi: 10.1073/pnas.0801338105. •• This paper describes the ribosomal origin of streptolysin S and demonstrates that it is a member of the thiazole/oxazole-containing peptides. Genome mining shows their biosynthetic gene clusters are widespread. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez DJ, Lee SW, Hensler ME, Markley AL, Dahesh S, Mitchell DA, Bandeira N, Nizet V, Dixon JE, Dorrestein PC. Clostridiolysin S, a post-translationally modified biotoxin from Clostridium botulinum. J Biol Chem. 2010;285:28220–28228. doi: 10.1074/jbc.M110.118554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cotter PD, Draper LA, Lawton EM, Daly KM, Groeger DS, Casey PG, Ross RP, Hill C. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008;4:e1000144. doi: 10.1371/journal.ppat.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haft DH. A strain-variable bacteriocin in Bacillus anthracis and Bacillus cereus with repeated Cys-Xaa-Xaa motifs. Biol Direct. 2009;4:15. doi: 10.1186/1745-6150-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haft DH, Basu MK, Mitchell DA. Expansion of ribosomally produced natural products: a nitrile hydratase- and Nif11-related precursor family. BMC Biol. 2010;8:70. doi: 10.1186/1741-7007-8-70. •• The authors used genome database mining to illustrate the widespread occurrence of the genes for lantipeptides and thiazole/oxazole containing peptides. In addition, they show that the genes encoding the precursor peptides are related to nitrile hydratase and Nif11 providing the first insights into the evolution of RiPP biosynthetic pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziemert N, Ishida K, Liaimer A, Hertweck C, Dittmann E. Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angew Chem, Int Ed Engl. 2008;47:7756–7759. doi: 10.1002/anie.200802730. [DOI] [PubMed] [Google Scholar]
- 40.Philmus B, Christiansen G, Yoshida WY, Hemscheidt TK. Post-translational modification in microviridin biosynthesis. ChemBioChem. 2008;9:3066–3073. doi: 10.1002/cbic.200800560. [DOI] [PubMed] [Google Scholar]
- 41.Ziemert N, Ishida K, Weiz A, Hertweck C, Dittmann E. Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Appl Environ Microbiol. 2010;76:3568–3574. doi: 10.1128/AEM.02858-09. • The authors explored the natural diversity of microviridins by PCR and report the identification and characterization of a novel molecule that was produced heterologously in E. coli. The study demonstrates great promise for production of cryptic microviridins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duquesne S, Destoumieux-Garzon D, Zirah S, Goulard C, Peduzzi J, Rebuffat S. Two Enzymes Catalyze the Maturation of a Lasso Peptide in Escherichia coli. Chem Biol. 2007;14:793–803. doi: 10.1016/j.chembiol.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Clarke DJ, Campopiano DJ. Maturation of McjA precursor peptide into active microcin MccJ25. Org Biomol Chem. 2007;5:2564–2566. doi: 10.1039/b708478a. [DOI] [PubMed] [Google Scholar]
- 44.Knappe T, Linne U, Zirah S, Rebuffat S, Xie XL, Marahiel M. Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J Am Chem Soc. 2008;130:11446–11454. doi: 10.1021/ja802966g. [DOI] [PubMed] [Google Scholar]
- 45.Wilkens M, Villanueva JE, Cofre J, Chnaiderman J, Lagos R. Cloning and expression in Escherichia coli of genetic determinants for production of and immunity to microcin E492 from Klebsiella pneumoniae. J Bacteriol. 1997;179:4789–4794. doi: 10.1128/jb.179.15.4789-4794.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nolan EM, Fischbach MA, Koglin A, Walsh CT. Biosynthetic tailoring of microcin E492m: post-translational modification affords an antibacterial siderophore-peptide conjugate. J Am Chem Soc. 2007;129:14336–14347. doi: 10.1021/ja074650f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vassiliadis G, Peduzzi J, Zirah S, Thomas X, Rebuffat S, Destoumieux-Garzon D. Insight into siderophore-peptide biosynthesis: Enterobactin is a precursor for microcin E492 post-translational modification. Antimicrob Agents Chemother. 2007;51:3546–3553. doi: 10.1128/AAC.00261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duquesne S, Destoumieux-Garzon D, Peduzzi J, Rebuffat S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep. 2007;24:708–734. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 49.Poey ME, Azpiroz MF, Lavina M. Comparative analysis of chromosome-encoded microcins. Antimicrob Agents Chemother. 2006;50:1411–1418. doi: 10.1128/AAC.50.4.1411-1418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vassiliadis G, Destoumieux-Garzon D, Lombard C, Rebuffat S, Peduzzi J. Isolation and characterization of two members of the siderophore-microcin family, microcins M and H47. Antimicrob Agents Chemother. 2010;54:288–297. doi: 10.1128/AAC.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Claesen J, Bibb M. Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc Natl Acad Sci USA. 2010;107:16297–16302. doi: 10.1073/pnas.1008608107. • This study demonstrates that cypemycin is not a member of the lantipeptides as its biosynthetic gene cluster lacks the characteristic biosynthetic genes. Genome mining shows that the genes involved in cypemycin biosynthesis are also found in other bacteria and the name linaridin is suggested for the new family of RiPPs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hallen HE, Luo H, Scott-Craig JS, Walton JD. Gene family encoding the major toxins of lethal Amanita mushrooms. Proc Natl Acad Sci USA. 2007;104:19097–19101. doi: 10.1073/pnas.0707340104. •• First demonstration that the lethal amatoxins and phallotoxins produced by mushrooms are not of non-ribosomal origin but are ribosomally synthesized and posttranslationally modified. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gillon AD, Saska I, Jennings CV, Guarino RF, Craik DJ, Anderson MA. Biosynthesis of circular proteins in plants. Plant J. 2008;53:505–515. doi: 10.1111/j.1365-313X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- 54.Herrmann A, Burman R, Mylne JS, Karlsson G, Gullbo J, Craik DJ, Clark RJ, Goransson U. The alpine violet, Viola biflora, is a rich source of cyclotides with potent cytotoxicity. Phytochemistry. 2008;69:939–952. doi: 10.1016/j.phytochem.2007.10.023. • This paper outlines a strategy using PCR techniques to sequence precursor genes that encode for cyclotides. Some of the molecules isolated from the producer plant showed potent cytotoxicity. [DOI] [PubMed] [Google Scholar]
- 55.Trabi M, Mylne JS, Sando L, Craik DJ. Circular proteins from Melicytus (Violaceae) refine the conserved protein and gene architecture of cyclotides. Org Biomol Chem. 2009;7:2378–2388. doi: 10.1039/b823020j. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J, Hu M, Li JT, Guan JP, Yang B, Shu WS, Liao B. A transcriptional profile of metallophyte Viola baoshanensis involved in general and species-specific cadmium-defense mechanisms. J Plant Physiol. 2009;166:862–870. doi: 10.1016/j.jplph.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 57.Burman R, Gruber CW, Rizzardi K, Herrmann A, Craik DJ, Gupta MP, Goransson U. Cyclotide proteins and precursors from the genus Gloeospermum: Filling a blank spot in the cyclotide map of Violaceae. Phytochemistry. 2010;71:13–20. doi: 10.1016/j.phytochem.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 58.Qin Q, McCallum EJ, Kaas Q, Suda J, Saska I, Craik DJ, Mylne JS. Identification of candidates for cyclotide biosynthesis and cyclisation by expressed sequence tag analysis of Oldenlandia affinis. Bmc Genomics. 2010;11:111. doi: 10.1186/1471-2164-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mulvenna JP, Mylne JS, Bharathi R, Burton RA, Shirley NJ, Fincher GB, Anderson MA, Craik DJ. Discovery of cyclotide-like protein sequences in graminaceous crop plants: Ancestral precursors of circular proteins? Plant Cell. 2006;18:2134–2144. doi: 10.1105/tpc.106.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olivera BM, Teichert RW. Diversity of the neurotoxic Conus peptides: A model for concerted pharmacological discovery. Mol Interv. 2007;7:251–260. doi: 10.1124/mi.7.5.7. [DOI] [PubMed] [Google Scholar]
- 61.Kaas Q, Westermann JC, Craik DJ. Conopeptide characterization and classifications: An analysis using ConoServer. Toxicon. 2010;55:1491–1509. doi: 10.1016/j.toxicon.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 62.Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. J Biol Chem. 2006;281:31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- 63.Bulaj G, West PJ, Garrett JE, Watkins M, Zhang MM, Norton RS, Smith BJ, Yoshikami D, Olivera BM. Novel conotoxins from Conus striatus and Conus kinoshitai selectively block TTX-resistant sodium channels. Biochemistry. 2005;44:7259–7265. doi: 10.1021/bi0473408. [DOI] [PubMed] [Google Scholar]
- 64.Ellison M, Haberlandt C, Gomez-Casati ME, Watkins M, Elgoyhen AB, McIntosh JM, Olivera BM. alpha-RgIA: A novel conotoxin that specifically and potently blocks the alpha 9 alpha 10 nAChR. Biochemistry. 2006;45:1511–1517. doi: 10.1021/bi0520129. [DOI] [PubMed] [Google Scholar]
- 65.Zhang MM, Fiedler B, Green BR, Catlin P, Watkins M, Garrett JE, Smith BJ, Yoshikami D, Olivera BM, Bulaj G. Structural and functional diversities among mu-conotoxins targeting TTX-resistant sodium channels. Biochemistry. 2006;45:3723–3732. doi: 10.1021/bi052162j. [DOI] [PubMed] [Google Scholar]
- 66.Biggs JS, Olivera BM, Kantor YI. alpha-conopeptides specifically expressed in the salivary gland of Conus pulicarius. Toxicon. 2008;52:101–105. doi: 10.1016/j.toxicon.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gowd KH, Twede V, Watkins M, Krishnan KS, Teichert RW, Bulaj G, Olivera BM. Conantokin-P, an unusual conantokin with a long disulfide loop. Toxicon. 2008;52:203–213. doi: 10.1016/j.toxicon.2008.04.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holford M, Zhang MM, Gowd KH, Azam L, Green BR, Watkins M, Ownby JP, Yoshikami D, Bulaj G, Olivera BM. Pruning nature: Biodiversity-derived discovery of novel sodium channel blocking conotoxins from Conus bullatus. Toxicon. 2009;53:90–98. doi: 10.1016/j.toxicon.2008.10.017. • The authors demonstrate a strategy that combines molecular biology techniques and phylogenetic analysis to discover novel conopeptides. After identification of precursor genes, the predicted peptides were chemically synthesized generating pharmacologically active compounds. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pi C, Liu J, Peng C, Liu Y, Jiang X, Zhao Y, Tang S, Wang L, Dong M, Chen S, et al. Diversity and evolution of conotoxins based on gene expression profiling of Conus litteratus. Genomics. 2006;88:809–819. doi: 10.1016/j.ygeno.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 70.Pi C, Liu Y, Peng C, Jiang X, Liu J, Xu B, Yu X, Yu Y, Wang L, Dong M, et al. Analysis of expressed sequence tags from the venom ducts of Conus striatus: focusing on the expression profile of conotoxins. Biochimie. 2006;88:131–140. doi: 10.1016/j.biochi.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 71.Watkins M, Hillyard DR, Olivera BM. Genes expressed in a turrid venom duct: Divergence and similarity to conotoxins. J Mol Evol. 2006;62:247–256. doi: 10.1007/s00239-005-0010-x. [DOI] [PubMed] [Google Scholar]
- 72.Bulaj G. Integrating the discovery pipeline for novel compounds targeting ion channels. Curr Opin Chem Biol. 2008;12:441–447. doi: 10.1016/j.cbpa.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]