Abstract
Regulators of G-Protein signaling (RGS) proteins are potent negative modulators of signal transduction through G-Protein coupled receptors. They function by binding to activated (GTP-bound) Gα subunits and accelerating the rate of GTP hydrolysis. Modulation of RGS activity by small molecules is an attractive mechanism to fine-tune GPCR signaling for therapeutic and research purposes. Here we describe the pharmacologic properties and mechanism of action of CCG-50014, the most potent small molecule RGS inhibitor to date. It has an IC50 for RGS4 of 30 nM and is >20-fold selective for RGS4 over other RGS proteins. CCG-50014 binds covalently to the RGS, forming an adduct on two cysteine residues located in an allosteric regulatory site. It is not a general cysteine alkylator as it does not inhibit activity of the cysteine protease papain at concentrations >3,000 fold higher than those required to inhibit RGS4 function. It is also >1,000-fold more potent as an RGS4 inhibitor than are the cysteine alkylators N-ethylmaleimide or iodoacetamide. Analysis of the cysteine reactivity of the compound shows that compound binding to Cys107 in RGS8 inhibits Gα- binding in a manner that can be reversed by cleavage of the compound-RGS disulfide bond. If the compound reacts with Cys160 in RGS8, the adduct induces RGS denaturation and activity cannot be restored by compound removal. The high potency and good selectivity of CCG-50014 make it a useful tool for studying the functional roles of RGS4.
Keywords: RGS protein, Small molecule protein-protein interaction inhibitor, GPCR
G-Protein Coupled Receptors (GPCRs) are important drug targets with profound clinical relevance. While direct modulation of GPCR activity with traditional pharmacological agents (orthosteric agonists/antagonists) has proven incredibly successful, compounds that provide more subtle modulation of receptor signaling may provide advantages over traditional GPCR-targeting drugs. Several approaches towards this goal have been proposed, including pathway-biased GPCR ligands (1), allosteric receptor modulators (2), or compounds that interfere with specific downstream modulators of G-Protein signaling (3). In this report we describe the properties and mechanism of a novel small molecule compound that potently inhibits a Regulator of G protein Signaling (RGS), an important modulator of GPCR signal transduction cascades.
The RGS proteins are negative modulators of many G-Protein Coupled Receptor (GPCR) signaling pathways (4). RGS proteins bind directly to the GTP-bound Gα subunit of activated heterotrimeric G-Proteins and increase the rate of GTP hydrolysis (5). By this mechanism, RGS proteins rapidly dampen GPCR signal transduction at the level of the active G protein subunits. This accelerated turnAoff of activated G-Proteins provides a cellular mechanism for enhanced temporal and spatial resolution of GPCR signaling (6, 7).
Genetic ablation of RGS activity either by deletion of a particular RGS gene or by expression of RGS-insensitive Gα subunits has dramatic physiological consequences (for review, see (6)). For example, RGS4-deficient mice display increased sensitivity to carbachol-potentiated glucose-stimulated insulin release (9). Deletion of RGS9 produces a variety of neurological effects, including sensitization to morphine analgesia and reward with decreased tolerance, deficits in working memory, and motor coordination defects (10, 11). The physiological effects of pan-RGS inhibition have also been studied. A knock- in mouse model has been developed that expresses a Gαi2 subunit with a single amino acid mutation (G184S) that prevents functional interactions with RGS proteins (7). These mice show dramatic phenotypes, including spontaneous antidepressant-like effects as well as resistance to diet-induced obesity (13, 14). Thus, modulating the activity of specific RGS proteins may provide significant therapeutic benefit (3, 8, 15). To this end, we have developed several classes of small molecule RGS inhibitors for use as pharmacological tools and as potential therapeutics (16, 17).
Inhibiting protein-protein interactions, such as the one between an RGS and a Gα subunit is particularly challenging (3, 18, 19). This is predominantly due to the lack of a suitable small molecule binding pocket at the protein-protein interaction interface. However, the GTPase accelerating activity of RGS4 is regulated by phosphotidylinositol 3,4,5 triphosphate at a site far removed from the Gα interaction interface (8, 9). Targeting this allosteric site (10) might be a more tractable approach towards inhibiting the RGS-Gα protein-protein interaction than attempting to orthosterically occlude the protein-protein interaction.
CCG-50014 was discovered in a high throughput biochemical screen designed to identify inhibitors of 5 different RGS proteins (11). This compound was the most potent inhibitor from this screen with a nanomolar IC50 value. In this article we confirm the potent and selective RGS inhibitory activity of this compound and describe its biochemical mechanism of action. Furthermore, we provide evidence that CCG-50014 is able to inhibit the RGS4-Gαo protein-protein interaction in a living cell. This compound represents the first of a class of small molecule RGS inhibitors that function in a cellular environment.
Experimental Procedures
Materials
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Hampton, NH) and were reagent grade or better. γ[32P]GTP (10 mCi/mL) and [35S]GTPγS (12.5 mCi/mL) were obtained from Perkin Elmer Life and Analytical Sciences (Boston, MA) and were isotopically diluted before use. Amylose resin was purchased from New England Biolabs (Ipswich, MA). Ni-NTA resin was purchased from Qiagen (Valencia, CA). Avidin-coated microspheres were purchased from Luminex (Austin, TX). CCG-50014 (4A[(4-[(4-fluorophenyl)methyl]-2-(4-methylphenyl)-1,2,4-thiadiazolidine-3,5-dione) and analogs were purchased from the Maybridge compound collection (Fisher Scientific, Waltham, MA), or were as described below using variation of previously published methods (12).
Synthesis
A Bruker Avance III 400 MHz NMR spectrometer was used to collect 1H and 13C NMR spectra. ESI mass spectra were recorded using a Bruker micrOTOF. Column chromatography was performed with a Combiflash Rf Companion, using Redisep Rf disposable columns containing 40–60 micron silica.
CCG-50014 (4-(4-fluorobenzyl)-2-p-tolyl-1,2,4-thiadiazolidine-3,5-dione)
N-chlorosuccinimide (4.0 g, 30.0 mmol) was added to a solution of 4-fluorobenzyl isothiocyanate (1.25 g, 7.5 mmol) and p-tolyl isocyanate (1.0 g, 7.5 mmol) in CHCl3 (70 mL) under nitrogen at room temperature. This solution was stirred for 18 h then opened to the air and stirred for an additional 30 min. The reaction was diluted with Et2O (30 mL) and filtered through a sintered funnel. The residue was washed with further Et2O (15 mL) and solvent was removed under reduced pressure. The crude product was purified by column chromatography (20% ethyl acetate/hexane), resulting in a pale yellow solid (970 mg, 41%) with spectral data identical to the Maybridge sample.
1H NMR (400 MHz, CDCl3) δ 2.35 (s, 3H), 4.87 (s, 2H), 7.01–7.05 (m, 2H), 7.20–7.22 (m, 2H), 7.35–7.38 (m, 2H), 7.48–l7.51 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 20.9, 45.3, 115.5, 115.7, 123.7, 130.0, 130.9, 131.0, 131.1, 133.0, 137.3, 151.0, 161.5, 163.9, 165.1.
CCG-203778 (3-(4-methylbenzyl)-1-p-tolylimidazolidine-2,4-dione)
Ethyl bromoacetate (3.3 g, 20 mmol) was added to a solution of p-toluidine (2.1 g, 20 mmol) and sodium acetate (2.1 g, 26 mmol) in ethanol (26 mL). The resulting solution was warmed to 80 oC and stirred for 1 h before being cooled to room temperature. The reaction was quenched with water (20 mL) and the aqueous fraction extracted with ethyl acetate (3 × 20 mL). The organic fractions were combined, dried (MgSO4), filtered and the solvent removed under reduced pressure. Silica chromatography (0–20% ethyl acetate/hexane) provided ethyl 2-(p-tolylamino)acetate as a pale oil (1.9 g, 50%).
Ethyl 2-(p-tolylamino)acetate (260 mg, 1.4 mmol) was dissolved in toluene (5 mL). Methyl benzylisocyanate (200 mg, 1.4 mmol) was added to this solution. The mixture was heated to reflux and stirred for 5 h, until TLC analysis demonstrated that the reaction was complete. The reaction was then cooled to room temperature and the solvent was removed under reduce pressure. Silica chromatography (0–20% ethyl acetate/hexane) provided ethyl 2-(3-(4-methylbenzyl)-1-p-tolylureido)acetate as a pale oil (455 mg, 98%).
Ethyl 2-(3-(4-methylbenzyl)-1-p-tolylureido)acetate (455 mg, 1.3 mmol) was dissolved in THF (10 mL) and added dropwise to a solution of NaH (68 mg, 2.8 mmol) in THF (10 mL) at 0 °C. The reaction was allowed to warm to rt slowly and stirred for 18 h before being quenched with water (5 ml). The aqueous fraction was then extracted with dichloromethane (3 × 20 mL). The organic fractions were combined, dried (MgSO4), filtered and the solvent removed under reduced pressure. Column chromatography (0–20% ethyl acetate/hexane) provided CCG-203778 (360 mg, 93%) as a white solid.
1H NMR (400 MHz, CDCl3) δ 2.33 (s, 6H), 4.25 (s, 2H), 4.71 (s, 2H), 7.13–7.15 (m, 2H), 7.16–7.18 (m, 2H), 7.36–7.38 (m, 2H), 7.40–7.43 (m, 2H). 13C NMR (100 MHz, CDCl3) δ 20.7, 21.1, 42.4, 50.0, 118.5, 128.9, 129.4, 129.8, 132.9, 134.2, 135.0, 137.9, 154.1. HRMS (ESI): [M+H]+ found 295.1448. C18H19N2O2 requires 295.1446.
Protein expression and purification
With the exception of RGS8 and mutants thereof, all RGS and G proteins were prepared as previously described (13). Briefly, an N-terminally truncated (Δ51) form of RGS4 was expressed with an N- terminal maltose binding protein fusion tag and a 10× histidine tag from the pMALC2H10 vector. Full-length RGS16 and a C-terminally truncated form of RGS19 (ΔC11) were also expressed as MBP/10× histidine tag fusions using the pMALC2H10 vector. The RGS7 homology domain was expressed as a GST fusion as previously described (14). The RGS8 homology domain was expressed as an N-terminal 6× histidine tag from the pQE-80 vector and was purified as previously described (15). This construct contains two cysteine residues from RGS8 – one at position 107 and 160. For the RGS8 cysteine → serine mutants, site directed mutagenesis was performed using the following primers for 107C RGS8 (C160S) (Sense: 5'-GCAGGAGCCATCCCTGACTAGCTTTGACCAAG-3'; Antisense: 5'-CGTCCTCGGTAGGGACTGATCGAAACTGGTTC-3'), and 160C RGS8 (C107S) (Sense: 5'-TGGAATTCTGGTTGGCCAGTGAGGAGTTCAAGAAG-3'; Antisense: 5'-ACCTTAAGACCAACCGGTCACTCCTCAAGTTCTTC-3'). Mutagenesis was performed using the QuickChange Multi-site Directed Mutagenesis kit (Agilent, La Jolla CA). Mutants were sequenced at the University of Michigan DNA Sequencing Core. Gαo was expressed with an N-terminal 6× histidine tag as previously described (16). G protein activity was determined by [35S]GTPγS binding (17). In all cases, proteins were purified to >90% homogeneity before use. See1 for a concise description of RGS8 cysteine mutant nomenclature.
Chemical labeling of purified Gαo and RGS proteins
RGS proteins were chemically biotinylated on free amines and Gαo was labeled with AlexaFluor-532 on free thiols as previously described (18).
Flow Cytometry Protein Interaction Assay (FCPIA) Dose Response and Reversibility experiments
FCPIA was performed as previously described using chemically biotinylated RGS proteins and AlexaFluor-532 labeled Gαo (18, 19). In brief, biotintylated RGS proteins were immobilized on LumAvidin microspheres (Luminex, Austin, Tx) and treated with compound or vehicle control for 15 minutes at room temperature. The beads were then mixed with fluorescently-labeled Gα in the presence of guanine diphosphate and aluminum fluoride and assessed for association with the RGS protein using a Luminex 200 flow cytometer.
Single Turnover GTPase Measurements
Compounds were tested for the ability to inhibit the RGS4 and RGS8-stimulated increase in GTP hydrolysis by Gαo as previously described (17, 26). Data are presented as the rate constant (k, sec−1) of GTP hydrolysis.
Thermal Stability Measurements
The thermal denaturation of RGS4, RGS8 and Gαo was measured using a ThermoFluor Instrument (Johnson & Johnson, Langhorne, PA). Protein (10 μM RGS4, 5 μM RGS8 or Gαo) was incubated with CCG-50014 or vehicle control for 15 minutes at room temperature in 50 mM HEPES pH 8.2, 500 mM NaCl, 5% glycerol in a volume of 15 μL in a black 384-well PCR microtiter plate (ThermoFisher Cat # TF-0384/K). To this mixture was added 200 μM 1-anilinonapthalene-8-sulfonic acid. The samples were overlaid with 5 μL of silicone oil and subjected to a temperature ramp with intervening measurements at 25°C. The experiments were performed using the following parameters: ramp temperature range: 30–90°C; temperature increment: 1°C; image collection temperature: 25°C; temperature holds: 30 seconds for ramp temperature, 15 seconds for image collection temperature. Melting temperatures (Tm) were calculated from the fluorescence data using the sigmoidal fitting procedure in the ThermoFluor++ software package (version 1.3.7).
Analysis of the protein adduct of RGS by ESI-LC/MS
The molecular mass of the RGS protein was analyzed by ESI-LC/MS using a LCQ ion-trap mass spectrometer (ThermoScientific, Waltham, MA). RGS8 wild-type or mutant proteins were diluted to 2 μM in 50 mM potassium phosphate buffer, pH 7.4 and CCG-50014 or an equivalent volume of DMSO was added to the sample. Following treatment with CCG-50014, an aliquot (~50 μL) of the protein solution was applied to a reverse-phase Zorbax 300-SB C3 column (2×150 mm, 5 μm) (Agilent Technologies, CA). The RGS protein was subjected to high performance liquid chromatography with a binary solvent system consisting of 0.1% TFA in water (Solvent A) and 0.1% TFA in acetonitrile (Solvent B) using the following gradient: 30% B for 5 min., linear increase to 90% B in 20 min., and 90% B for 30 min. The flow rate was 0.25 mL/min. The mass spectrometer was tuned with horse heart cytochrome c and the instrumental settings for the mass spectrometer were: spray voltage, 3.5 kV; capillary temperature, 220°C; sheath gas flow, 80 (arbitrary units); auxiliary gas flow, 20 (arbitrary units). The molecular masses of the unmodified and inhibitor-modified RGS proteins were determined by deconvolution of the apoprotein charge envelopes using the Bio-works software (Thermo Scientific, Waltham, MA).
Papain Activity Assay
Papain (Sigma-Aldrich, St. Louis, MO) activity was monitored by the increase in fluorescence produced by the liberation of fluorescein from auto-quenched fluorescein-conjugated casein (FITC-casein, AnaSpec, San Jose, CA). Papain (0.625 U) was diluted into a final reaction volume of 100 μL in 20 mM sodium acetate pH 6.5 supplemented with 2 mM EDTA. The enzyme was treated with iodoacetamide, N-ethyl maleimide, CCG-50014, or vehicle control for 30 minutes at ambient temperature. To this, FITC-casein was added to a final concentration of 250 nM. The reaction was allowed to proceed at ambient temperature in the dark. At various times, the fluorescence intensity (ex. 485 nm, em. 520 nm) was measured using a Victor II plate reader (Perkin Elmer, Boston, MA). As a control, CCG-50014 was tested using FCPIA at pH 6.5 and it retains full inhibitory activity against the RGS4-Gαo protein-protein interaction.
Docking of CCG-50014 to RGS8
The energy-based docking software Autodock (ver. 4.0) was used to explore potential binding sites of CCG-50014 on RGS8. The coordinates of RGS8 were obtained from the Protein Data Bank (PDB ID 2IHD). Water and a chloride ion were removed from the structure prior to docking. The coordinates of the CCG-50014 ligand were built using the ChemBioOffice 2008 software suite (CambridgeSoft, Cambridge, MA) and the geometry of CCG-50014 was optimized using the semi-empirical quantum PM3 method included in the ChemBioOffice 2008 software suite. For unbiased docking, the grid box of the RGS was set at 60×60×60 Å3 to encompass the entire RGS protein. The flexible CCG-50014 ligand was docked to the rigid RGS using a Lamarckian Genetic Algorithm with the following parameters: mutation rate, 0.02; cross-over rate, 0.8; maximal number of generations, 2.7×105.
RGS4 Membrane Translocation
HEK-293T cells were grown to 80–90% confluency in 6-well dishes in DMEM supplemented with 10% fetal bovine serum and Penicillin (100 units/ml)-Streptomycin (100 μg/ml) under 5% CO2 at 37 °C. RGS and Gαo expression was induced by transient co-transfection with 250 ng of full-length human RGS4 with an N-terminal GFP tag (RGS4pEGFP-C1), 250 ng of pcDNA3.1 or pcDNA3.1 encoding wildtype human Gαo or both. Cells were seeded onto poly-D-lysine coated glass coverslips and cultured for 24–48 hours before live cell imaging. Images were acquired on an Olympus Fluoview 500 confocal microscope with a 60 × 1.40 numerical aperture oil objective. Images were obtained by taking a series of stacks every 0.5 μM through the cell. The light source for the fluorescent studies was a 488 nm laser with a 505–525 nm bandpass filter. Images were quantified using NIH ImageJ software version 1.43r and GraphPad Prism v. 5.04.
Results
FCPIA characterization of RGS inhibitory activity
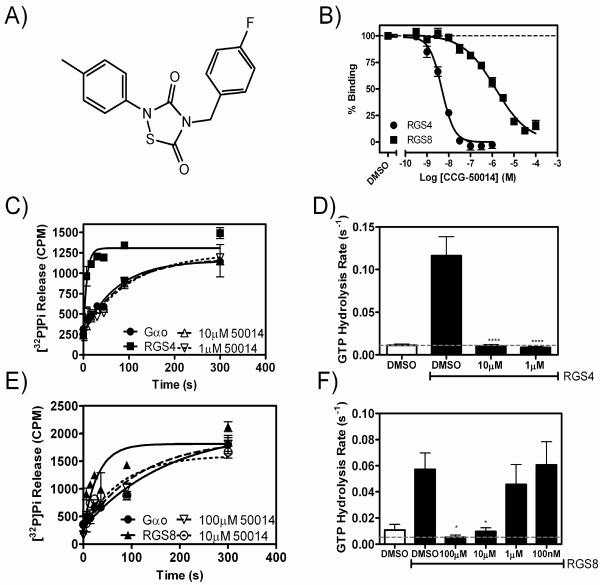
CCG-50014 (Fig. 1A) was originally identified as an inhibitor of RGS4, 8 and 16 in a polyplex high throughput screen to identify inhibitors of the RGS-Gα interaction (11). Here we confirm that CCG-50014 is highly potent at inhibiting RGS4 (IC50 30 nM) and show that it fully inhibits several other RGS proteins including RGS8, 16, and 19, but did not have activity on RGS7 (Fig. 1 and Table 1). Furthermore, the compound has no activity on a mutated form of RGS4 that lacks cysteine residues in the RGS homology domain (RGS4Cys−), suggesting that CCG-50014 might be thiol-reactive (Table 1).
Figure 1. The chemical structure and RGS inhibitory activity of CCG-50014.
A) The chemical structure of CCG-50014 (4-[(4-fluorophenyl)methyl]-2-(4-methylphenyl)-1,2,4-thiadiazolidine-3,5-dione). B) CCG-50014 concentration-dependently inhibits the binding between aluminum fluoride-activated Gαo and RGS4 or RGS8. Data shown are an average of three independent experiments. This experiment has been independently repeated 28 times, producing average IC50 values of 30±6 nM against RGS4 and 11±2 μM against RGS8. C,D) CCG-50014 also inhibits the GAP activity of RGS4 and E,F) RGS8. Using a single-turnover GAP assay, CCG-50014 concentration-dependently inhibits the GAP activity of both RGS4 and RGS8. * P <0.05, *** P <0.0001. All experiments were independently repeated a minimum of three times.
Table 1.
RGS specificity for inhibition of Gα/RGS binding.
| RGS | IC50 (μM) ± SEM | Hill Slope |
|---|---|---|
| RGS4 wild Type | 0.030 ± 0.006 | −1.53 |
| RGS4Cys−* | N/A | N/A |
| RGS7 | N/A | N/A |
| RGS8 | 11 ± 2 | −0.99 |
| RGS16 | 3.5 ± 2.4 | −1.33 |
| RGS19 | 0.12 ± 0.02 | −0.61 |
IC50 values in the FCPIA assay show that CCG-50014 is >40 fold more potent for RGS4 than other RGS proteins. Data are presented as: mean IC50 values ± SEM from at least three independent experiments (for RGS4 and RGS8, n >28). N/A: No inhibition below the aqueous solubility limit of the compound (~200 μM).
RGS4Cys− is a mutated form of RGS that contains no cysteine residues in the RGS homology domain (13).
CCG-50014 inhibits the catalytic GTPase accelerating activity of RGS8 and RGS4
In a single turnover GTPase assay, CCG-50014 inhibited the GTPase accelerating activity of RGS8 and RGS4 on Gαo (Fig. 1C–F). Under these assay conditions, RGS8 and RGS4 accelerate the rate of Gαo-mediated GTP hydrolysis by approximately 5- and 10-fold, respectively. CCG-50014 inhibited the activity of both RGS proteins. At a maximal concentration (100 μM), CCG-50014 did not alter the intrinsic rate of GTP hydrolysis by Gαo, proving that the compound does not act by altering the enzymatic activity of the G protein under single-turnover conditions (Fig. S1).
CCG-50014 Binds to RGS Proteins but not to Gαo
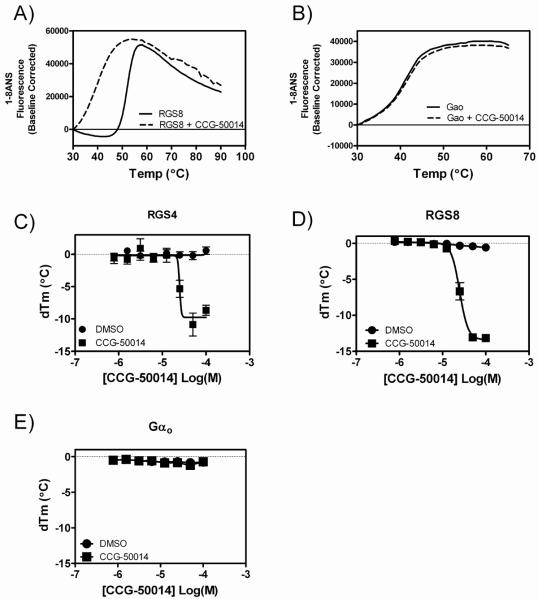
The melting temperature of a protein can be influenced by the binding of small molecules (27, 28). The thermal denaturation of RGS4, RGS8 and Gαo was characterized in the presence and absence of CCGA-50014 using a ThermoFluor® instrument. CCGA-50014 concentration-dependently destabilized RGS4 and RGS8 (Fig. 2A,C,D) but had no effect on Gαo (Fig. 2B,E). This suggests that the compound is interacting exclusively with the RGS protein. Compound binding to RGS8 was further confirmed by Liquid Chromatography-Mass Spectral (LC-MS) analysis as described below.
Figure 2. CCG-50014 thermally destabilizes RGS8 in a concentration-dependent manner, but has no effect on the thermal stability of Gαo.
Representative melting traces of A) RGS8 and B) Gαo in the absence (solid trace) and presence (dashed trace) of 100 μM CCG-50014. Concentration-response curves showing the thermal destabilization effects of CCG-50014 on C) RGS4, D) RGS8 and E) Gαo. Data are presented as the mean±SEM of three independent experiments.
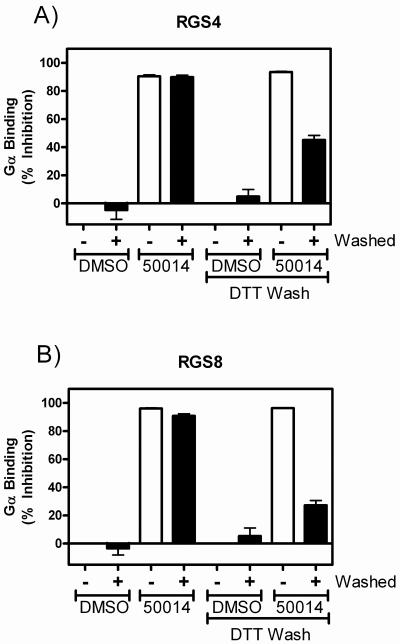
CCG-50014 irreversibly inhibits RGS proteins
FCPIA-based reversibility experiments were performed to probe the mechanism of action of the compound on both RGS4 and RGS8 (Fig. 3). RGS-coated polystyrene beads were incubated with a saturating concentration (100 μM) of CCG-50014 for 15 minutes before being thoroughly washed by repeated centrifugation and resuspension. These beads were then analyzed for Gαo binding using FCPIA. Washing of the beads did not restore Gαo binding activity by the RGS proteins. This irreversible inhibition was partially overcome by washing the beads with buffer containing 1 mM dithiothreitol (DTT), suggesting that the mechanism of reactivity could be through sulfhydryl modification, a mechanism in common with the previously described RGS inhibitor, CCG-4986 (17, 24).
Figure 3. CCG-50014 is an irreversible inhibitor of RGS4 and RGS8 and its effects are partially reversed by the thiol reductant DTT.
A) RGS4 and B) RGS8 containing beads were treated for 15 minutes with 100 μM CCG-50014 prior to vigorous washing to remove any unbound compound. To determine if the compound was reacting in a thiol-sensitive manner, washing was performed in the absence or presence of 1 mM DTT. Data are presented as the mean±SEM from at least three independent experiments.
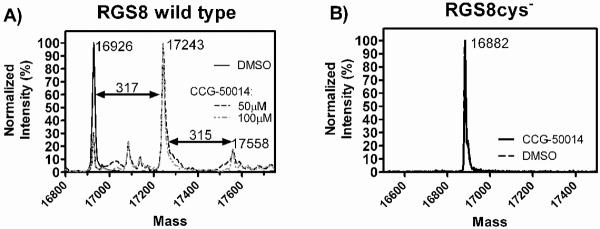
CCG-50014 is a covalent sulfhydryl modifier of RGS8
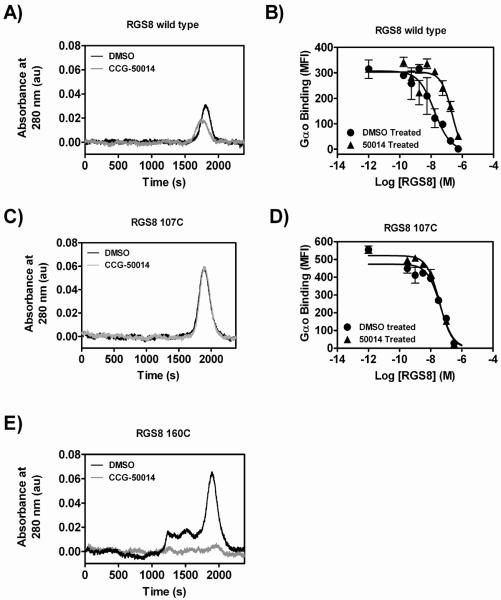
The data thus far suggest that CCG-50014 covalently modifies both RGS4 and RGS8. To test this hypothesis directly, we performed high performance liquid chromatography–mass spectral analysis on RGS8 samples treated with CCG-50014 (Fig. 4A). RGS8 was chosen to simplify the analysis since it only contains two cysteine residues in the RGS homology (RH) domain. After compound treatment, there was a peak shift of the RGS8 corresponding to a full mass adduct of CCG-50014. When WT RGS8 was treated with high concentrations of CCG-050014 (100 μM), a second minor peak corresponding to two full adducts was also observed. To confirm that this action was via cysteine reactivity, the mutant RGS8 where the two cysteines in the RGS homology (RH) domain were mutated to serine (RGS8cys−) was also analyzed and no adduct was observed (Fig 4B).
Figure 4. CCG-50014 forms a covalent adduct on RGS8.
A) WT RGS8 protein (2 μM) was treated with the indicated concentrations CCG-50014 before analysis via LC-MS. After treatment with compound a predominant peak appeared with a mass shift of 317 as compared to the vehicle-treated protein, correlating with a full compound mass adduct (CCG-50014 MW: 316.4). A second minor peak with an additional mass shift of 315 was observed, which correlates to the addition of a second adduct of CCG-50014. B) No adducts are observed on RGS8Cys− mutant, in which both C107 and C160 have been mutated to serine.
CCG-50014 depends on cysteine residues to inhibit the AlF4-Gαo/RGS interaction
To identify the potential cysteine targets of CCG-50014, we studied the compound's effects on RGS8. This protein only contains two cysteines in the RGS homology domain, Cys107 and Cys160, making it a more tractable model system than RGS4. Each cysteine from the RGS8 RH domain was mutated to serine and the activity of the compound was analyzed via FCPIA (Fig. 5). These mutants have been named according to the cysteine residue that they retain1. Neither cysteine was required for sensitivity to the compound, but mutating both cysteines to serine (RGS8cys−) reduced the potency of CCG-50014 by >100 fold. The insensitivity of the RGS8cys− as well as the insensitivity of the RGS4 cysteine null mutant (Table 1), suggests a similar mechanism of action of CCG-50014 on the two proteins.
Figure 5. CCG-50014 requires at least one cysteine residue on RGS8 for full activity.
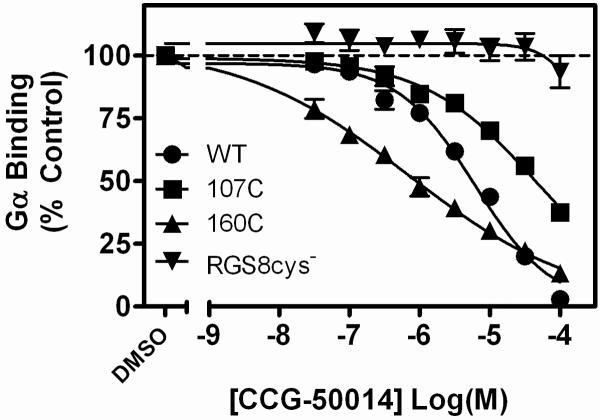
WT or mutant RGS8 was biotinylated, loaded on beads, and AF532-Gαo binding was measured by FCPIA as described in Materials and Methods. Mutating both cysteines to serine (RGS8cys−) produced a protein that was completely insensitive to the effect of CCG-50014. RGS8 mutants with only one cysteine, either cys107 (107C) or cys160 (160C), provided sensitivity to CCG-50014. The inhibition parameters (IC50 (μM), Hill Coefficient) for CCG-50014 on these proteins were as follows: wildtype RGS8 (wt): 6.1 μM, −0.79; 107C: 46.5 μM, −0.54; 160C: 0.71 μM, −0.36; RGS8cys−: >100 μM. Data are presented as the mean±SEM of three independent experiments.
To determine the relative contribution of each cysteine residue in the RGS8 RH domain in the mechanism of action of CCG-50014, WT RGS8 and the two individual cysteine mutants were treated with a saturating concentration (100 μM) of CCG-50014 before removal of the compound and analysis of the treated protein by gel filtration chromatography and FCPIA (Fig. 6). CCG-50014 treatment of WT RGS8 induced a minor mobility shift of the protein on gel filtration and the recovered protein (with it's covalently attached CCG-50014) was analyzed for its ability to compete with untreated bead-bound RGS8 for binding to Gαo. In these experiments, untreated RGS8 coated beads were incubated with increasing amounts of the vehicle- or CCG-50014-treated protein that was recovered from the gel filtration chromatography. To this mixture, fluorescently tagged Gαo was added and allowed to equilibrate with the RGS mixtures. If CCG-50014 was forming a stable, irreversible complex with the RGS, it would be expected to compete for Gαo binding less robustly than the vechicle-treated protein. Under these conditions, wild-type RGS8 that was treated with CCG-50014 was 14-fold less potent at competing for Gα binding than was vehicle-treated protein consistent with residual inhibition of >90%. The CCG-50014- and vehicle-treated 107C RGS8 migrated through the column in a manner identical to that of WT RGS8 and no discernable difference in competition for Gαo binding was observed. The 160C RGS8 mutant, however, formed aggregates upon treatment with CCG-50014 and no monomeric, soluble protein was recovered.
Figure 6. CCG-50014 induced protein aggregation is dependent on the presence of 160C.
A,B) Wild type, C,D) 107C, or E) 160C RGS8 was treated with a 5-fold excess of CCG-50014 before removal of the compound via gel filtration on a 20 mL S75 superdex column. A,C,E) Shown are representative UV chromatogram traces. B,D) Protein recovered from the peak was tested for Gαo binding in an FCPIA competition assay with AF532-Gao binding to WT RGS8. The wild-type RGS8 chromatogram shows a slightly left shifted and suppressed peak after CCG-50014 treatment, which coincides with a 14-fold decrease in Gαo binding. The 107C mutant protein showed no CCG-50014-induced change in migration on gel filtration and any inhibition of Gαo binding activity was reversed by the gel filtration procedure. The 160C mutant protein completely (and visually) aggregates upon compound treatment and is removed by the prefiltration of the samples.
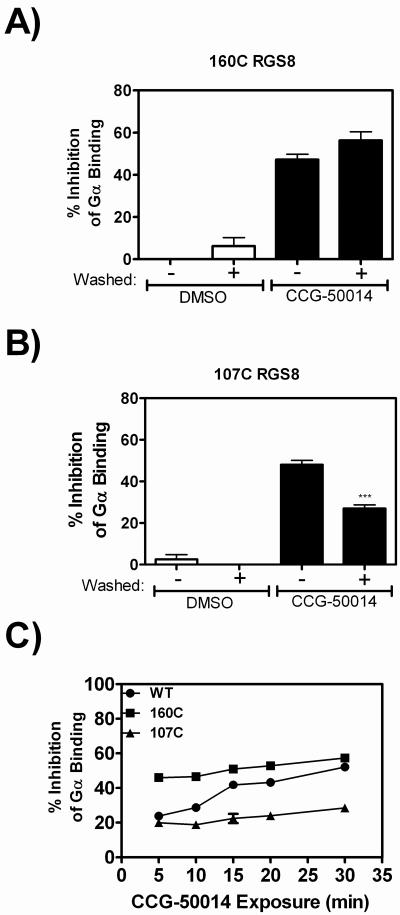
To further probe the mechanism of action of this compound, we studied the time course of the development of irreversible inhibition of the RGS8 cysteine mutants. Bead-bound proteins were treated with 20 μM CCG-50014 and then probed for Gα binding using FCPIA. These experiments revealed that the effect of CCG-50014 on 160C RGS8 was completely irreversible (Fig. 7A), while the effect on 107C RGS8 could be partially reversed by washing away the compound (Fig. 7B).
Figure 7. Irreversible inhibition of RGS8 is predominantly mediated by Cys160.
Mutant proteins A) 107C RGS8 and B) 160C RGS8 were pre-bound to beads and exposed to 20 μM CCG-50014, after which reversibility experiments were performed. C) Development of irreversible inhibition after exposure to CCG-50014 differs between the individual cysteine mutants and provides a means to understand the compound's mechanism of action. Wild-type, 160C or 107C RGS8 was treated with 20 μM CCG-50014 for the indicated amount of time before compound removal by extensive washing. The amount of irreversible inhibition was quantified by comparing the G-protein binding to CCG-50014 treated beads to DMSO treated beads. The total amount of inhibition (without a wash step) at this concentration of CCG-50014 for each protein was as follows: WT RGS8: 64±2%; 107C RGS8: 45±2%; and 160C RGS8: 56±1%. Data are presented as the mean±SEM from three independent experiments. ***P<0.0001 using an unpaired t test.
Because Cys160 is buried in the core of RGS8 and Cys107 is closer to the surface of the protein, we hypothesized that the compound might interact more rapidly with Cys107 than Cys160 in the context of wild-type protein. Capitalizing on the fact that there are differential effects of CCG-50014 (partially reversible inhibition vs protein aggregation) depending on which cysteine is labeled, we tested this hypothesis using FCPIA reversibility experiments. The experiment was designed to monitor the development of irreversible inhibition of wild-type RGS8 and the two RGS8 mutants by CCG-50014 in a time-dependent manner (Fig 7C). Wild-type, 107C, or 160C RGS8 were immobilized on beads and treated for varying periods of time with 20 μM CCG-50014 before extensive washing. The beads were then probed for Gαo binding using FCPIA and compared to RGS-coated beads that had been treated with DMSO alone. At this concentration of CCG-50014, 107C RGS8 binding to Gα was inhibited by ~20% and 160C RGS8 binding to Gα was inhibited by ~50% at all time points tested. In both cases, the compound rapidly exerted its effect on the RGS protein. The Wild-type protein showed a delayed development of irreversible inhibition; at early time points, the inhibition was ~20% and increased to, but did not exceed, ~50% over 30 minutes. This suggests that there is a differential mechanism of action of the compound on the two individual mutants that is combined in the wild type protein with a kinetic lag that is only seen when both cysteines are present.
CCG-50014 is not a general cysteine alkylator
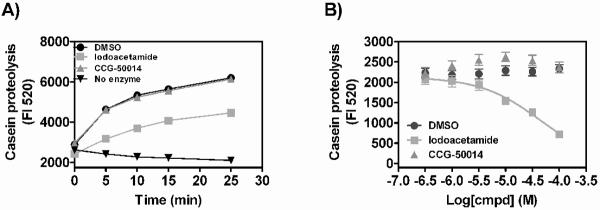
Cysteine reactive compounds might be expected to have more off-target effects than non-reactive compounds. To determine if this compound could bind to and inhibit reactive cysteines in non-RGS proteins, we tested the ability of CCG-50014 and a known cysteine alkylator (iodoacetamide, IA) to inhibit the cysteine protease papain (Fig. 8). IA inhibited the proteolytic activity of papain in a concentration-dependent manner. However, even at high concentrations (100 μM), CCG-50014 had no effect on papain activity. This suggests that there is selectivity of this class of compounds for cysteines in the RGS over other reactive cysteines.
Figure 8. CCG-50014 does not inhibit the cysteine protease, papain.
A) Papain (0.625 U) was mixed with self-quenching FITC-conjugated casein and the liberated fluorescence that results from casein-dependent proteolysis was observed as a function of time in the presence of different cysteine alkylators. Iodoacetamide (100 uM) markedly inhibits casein proteolysis by papain while 100 μM CCG-50014 has no effect. B) Concentration dependence of the effect of compounds on casein proteolysis (5 min). Data are presented as the mean±SEM from three independent experiments.
General cysteine alkylators do not inhibit RGS proteins
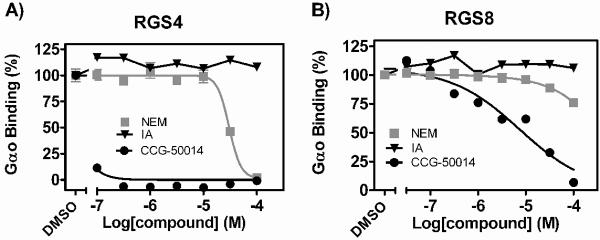
One potential explanation for the observed RGS selectivity of CCG-50014 could be that RGS proteins are particularly sensitive to thiol modification. We tested the ability of two general cysteine alkylators, N-ethyl maleimide (NEM) and IA, to inhibit the RGS/Gαo interaction (Fig. 9). IA had no effect on Gαo binding to any of the RGS proteins tested. At high concentrations (IC50: 30 μM), NEM inhibited RGS4/Gαo binding, however it had no effect on RGS8 (Fig. 9) or papain (data not shown). These data show that CCG-50014 is more than 3.5 orders of magnitude more potent on RGS4 than either of the general cysteine alkylators tested. This strongly suggests that RGS proteins are not particularly sensitive to cysteine modification and the effect observed by CCG-50014 is more than just random thiol alkylation.
Figure 9. CCG-50014 is a much more potent RGS inhibitor than two general cysteine alkylators N-ethyl maleimide (NEM) and iodoacetamide (IA).
Dose response curves for NEM, IA, and CCG-50014 against A) RGS4 and B) RGS8. The only protein that displayed any sensitivity to the alkylators tested was RGS4, which was inhibited by NEM with an IC50 value >3.5 Log higher than that of CCG-50014. Data are presented as the mean±SEM from three independent experiments.
Computational modeling of the CCG-50014-RGS8 interaction
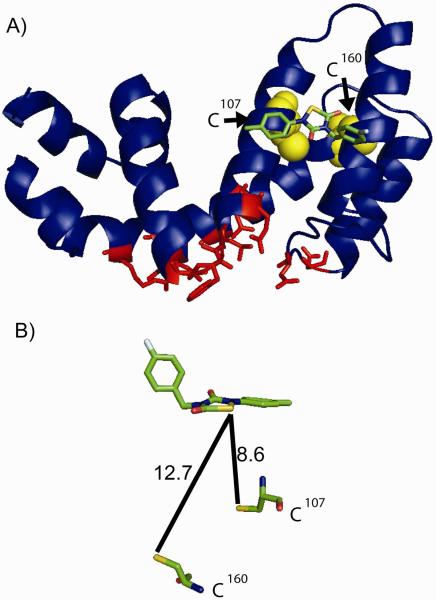
To identify potential binding sites for CCG-50014 on RGS8, we performed an unbiased molecular docking simulation over the whole RGS8 molecular surface. Of the 100 docking poses obtained from the simulation, CCG-50014 docked solely in one pocket on the RGS. This is located near the region of the surface of RGS8 the corresponding “B”-site of RGS4 (Fig. 10). The “B” site (10) has been suggested as the locus of allosteric inhibition of RGS4 family proteins by acidic lipids (8). This places the compound a considerable distance from the two cysteine residues known to play a role in the compound's inhibitory activity (Fig. 10B). It would require a substantial change in the conformation of the protein for the compound to dock at this site and react with a cysteine residue.
Figure 10. Hypothesized binding site of CCG-50014 on RGS8.
A) CCG-50014 was docked to RSG8 using Autodock software (ver 4.0) as described in Materials and Methods and one docking site showed the greatest predicted affinity (18 μM Ki). This site is far from the Gα binding interaction interface and is near the RGS4 which is important for RGS regulation by calmodulin and acidic phospholipids. Conserved residues between RGS4 and RGS8 that directly contact Gαi in the RGS4-Gαi1 structure (PDB 1AGR (24)) are shown in red. B) Assuming a static protein, this binding site places the compound close to the two cysteine residues in RGS8, but not within a distance compatible with direct covalent reaction. A conformational change must occur in the RGS to allow compound intercalation into the helix bundle. Distances are shown in angstroms. RGS8 structure from 2IHD.
Limiting the reactivity of CCG-50014 diminishes potency
To determine the importance of the cysteine reactivity in the mechanism of action of this compound, an analog of CCG-50014 in which the sulfur was replaced with a methylene, was synthesized and tested for activity (Table S1). This compound has limited, if any, activity in the FCPIA assay, suggesting that the main mechanism of action of CCG-50014 is through covalent reactivity with one or more cysteine residues on the RGS.
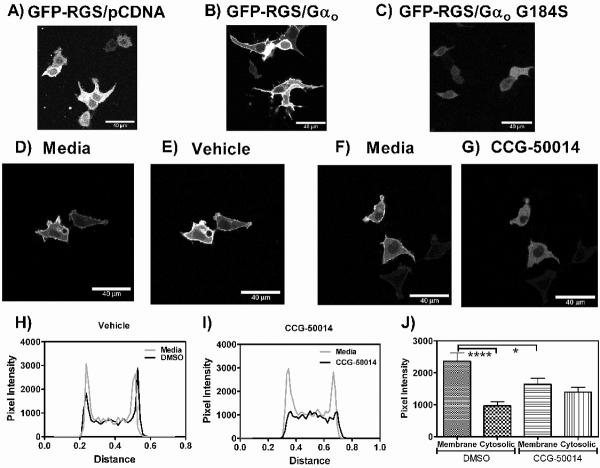
CCG-50014 inhibits the Gαo-dependent membrane translocation of RGS4 in living cells
RGS4 is a cytosolic protein that can be recruited to the plasma membrane by overexpression of certain Gα subunits (Fig. 11 A/B/C) or GPCRs (20). CCG-50014 inhibited the Gαo-dependent membrane translocation of GFP-RGS4 in living HEK293T cells (Fig. 11). Treatment of cells that were transiently transfected with GFP-RGS4 and Gαo with vehicle (0.1% DMSO, Fig. 11 D/E/H) does not alter the membrane localization of RGS4, however, 100 μM CCG-50014 reversed the membrane localization of the RGS (Fig. F/G/I). Through line scan analysis, we observed a significant decrease in membrane localization of the RGS after treatment with CCG-50014. There was also a trend towards an increase in the cytosolic signal of GFP-RGS4 after compound treatment, suggesting the observed phenomenon was not due to non-specific diminishment of the GFP signal.
Figure 11. CCG 50014 inhibits the Gαo dependent membrane localization of RGS4.
A) When overexpressed in HEK293T cells, GFP-RGS4 is localized to the cytosol. B) Co-expression with Gαo induces a subcellular translocation of the GFP-RGS4 to the plasma membrane. C) This translocation does not occur in response to co-expression with the RGS-insensitive Gαo mutant (G184S). D/E) Cells expressing Gαo and GFP-RGS4 show no change in the plasma membrane localization of the RGS when treated with vehicle (0.1% DMSO), however treatment with F/G) CCG-50014 (100μM) rapidly induces a loss of the plasma membrane localization of the RGS without diminishing the overall signal. Representative line scans across a cell treated with H) vehicle and I) CCG-50014 also show this effect. J) Quantification of this effect from 10 cells treated with vehicle or CCG-50014 (100 μM) shows a significant decrease in the amount of RGS-GFP located at the membrane after compound addition. A trend towards increased cytosolic localization after compound treatment is also observed, suggesting that the treatment doesn't just diminish the GFP signal. ****p<0.0001, *p<0.05.
Discussion
Molecules disrupting the RGS/Gα interaction should increase the magnitude and/or duration of G-protein signaling responses, leading to pronounced physiological effects. Genetic ablation of RGS expression produces dramatic phenotypes, suggesting that a small molecule RGS inhibitor might provide similar actions in vivo.
CCG-50014 is the most potent small molecule RGS inhibitor identified to date. It inhibits the in vitro interaction between RGS4 and Gαo with a 30 nanomolar IC50 value. It is >100 fold selective for RGS4 over two closely related RGS proteins, RGS8 and RGS16 (Fig. 1, Table 1). CCG-50014 is a covalent modifier of cysteine residues (Fig. 4), raising concerns about the therapeutic potential of this class of compounds. Even so, this compound has provided significant insight into the mechanism of allosteric RGS inhibition; furthermore it is the first RGS inhibitor to block the RGS4-Gαo protein-protein interaction in living cells.
RGS sensitivity to CCG-50014 requires at least one cysteine residue – a hallmark of a sulfhydryl-reactive irreversible inhibitor (11). This observation was confirmed by FCPIA reversibility experiments (Fig. 3) and subsequent mass spectral analysis of CCG-50014 treated RGS8 (Fig. 4). Based on the chemical structure of the compound and the full molecular weight adduct observed in the LC-MS experiments, it is likely that the mechanism of reaction of CCG-50014 with a cysteine residue on an RGS protein is by nucleophilic attack of the cysteine thiol onto the sulfur atom of the central heterocycle causing ring opening through cleavage of the sulfur-nitrogen bond. This would be consistent with the reported reactivity of 1,2,4-thiadiazolidine-3,5-diones with triphenylphosphine (12). The resultant disulfide linkage is sensitive to reductants, which is consistent with the DTT-induced reversibility of CCG-50014 inhibition (Fig. 3).
Interestingly, CCG-50014 interacts with cysteine residues in RGS8 that are distant from the Gα interaction interface (Fig 10), suggesting an allosteric mechanism of action similar to previously reported effects of two other RGS inhibitor compounds (13, 21). Unbiased computational modeling predicts that CCG-50014 could bind non-covalently to a site on RGS8 that is near to the acidic phospholipid binding site on RGS4 (8, 9). Binding in this site would place the reactive group of CCG-50014 within 8–13 Å of the two cysteines in the RGS8 RH domain. While at this distance, it is unlikely that a covalent bond could be formed; the compound may initially bind to this pocket and a subsequent conformational change in the protein could provide access to the cysteine thiol. Assuming that the compound docks as modeled, this conformational change is likely to be the fundamental mechanism by which the allosteric modulation of G protein binding activity is conferred.
The differential sensitivities of the cysteine mutants to CCG-50014 would also be explained by this binding modality. The decreased sensitivity to and increased reversibility of CCG-50014 on 107C RGS8 (Fig. 5) is in accord with the fact that Cys107 is more solvent accessible and is closer to the hypothesized binding site of the compound. Compound reacting with Cys160 causes drastic protein unfolding (Fig. 6), which also fits with this model.
The data presented here allow us to postulate a potential mechanism of action for CCG-50014. We propose that the compound initially interacts with Cys107, possibly because the compound may dock at a site close to this residue. Upon reacting with this cysteine, CCG-50014 can trap the RGS in a conformation that is incapable of binding to Gα. Reversal of this reaction is possible, leading to reactivation of the RGS on washing at early times. If the compound interacts with the more deeply buried cysteine (Cys160) it causes a dramatic conformational change in the protein (likely a disruption of the hydrophobic core), leading to protein unfolding. Fitting with this hypothesis, our data suggest that Cys107 is labeled more rapidly than Cys160 in the wild type protein (Fig. 7C). Then, either the Cys107-bound compound transfers to Cys160 or a second CCG-50014 molecule binds to Cys160 to produce the completely irreversible protein denaturation observed in gel filtration experiments (Fig. 6). Therefore, the mechanism behind the irreversible inhibition after labeling of Cys160 is likely due to a massive destabilization of the hydrophobic core of the RH domain that would occur by the insertion of CCG-50014.
The development of cysteine-reactive small molecule inhibitors into useful research probes and therapeutic agents is challenging, yet surmountable. There are a few successful therapeutics that function by covalently binding to cysteine residues. For example, the acid-reflux drug omeprazole operates in the stomach by covalently modifying a proton exchanger (22). There is also a class of cysteine-reactive irreversible tyrosine kinase inhibitors, typified by CI-1033, that are currently in clinical trials (23). Cysteine reactive compounds thus have a place in modern pharmacology, however they must be closely studied to determine their selectivity profile. While a comprehensive analysis of CCG-50014 effects upon all cysteine-dependent processes in a cell is clearly impossible, we show that CCG-50014 does not inhibit the activity of the cysteine protease papain at concentrations over 3000 times higher than that required for RGS inhibition (Fig. 8). In contrast, the cysteine alkylator iodoacetamide concentration-dependently inhibited the activity of this protease but had no effect on RGS4 or RGS8. This suggests that there is some selectivity of this class of compounds for cysteines in the RGS over other reactive cysteines. It is possible that the compound cannot enter the active site of papain and therefore it would be prudent to extend these studies to a panel of physiologically relevant thiol-dependent processes.
Furthermore, we have shown that CCG-50014 is able to inhibit the Gαo-dependent membrane localization of RGS4 in living cells (Fig. 11) representing the first small molecule RGS inhibitor with cellular activity. These data suggest that CCG-50014 or related analogs should be useful pharmacological probes to study the physiological roles of RGS proteins in biology.
In this study we characterize the mechanism of action of the most potent RGS inhibitor identified to date. While this compound has significant liabilities as a potential drug candidate, it does highlight the fact that it is possible for a small molecule to inhibit the RGS/Gα protein-protein interaction with nanomolar potency. Furthermore, we have shown that CCG-50014 is able to inhibit the RGS4-Gαo protein-protein interaction in living cells. Current work is focused on characterizing the structure-activity landscape of this compound class and transitioning these studies to physiological models of RGS activity.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. John Tesmer and Dr. Roger Sunahara and their laboratories for insightful scientific discussions and collaborations. The authors acknowledge Andrew Storaska for preparation of some of the proteins used in this study. We would also like to thank University of Michigan comprehensive Cancer Center for subsidizing the cost of DNA sequencing. This work utilized the Cell and Molecular Biology Core of the Michigan Diabetes Research and Training Center, funded by DK020572 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Abbreviations
- GPCR
G-Protein Coupled Receptor
- RGS
Regulator of G-Protein Signaling
- FCPIA
Flow Cytometry Protein Interaction Assay
- DTT
Dithiothreitol
- IA
Iodoacetamide
- NEM
N-Ethyl Maleimide
- Tm
Melting Temperature
- ESI-LC-MS
Electrospray Ionization Liquid Chromatography Mass Spectrometry
- TFA
Trifluoroacetic acid
- EDTA
Ethylenediaminetetraacetic acid
- FITC
Fluorescein isothiocyanate
- AF532
AlexaFluor© 532
- RH
RGS homology
Footnotes
The RGS8 construct used in this study contains two cysteine residues, C107 and C160. Individual cysteine → serine mutants were generated and named based upon the cysteine residue that was maintained in the primary sequence. Therefore, C160S RGS8 is called 107C RGS8 and C107S is called 160C RGS8. The double mutant (C160S/C107S) is called RGS8Cys−.
Supplemental Information Available: A table containing the chemical structure and RGS inhibitory activity of CCG-50014 and CCG-303778, an analog of CCG-50014 in which the sulfur atom is replaced with a methylene. A figure depicting the rate constants of GTP hydrolysis by Gαo in the presence and absence of 100 μM CCG-50014. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by NIH RO1 R01DA023252 (RRN)
References
- 1.Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis JA, Lebois EP, Lindsley CW. Allosteric modulation of kinases and GPCRs: design principles and structural diversity. Curr Opin Chem Biol. 2008;12:269–280. doi: 10.1016/j.cbpa.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Blazer LL, Neubig RR. Small molecule protein-protein interaction inhibitors as CNS therapeutic agents: current progress and future hurdles. Neuropsychopharmacology. 2009;34:126–141. doi: 10.1038/npp.2008.151. [DOI] [PubMed] [Google Scholar]
- 4.Siderovski DP, Willard FS. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 2005;1:51–66. doi: 10.7150/ijbs.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman DM, Kozasa T, Gilman AG. The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J Biol Chem. 1996;271:27209–27212. doi: 10.1074/jbc.271.44.27209. [DOI] [PubMed] [Google Scholar]
- 6.Sjogren B, Blazer LL, Neubig RR. Regulators of G protein signaling proteins as targets for drug discovery. Prog Mol Biol Transl Sci. 2010;91:81–119. doi: 10.1016/S1877-1173(10)91004-1. [DOI] [PubMed] [Google Scholar]
- 7.Huang X, Fu Y, Charbeneau RA, Saunders TL, Taylor DK, Hankenson KD, Russell MW, D'Alecy LG, Neubig RR. Pleiotropic phenotype of a genomic knock-in of an RGS-insensitive G184S Gnai2 allele. Mol Cell Biol. 2006;26:6870–6879. doi: 10.1128/MCB.00314-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishii M, Fujita S, Yamada M, Hosaka Y, Kurachi Y. Phosphatidylinositol 3,4,5-trisphosphate and Ca2+/calmodulin competitively bind to the regulators of G-protein-signalling (RGS) domain of RGS4 and reciprocally regulate its action. Biochem J. 2005;385:65–73. doi: 10.1042/BJ20040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popov SG, Krishna UM, Falck JR, Wilkie TM. Ca2+/Calmodulin reverses phosphatidylinositol 3,4, 5-trisphosphate-dependent inhibition of regulators of G protein-signaling GTPase-activating protein activity. J Biol Chem. 2000;275:18962–18968. doi: 10.1074/jbc.M001128200. [DOI] [PubMed] [Google Scholar]
- 10.Zhong H, Neubig RR. Regulator of G protein signaling proteins: novel multifunctional drug targets. J Pharmacol Exp Ther. 2001;297:837–845. [PubMed] [Google Scholar]
- 11.Roman DL, Ota S, Neubig RR. Polyplexed Flow Cytometry Protein Interaction Assay: A Novel High-Throughput Screening Paradigm for RGS Protein Inhibitors. J Biomol Screen. 2009 doi: 10.1177/1087057109336590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasim S, Crooks PA. N-Chlorocuccinimide is a convenient oxidant for the synthesis of 2,4-disubstituted 1,2,4-thiadiazolidine-3,5-diones. Tet Lett. 2009;50:257–259. [Google Scholar]
- 13.Roman DL, Blazer LL, Monroy CA, Neubig RR. Allosteric inhibition of the regulator of G protein signaling-Galpha protein-protein interaction by CCG-4986. Mol Pharmacol. 2010;78:360–365. doi: 10.1124/mol.109.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan KL, Sarvazyan NA, Taussig R, Mackenzie RG, DiBello PR, Dohlman HG, Neubig RR. A point mutation in Galphao and Galphai1 blocks interaction with regulator of G protein signaling proteins. J Biol Chem. 1998;273:12794–12797. doi: 10.1074/jbc.273.21.12794. [DOI] [PubMed] [Google Scholar]
- 15.Soundararajan M, Willard FS, Kimple AJ, Turnbull AP, Ball LJ, Schoch GA, Gileadi C, Fedorov OY, Dowler EF, Higman VA, Hutsell SQ, Sundstrom M, Doyle DA, Siderovski DP. Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits. Proc Natl Acad Sci U S A. 2008;105:6457–6462. doi: 10.1073/pnas.0801508105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee E, Linder ME, Gilman AG. Expression of G-protein alpha subunits in Escherichia coli. Methods Enzymol. 1994;237:146–164. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- 17.Sternweis PC, Robishaw JD. Isolation of two proteins with high affinity for guanine nucleotides from membranes of bovine brain. J Biol Chem. 1984;259:13806–13813. [PubMed] [Google Scholar]
- 18.Blazer LL, Roman DL, Muxlow MR, Neubig RR. Use of flow cytometric methods to quantify protein-protein interactions. Curr Protoc Cytom. 2010;Chapter 13(Unit 13):11, 11–15. doi: 10.1002/0471142956.cy1311s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol. 2007;71:169–175. doi: 10.1124/mol.106.028670. [DOI] [PubMed] [Google Scholar]
- 20.Roy AA, Lemberg KE, Chidiac P. Recruitment of RGS2 and RGS4 to the plasma membrane by G proteins and receptors reflects functional interactions. Mol Pharmacol. 2003;64:587–593. doi: 10.1124/mol.64.3.587. [DOI] [PubMed] [Google Scholar]
- 21.Blazer LL, Roman DL, Chung A, Larsen MJ, Greedy BM, Husbands SM, Neubig RR. Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins. Mol Pharmacol. 2010;78:524–533. doi: 10.1124/mol.110.065128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sachs G, Prinz C, Loo D, Bamberg K, Besancon M, Shin JM. Gastric acid secretion: activation and inhibition. Yale J Biol Med. 1994;67:81–95. [PMC free article] [PubMed] [Google Scholar]
- 23.Ocana A, Amir E. Irreversible pan-ErbB tyrosine kinase inhibitors and breast cancer: current status and future directions. Cancer Treat Rev. 2009;35:685–691. doi: 10.1016/j.ctrv.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.