Abstract
The CHEK2-1100delC mutation is recurrent in the population and is a moderate risk factor for breast cancer. To identify additional CHEK2 mutations potentially contributing to breast cancer susceptibility, we sequenced 248 cases with early-onset disease; functionally characterized new variants and conducted a population-based case–control analysis to evaluate their contribution to breast cancer risk. We identified 1 additional null mutation and 5 missense variants in the germline of cancer patients. In vitro, the CHEK2-H143Y variant resulted in gross protein destabilization, while others had variable suppression of in vitro kinase activity using BRCA1 as a substrate. The germline CHEK2-1100delC mutation was present among 8/1,646 (0.5%) sporadic, 2/400 (0.5%) early-onset and 3/302 (1%) familial breast cancer cases, but undetectable amongst 2,105 multiethnic controls, including 633 from the US. CHEK2-positive breast cancer families also carried a deleterious BRCA1 mutation. 1100delC appears to be the only recurrent CHEK2 mutation associated with a potentially significant contribution to breast cancer risk in the general population. Another recurrent mutation with attenuated in vitro function, CHEK2-P85L, is not associated with increased breast cancer susceptibility, but exhibits a striking difference in frequency across populations with different ancestral histories. These observations illustrate the importance of genotyping ethnically diverse groups when assessing the impact of low-penetrance susceptibility alleles on population risk. Our findings highlight the notion that clinical testing for rare missense mutations within CHEK2 may have limited value in predicting breast cancer risk, but that testing for the 1100delC variant may be valuable in phenotypically- and geographically-selected populations.
Keywords: CHEK2, susceptibility, breast, cancer, mutation
The highly penetrant breast cancer susceptibility genes BRCA1 and BRCA2 have been linked to ~80% of familial breast cancer,1,2 but to only ~10% of cases without a strong family history.3–5 While heritability studies demonstrate a substantial genetic contribution to this more common form of breast cancer, the risk loci remain largely unidentified.6 It has been suggested that low penetrance or modifier genetic alleles, that are more common in the population, may account for the underlying genetic predisposition to breast cancer risk.7
In studying cancer-prone families, we initially detected an inactivating mutation in the cell cycle checkpoint kinase gene CHEK2,8 CHEK2-1100delC, which has now been identified as a recurrent mutation associated with increased risk for breast and perhaps prostate cancers.9–14 In some European populations, CHEK2-1100delC is present at a frequency of 1% and it confers a relative risk of 2-fold for female breast cancer and 10-fold for bilateral breast cancer and for male breast cancer in non-BRCA1/BRCA2 -linked families,9 as well as 2-fold increased risk of developing a second breast cancer.15 In addition to familial cases, CHEK2-1100delC has also been found to confer a 2- to 3-fold increased risk of breast cancer within the general population.10,14 While tumors from CHEK2-1100delC carriers have no consistently distinguishable features from those arising in noncarriers, carriers have both poorer disease-free (p = 0.006) and overall survival (p = 0.072) than noncarriers.15,16
The role of other CHEK2 population variants in cancer predisposition is less well established: the missense mutation CHEK2-I157T, for instance, was first detected among cancer-prone families8 and subsequently shown to be present in ~6% of the Finnish population,11,12 but at appreciably lower frequencies (<2%) within German and North American populations.17,18 Epidemiological studies have suggested that this variant may be associated with a moderate increase in prostate cancer risk in the Finnish and Polish populations.13,19 In vitro studies indicate that the CHEK2-I157T protein may be defective in some, but not all, CHEK2 functions.20–24 These observations support the notion that missense mutations with partial loss-of-function may contribute to oncogenesis and reach appreciable frequencies in one or more populations.
The functional properties of CHEK2 were first defined for its yeast orthologs, the G2/M checkpoint proteins cds1 (S. pombe) and rad53 (S. cerevisiae).25–29 In mammalian cells, CHEK2 (also referred to as CHK2) modulates multiple checkpoints following ionizing radiation, in an ATM-dependent manner.30 Phosphorylation targets of CHEK2 include p53, CDC25A and CDC25C, resulting in activation of the G1/S, S and G2/M checkpoints, respectively.30–33 In addition, BRCA1 itself is phosphorylated by CHEK2 following DNA damage, an effect linked to its altered subnuclear localization.34 Mouse models of Chk2 inactivation have supported an important, but apparently redundant role in the maintenance of genomic stability. Cells from Chk2-null mice have defects in p53-dependent checkpoints,35,36 and the mice themselves show increased tumorigenesis following treatment with the carcinogen DMBA.37 Mouse embryo fibroblasts derived from a Chk2-1100delC knock-in animal have an altered cell cycle profile, and demonstrate a constitutively activated DNA damage response.38 Inactivation of both Chk2 and Brca1 in the mouse leads to dramatic synergy in mammary tumorigenesis,39 suggesting that Chk2 inactivation may relieve p53 and other checkpoints that normally suppress Brca1-mediated tumorigenesis. In addition, mice with a knock-in mutation disrupting the Ser988 Brca1 residue targeted by Chk2 show some increased tumorigenicity, pointing to a role for Chk2-mediated phosphorylation of Brca1 in cancer susceptibility.40
The complex biological properties of CHEK2, together with the emergence of genetic variation in different ethnic populations, pose an important challenge for its characterization as a model low penetrance breast cancer gene. A recent case–control study, using haplotype-tagged SNPs, found no association between common variants within CHEK2 and postmenopausal breast cancer risk in the Swedish population.41 Here we describe the resequencing of the CHEK2 coding region from the germline of cancerprone breast and prostate cancer cases to identify additional allelic variants, coupled with in vitro biochemical analysis of these mutants to classify their functional properties, and finally genotyping of the variants in early onset, familial and sporadic breast cancers spanning multiple ethnic groups.
Material and methods
Early-onset, familial and unselected breast cancer cohorts
EBV-immortalized lymphoblastoid cell-lines were derived from 400 patients with early-onset breast cancer (EOBC), diagnosed at age 40 or earlier, at Massachusetts General Hospital and have been described in detail elsewhere.3 Probands of 302 breast cancer families, including 155 families linked to BRCA1 or BRCA2, were accrued through the Family Risk Assessment Program at Fox Chase Cancer Center, Philadelphia. A significant proportion (122/ 302, 40%) of these families were of Ashkenazi Jewish descent. Forty-seven African–American breast cancer families were recruited through the University of Chicago.
Germline DNA was also available for analysis from 1,646 ethnically diverse women (345 African–Americans, 393 Caucasians, 118 Hawaiians, 428 Japanese and 362 Latinas) diagnosed with sporadic breast cancer. These cases are part of a larger multiethnic cohort described previously.42 Consent to obtain, store and analyze germline material was obtained on all patients according to institutional review board guidelines.
Early-onset prostate cancer cohort
We established EBV-immortalized lymphoblastoid cell-lines from 79 men with early-onset prostate cancer, defined as having a diagnosis of disease before age 50 irrespective of family history of prostate cancer, at Massachusetts General Hospital. Four cases were diagnosed before age 41; 21 cases between ages 41–45 and 54 cases between ages 46–49. Within this cohort, 6 cases (7%) had one or more relatives diagnosed with prostate cancer before age 55; 11 cases (14%) had one or more relatives diagnosed with prostate cancer between ages 56–65 and 22 cases (27%) had one or more relatives diagnosed with prostate cancer age 66 or over.
Control populations
A multiethnic control group was comprised of 1,905 unaffected individuals (431 African–Americans, 433 Caucasians, 279 Hawaiians, 378 Japanese and 384 Latinas).42 In addition, 200 EBV-immortalized lymphoblastoid cell-lines derived from healthy blood donors at Massachusetts General Hospital served as matched controls for both the early-onset breast and early-onset prostate cancer cohorts. DNA from 800 unaffected, BRCA1 and BRCA2 founder mutation-negative, Ashkenazi Jewish individuals collected at Fox Chase Cancer Center was also analyzed.
Mutation detection
The entire coding region of CHEK2 was analyzed for germline mutations within 169 early-onset breast and 79 early-onset prostate cancer cases. Briefly, RNA was isolated from EBV-immortalized lymphoblastoid cell-lines using RNA-Stat60 (Tel-Test, Friendswood, TX) and cDNA was generated using the first-strand synthesis kit (Roche, Germany) according to the manufacturer’s instructions. The open reading frame of CHEK2 was amplified by RT-PCR in 4 overlapping fragments (PCR conditions are available upon request). Amplicons were purified using exonuclease I and shrimp alkaline phosphatase (United States Biochemical, Cleveland, OH). Bidirectional capillary sequencing was performed using the ABI BigDye Terminator kit (version 1.1) (Applied Biosystems, Foster City, CA) and an ABI3100 instrument. Sequences were reviewed for the presence of double peaks, indicative of heterozygous germline mutations, using Sequence Navigator software in combination with Factura to mark heterozygous positions (Applied Biosystems, Foster City, CA).
Genotyping of sequence variants
Sequence variants were genotyped using a Sequenom MassArray system. In brief, primers and probes were designed for each SNP by the SpectroDesign software and are available on request. Multiplex PCR was performed in 5-µl volumes that contain 0.1 U of Taq polymerase (Amplitaq Gold, Applied Biosystems [ABI]), 5 ng genomic DNA, 2.5 pmol of each PCR primer and 2.5 mol of dNTP. Thermocycling was at 95°C for 15 min, followed by 45 cycles of 95°C for 20 sec, 56°C for 30 sec and 72°C for 30 sec. Unincorporated dNTPs were deactivated using 0.3 U of shrimp alkaline phosphatase (Roche, Germany), followed by primer extension by use of 5.4 pmol of each primer extension probe, 50 µM of the appropriate dNTP/ddNTP combination and 0.5 units of Thermosequenase (Amersham Pharmacia, Piscataway, NJ). Reactions were cycled at 94°C for 2 min, followed by 40 cycles of 94°C for 5 sec, 50°C for 5 sec and 72°C for 5 sec. After addition of a cation-exchange resin to remove residual salt from the reactions, ~7 nl of the purified primer-extension reaction was loaded onto a matrix pad (3-hydroxypicolinic acid) of a Spectro-CHIP (Sequenom). SpectroCHIPs were analyzed using a Bruker Biflex III MALDI-TOF mass spectrometer (SpectroREADER [Sequenom]) and the spectra processed using SpectroTYPER (Sequenom). The genotyping percentage exceeded 90% for each assay. Given the low frequency of the alleles in the general population, a known heterozygote was run with the samples. Error rates as previously assessed by duplicate samples on this platform have been estimated at 0.3%. A 2-sided Fisher’s Exact test was applied to determine significance.
Expression constructs and generation of inducible cell-lines
A full-length CHEK2 cDNA was constructed by ligation of a PCR-generated 5′ end to EST clone AI809500 (American Type Culture Collection, Manassas, VA). Site-directed mutagenesis was used to generate mutant CHEK2 constructs encoding the naturally occurring R3W, P85L, R137Q, H143Y, R145W, I157T, A247D, 589delA and 1100delC mutations and the synthetic catalytically inactive D347A mutation. Constructs were cloned into CMV-driven vectors pCDNA3 or Gateway, encoding a Flag epitope (Invitrogen, Carlsbad, CA) and into the tetracycline-regulated vector pUHD10-3. The integrity of all constructs was confirmed by nucleotide sequencing. The TNT coupled transcription–translation kit (Promega, Madison, WI) was used for in vitro translation. For transient transfection, U2OS, 293 and COS-7 cells were transfected with 5 µg of pCDNA3Flag CHEK2 and 0.5 µg of pEGFP-C1 (Clontech, Mountain View, CA) plasmids using the calcium phosphate method. Cell lysates were analyzed 40 hr after transfection by immunoblotting, using either anti-Flag (SigmaAldrich, St. Louis, MO) or anti-GFP antibodies (Clontech, Mountain View, CA). Inducible, tetracycline-repressible expression of CHEK2 constructs was achieved by cotransfecting a U2OS founder cell line with various pUHD10-3 CHEK2 constructs, along with a plasmid encoding hygromycin resistance. Hygromycin-resistant clones were picked and analyzed for similar levels of inducible CHEK2 expression by Northern blot analysis, and at least 2 independent clones were selected for measurements of protein half-life.
In vitro kinase assays and determination of protein expression
In vitro kinase activity of anti-Flag-immunoprecipitated CHEK2 proteins from U2OS cells with inducible expression was determined in unirradiated cells or 1 hr after γ-irradiation (10 Gy). GST-BRCA1 (amino acids 758–1064), as described by Lee et al.,34 was used as a substrate, in a 30-min incubation using 20 mM HEPES (pH 7.4), 10 mM MgCl2, 10 mM MnCl2, 40 µM ATP and 15 µCi [γ-32P]ATP at 30°C.
The expression of wild-type and mutant CHEK2 proteins was compared. U2OS cells were transiently transfected with 5 µg of each Flag-tagged CHEK2 expression construct and 0.5 µg of pEGFP-C1 (Clontech, Mountainview, CA) using Lipofectamine-Plus according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Steady state levels of CHEK2 were measured by immunoblotting 40 hr posttransfection, using an anti-Flag antibody or an anti-GFP antibody. CHEK2 mRNA levels were determined by Northern blotting.
Results and discussion
Germline CHEK2 variants in early-onset breast and prostate cancer cases
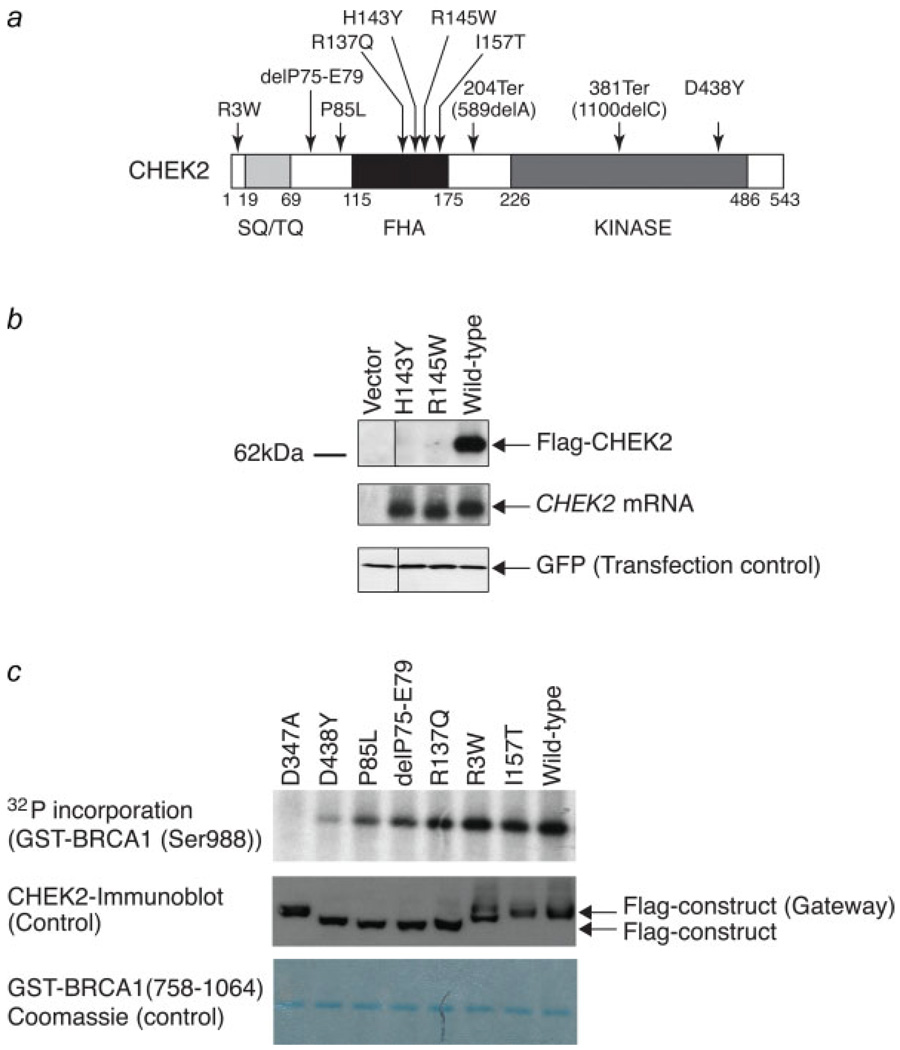
To define the spectrum of CHEK2 variants in cancer-prone populations, we undertook a sequential analysis, involving initial resequencing of the entire coding region in a panel of high-risk patients, followed by genotyping of specific variants in independent population-based cohorts. These results were then correlated with in vitro assays of CHEK2 function. Given the link between CHEK2 mutations and susceptibility to either breast or prostate cancers, we sequenced CHEK2 from germline DNA of 169 individuals diagnosed with EOBC (diagnosed < age 40), and 79 individuals with early-onset prostate cancer (diagnosed < age 50). Sequence variants were identified in 5 (3%) breast cancer and 2 (2.5%) prostate cancer cases. The mutations consisted of 1 case each of the known truncating mutation 1100delC; previously reported missense mutations P85L,43 R137Q44 and in-frame deletion delP75-E7917,45; a novel truncating mutation 589delA and novel missense variants H143Y and D438Y (Table I) (Fig. 1a).
Table I.
Germline Chek2 Mutations in Early-Onset Breast and Prostate Cancer Cohorts
| Index case | Nucleotide change | Amino acid change | Domain |
|---|---|---|---|
| EOPC | Deletion of nucleotides 223–237 | delP75-E79 | Unknown |
| EOBC | C to T at nucleotide 254 | P85L | Unknown |
| EOBC | G to A at nucleotide 410 | R137Q | FHA |
| EOBC | C to T at nucleotide 427 | H143Y | FHA |
| EOBC | 589delA | 204Ter | Kinase |
| EOBC | 1100delC | 381Ter | Kinase |
| EOPC | G to T at nucleotide 1312 | D438Y | Kinase |
EOPC, early-onset prostate cancer; EOBC, early-onset breast cancer.
FIGURE 1.
Impaired stability and kinase activity of mutant CHEK2 proteins. (a) location of naturally occurring CHEK2 variants, identified within the germline of cancer-prone individuals, in relation to the SQ/TQ, FHA and kinase domains of the protein. (b) CHEK2-H143Y encodes an unstable protein. Expression levels of various mutant and wild-type CHEK2 proteins were measured by immunoblotting, following transient transfection of U2OS cells with flag-tagged expression constructs (top). CHEK2 transcript levels were determined by Northern blotting (middle). Transfection efficiencies were evaluated by immunoblotting using an anti-GFP antibody (bottom). (c) In vitro kinase activity of naturally occurring CHEK2 mutants, as well as a synthetic mutant (CHEK2-D347A), compared with the activity of the wild-type protein. CHEK2 proteins, expressed under the control of a tetracycline-inducible promoter in U2OS cells, were immunoprecipitated using anti-Flag antibodies after treatment with γ-irradiation. In vitro kinase activity of CHEK2 proteins was measured using a BRCA1 fragment (residues 758–1064) as a substrate (top). Total amounts of CHEK2 proteins were determined by immunoblotting with anti-Flag antibody (middle). Coomassie blue staining was used to confirm equal amounts of substrate loading (bottom).
Functional properties of CHEK2 variants
The 2 truncating mutations, 1100delC and 589delA disrupt the catalytic domain of CHEK2, resulting in an inactive kinase, as previously formally demonstrated for CHEK2–1100delC.21 Two uncharacterized missense mutations, CHEK2-R137Q and CHEK2-H143Y localized within the forkhead associated (FHA) domain, implicated in protein–protein interactions and previously linked to protein destabilizing mutations.20 Interestingly, the arginine at position 137 has been postulated to form a hydrogen bond with a peptide substrate.24 A third missense mutation, CHEK2-D438Y, mapped within the catalytic domain of the protein. To test the functional characteristics of novel CHEK2 variants, we engineered these mutations into expression constructs and tested protein stability, as well as their ability to mediate in vitro phosphorylation of a BRCA1-derived peptide. Transient transfection studies showed that CHEK2-H143Y, but not the other mutant constructs, encodes a grossly unstable protein, and is therefore likely to be a loss-of-function mutation (Fig. 1b). This mutation affects an invariant histidine, not only among all CHEK2 species but also within all FHA domain-containing proteins described thus far (reviewed in Ref. 46), suggesting a critical role for this residue in protein function. Interestingly, mutation of the equivalent residue within each of the 2 FHA domains of S. cerevisae Rad53, abolished binding of scRad53 to scRad9 thus abrogating the normal DNA damage response.47 The protein instability associated with the H143Y missense mutation in the FHA domain is consistent with previous reports that the CHEK2-R145W mutation within the FHA domain appears to disrupt the predicted α-helical folds, resulting in an unstable protein.24 Indeed, the predicted secondary structure of the protein encoded by CHEK2-H143Y mutation shows that this single substitution disrupts the β-sheet and forms an extended α-helix in the FHA domain, which may disrupt the substrate binding surface.48
To extend our functional analysis, we measured the in vitro kinase activity of transfected proteins immunoprecipitated from cells following their activation by ionizing irradiation (Fig. 1c). Phosphorylation of a BRCA1-derived peptide containing the Ser988 residue targeted by CHEK2 was quantified for mutant and wild-type constructs. In multiple experiments, CHEK2-D438Y showed a 70% reduction in activity compared with wild-type CHEK2, while the P85L and delP75-E79 mutants exhibited 40– 50% of wild-type activity. These mutations may therefore encode proteins with attenuated, rather than complete loss-of-function. In contrast, CHEK2-R137Q had no effect on enzymatic activity. Taken together, complete loss-of-function may be presumed for the 1100delC and 589delA truncating mutations, while a partial loss-of-function is suggested by in vitro analysis of the H143Y, P85L, D438Y and delP75-E79 mutations, but not R137Q. This is the first study to measure the ability of mutant CHEK2 proteins to phosphorylate BRCA1.
Frequency of CHEK2 variants in sporadic and high-risk breast cancer populations
To determine the population frequency of the CHEK2 germline variants and their potential link to breast cancer risk, we genotyped each mutation in expanded cohorts of early-onset and familial breast cancer, as well as in a case control population-based cohort of sporadic breast cancer (Table II). In addition to the variants identified above, we also genotyped 3 previously reported mutations: an apparently neutral mutation R3W identified in a variant LFS family8; an FHA domain mutation, R145W, encoding a grossly unstable protein found in a variant LFS family and an unrelated colorectal cancer cell-line20 and I157T, a recurrent variant observed in less than 2% of German and North American populations but in up to 6% of the Finnish population, and reported to result in variable defects in CHEK2-mediated phosphorylation.8,11–21 Of all mutations tested, only two, CHEK2-1100delC and CHEK2-P85L were found to be recurrent in the populations studied here. In theory, an analysis of 1,646 cases and 1,905 controls has 80% power to detect an odds ratio of 1.8, assuming an allele frequency of 0.01 or greater and an α level of 0.05.
Table II.
Population Frequencies of Germline Chek2 Variants
| CHEK2 variant | Frequency of germline CHEK2 variant |
|||
|---|---|---|---|---|
| Breast cancer cohorts | ||||
| Controls (n = 2,105) | Early-onset (n = 400) | Familial (n = 300) | Sporadic (n = 1,646) | |
| delP75-E79 | nd | 0 (<0.03%) | 0 (<0.03%) | nd |
| P85L | 19 (0.9%) | 1 (0.25%) | 2 (0.67%) | 2 (0.12%) |
| R137Q | 2 (0.09%) | 0 (<0.03%) | 0 (<0.03%) | 0 (<0.06%) |
| H143Y | 0 (<0.05%) | 1 (0.25%) | 0 (<0.03%) | 0 (<0.06%) |
| 589delA | 0 (<0.05%) | 0 (<0.03%) | 0 (<0.03%) | 0 (<0.06%) |
| 1100delC | 0 (<0.05%) | 2 (0.5%) | 3 (1.0%) | 8 (0.48%) |
| D438Y | 1 (0.05%) | 0 (<0.03%) | 0 (<0.03%) | 0 (<0.06%) |
| R3W1 | 0 (<0.05%) | 0 (<0.03%) | 1 (0.3%) | 0 (<0.06%) |
| R145W1 | 0 (<0.05%) | 0 (<0.03%) | 0 (<0.03%) | 0 (<0.06%) |
| I157T1 | 2 (0.09%) | 0 (<0.03%) | 0 (<0.03%) | 0 (<0.06%) |
The index case in which the mutation was first identified is excluded. nd, not determined.
Previously described variants (8, 20).
The previously described 1100delC mutation was present in 8/ 1,646 (0.5%) sporadic breast cancer, 2/400 (0.5%) EOBC and 3/ 302 (1%) familial breast cancer cases. In contrast, this mutation was not detected in 2,105 multiethnic controls, including 633 from the US. The frequency of CHEK2-1100delC in breast cancer cases is comparable to that observed in a New York-based population,49 and somewhat lower than that reported in a European population.9 However, the prevalence of this mutation in healthy controls appears to be considerably lower than the 0.3% frequency reported for the New York population, presumably reflecting genetic diversity within the tested populations. Of note, in the multiethnic study, increased prevalence of CHEK2-1100delC was only detectable in African–American breast cancer cases (3/345 cases versus 0/431 controls, p = 5 0.087 by a two-sided Fisher’s Exact test) and Whites (4/393 cases versus 0/433 controls, p = 5 0.05) but did not reach statistical significance amongst Latinas (1/362 cases versus 0/384 controls, p = 5 0.485) and was undetectable in the Japanese and native Hawaiian populations.
Previous studies have indicated that the frequency of CHEK2-1100delC is increased in familial breast cancer cases lacking BRCA1 mutations, but not in families where breast cancer predisposition is attributable to an inherited BRCA1 mutation.9 These observations have been taken as evidence that CHEK2 and BRCA1 may function in the same biological pathway, making concomitant mutations functionally redundant. This conclusion has been challenged by mouse models in which Chk2 and Brca1 inactivation appear highly synergistic in promoting mammary tumorigenesis.39 In our analysis of familial breast cancer cases, we identified 3 carriers of CHEK2-1100delC among 155 BRCA1 mutation-positive cases, but none among 147 non-BRCA families (p = 0.25). We therefore cannot confirm the specific exclusion of CHEK2-1100delC mutations in BRCA1-linked breast cancer families. Two attenuated variants of CHEK2, P85L and H143Y were also observed in BRCA1 mutation carriers.
P85L is an ancient CHEK2 allelic variant
The second recurrent CHEK2 mutation, P85L, was observed in African–American and Ashkenazi Jewish populations, but it was not associated with an increased risk to breast cancer in either group. Among African–Americans, 2/345 (0.6%) breast cancer cases and 13/431 (3.0%) controls carried this allele (p = 0.02), while the corresponding frequency among Ashkenazi Jews was 4/167 (2%) breast cancer cases and 5/528 (1%) controls (p = 0.901). A single Latina control also carried this variant (1/384, 0.3%), but it was not detected among any sporadic cases within this ethnic subgroup.
Of note, P85L has also been recently reported as a neutral polymorphism in the Ashkenazi Jewish population.50 The relatively high frequency of P85L in African–American and Ashkenazi Jewish populations is remarkable, given its absence in other ethnic groups. To define whether this mutation is present on the same haplotype in these 2 populations, we undertook a genotypic analysis of 9 SNPs encompassing codon 85 of CHEK2, and spanning a distance of 4.5 kb on chromosome 22q. The mutant CHEK2 allele (T) exists on the same haplotype (TTCTTTTGGG) in both African–Americans and Ashkenazim. The frequency of the background haplotype (4–14%) is comparable in all ethnic groups studied, including Whites, native Hawaiians, Japanese and Latinas. These observations raise the possibility that CHEK2-P85L is an ancestral mutation that arose within this shared haplotype, but has been retained in African–American and Ashkenazi Jewish populations, while disappearing because of genetic drift in other populations.
Conclusion
The CHEK2-1100delC mutation was initially identified by sequencing of high risk individuals, followed by genotypic analysis of larger patient cohorts, defining its prevalence in both control and cancer populations.8,9 Additional variants of CHEK2, typically missense mutations, have been identified in patients with breast, prostate and other cancers (reviewed in Ref. 50), but the clinical significance of these variants has not been readily defined.
CHEK2-1100delC encodes a kinase dead protein,21 and germline transmission of this allele is associated with somatic LOH in tumor specimens.20 As such, CHEK2-1100delC provides the standard for CHEK2 loss-of-function mutations. We observed the frequency of CHEK2-1100delC as 1% in familial breast cancer and 0.5% in sporadic and early-onset cases. These estimates are consistent with a recent New York-based analysis, but considerably below European studies.9,49 Also, while the New York studies defined a population frequency of 0.3%, we did not detect any 1100delC variants among 2,105 multiethnic controls. Among cases, CHEK2-1100delC was restricted to African–Americans, Caucasians and Latino populations, but it was not detected among Japanese or native Hawaiian groups. This is consistent with previous findings that the population frequency of this allele is variable according to geographic location ranging from 0.3 to 0.7% within the US49,51,52 to 1.2–1.6% within the Netherlands.9 To our knowledge this is the first report to show an increased risk of breast cancer associated with the 1100delC allele in the African–American population, as well as to demonstrate that it is infrequent among Asian, Latina and Hawaiian populations underscoring the importance of studying variants across multiple populations. In contrast to 1100delC, the prevalence of a second truncating mutation predicted to encode an inactive kinase, CHEK2-589delA, was not observed in our genotyping studies. Other bona fide dysfunctional mutations encoding unstable proteins (R145W and H143Y) were also not recurrent in the population.
A second CHEK2 mutation that has generated considerable interest is I157T, a missense variant encoding a protein that appears capable of phosphorylating and inactivating CDC25C leading to G2 arrest, but with reduced in vitro catalytic activity towards CDC25A22 and an impaired ability to bind to p5323 or BRCA1.24 A number of studies have reported an increased risk to prostate cancer associated with this variant CHEK2 allele.13,17,19 The I157T allele is present in 4.8% of controls compared with 7.8% of unselected prostate cancer cases and 16% of familial prostate cancer cases from Poland.19 Similarly, within the Finnish population, 5.4% of healthy individuals are carriers of this variant compared with 10.8% of inherited prostate cancer cases (p = 0.04, OR 2.12).13 However, we did not detect I157T in any populations in the US, thus pointing to its geographic pattern of variation.
A third interesting CHEK2 variant emerging from our analysis is P85L. This mutation has also recently been reported as a neutral polymorphism in the Ashkenazi population.50 While P85L was able to rescue the lethality of a Rad53-deficient strain of S. cerevisiae, suggesting that it does not represent a loss-of-function mutation,50 our own studies of in vitro kinase activity do show some degree of attenuated function. This functional defect does not appear to be physiologically relevant with respect to breast cancer predisposition, since neither we nor Shaag et al.,50 observed an increased frequency of P85L among breast cancer cases. As noted above, the unusual frequency of P85L, arising on a common ancestral haplotype, and subsequently lost in most populations but retained at high frequency in the Ashkenazi Jewish and African– American populations, points to the random effect of genetic drift, and demonstrates that particular variants will not be present across all populations. Therefore, to comprehensively evaluate the contribution of particular variants to disease risk, attempts should be made to study diverse ethnic populations.
In conclusion, while a number of dysfunctional alleles of CHEK2 may be associated with individual breast cancer predisposition, the absence of recurrent mutations, combined with the weak predictive power of low penetrance mutations limit the clinical utility of such variants. In contrast, the 1100delC allele appears to be the only CHEK2 mutation whose frequency in the population allows reliable estimates of increased cancer risk. The population prevalence of other variants, with more subtle functional defects, is so low that they do not appear to confer an appreciable contribution to breast cancer risk in the general population. As additional low penetrance cancer predisposition genes are identified in the population, insights from the analysis of CHEK2 may be important in defining the contribution of other genetic variants to cancer risk.
Acknowledgements
Supported in part by NIH grants CA87691 (to D.A.H.) and CA63464 (to D.A.), the National Cancer Institute Specialized Programs of Research Excellence on breast cancer at Massachusetts General Hospital (to D.A.H.), the Avon Products Foundation (to D.W.B.), the AACR-National Foundation for Cancer Research Professorship in Basic Cancer Research (to D.A.H.) and the Doris Duke Foundation (D.A.H.). MLF was supported by a fellowship from the Howard Hughes Medical Institute. DWB acknowledges the Intramural Program of the National Human Genome Research Institute at NIH. We acknowledge the help of Ms. JoEllen Weaver and the Biosample Repository at Fox Chase Cancer Center for the processing of DNA samples and Dr. Betsy Bove and the Clinical Molecular Genetics Laboratory at Fox Chase Cancer Center for BRCA1 and BRCA2 mutation data. We thank Ms. Kathryn Penney, Ms. Maria Sun and Ms. Zofia Gajdos for technical assistance.
References
- 1.Couch FJ, Farid LM, DeShano ML, Tavtigian SV, Calzone K, Campeau L, Peng Y, Bogden B, Chen Q, Neuhausen S, Shattuck-Eidens D, Godwin AK, et al. BRCA2 germline mutations in male breast cancer cases and breast cancer families. Nat Genet. 1996;13:123–125. doi: 10.1038/ng0596-123. [DOI] [PubMed] [Google Scholar]
- 2.Phelan CM, Lancaster JM, Tonin P, Gumbs C, Cochran C, Carter R, Ghadirian P, Perret C, Moslehi R, Dion F, Faucher MC, Dole K, et al. Mutation analysis of the BRCA2 gene in 49 site-specific breast cancer families. Nat Genet. 1996;13:120–122. doi: 10.1038/ng0596-120. [DOI] [PubMed] [Google Scholar]
- 3.FitzGerald MG, MacDonald DJ, Krainer M, Hoover I, O’Neil E, Unsal H, Silva-Arrieto S, Finkelstein DM, Beer-Romero P, Englert C, Sgroi DC, Smith BL, et al. Germ-line BRCA1 mutations in Jewish and non-Jewish women with early-onset breast cancer. N Engl J Med. 1996;334:143–149. doi: 10.1056/NEJM199601183340302. [DOI] [PubMed] [Google Scholar]
- 4.Krainer M, Silva-Arrieta S, FitzGerald MG, Shimada A, Ishioka C, Kanamaru R, MacDonald DJ, Unsal H, Finkelstein DM, Bowcock A, Isselbacher KJ, Haber DA. Differential contributions of BRCA1 and BRCA2 to early-onset breast cancer. N Engl J Med. 1997;336:1416–1421. doi: 10.1056/NEJM199705153362003. [DOI] [PubMed] [Google Scholar]
- 5.Langston AA, Malone KE, Thompson JD, Daling JR, Ostrander EA. BRCA1 mutations in a population-based sample of young women with breast cancer. N Engl J Med. 1996;334:137–142. doi: 10.1056/NEJM199601183340301. [DOI] [PubMed] [Google Scholar]
- 6.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Kos-kenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 7.Antoniou AC, Pharoah PD, McMullan G, Day NE, Ponder BA, Easton D. Evidence for further breast cancer susceptibility genes in addition to BRCA1 and BRCA2 in a population-based study. Genet Epidemiol. 2001;21:1–18. doi: 10.1002/gepi.1014. [DOI] [PubMed] [Google Scholar]
- 8.Bell DW, Varley JM, Szydlo TE, Kang DH, Wahrer DC, Shannon KE, Lubratovich M, Verselis SJ, Isselbacher KJ, Fraumeni JF, Birch JM, Li FP, et al. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- 9.Meijers-Heijboer H, Wijnen J, Vasen H, Wasielewski M, Wagner A, Hollestelle A, Elstrodt F, van den Bos R, de Snoo A, Fat GT, Brekelmans C, Jagmohan S, et al. CHEK2-Breast Cancer Consortium. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 10.CHEK2 Breast Cancer Case-Control Consortium. CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74:1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allinen M, Huusko P, Mantyniemi S, Launonen V, Winqvist R. Mutation analysis of the CHK2 gene in families with hereditary breast cancer. Br J Cancer. 2001;85:209–212. doi: 10.1054/bjoc.2001.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kilpivaara O, Vahteristo P, Falck J, Syrjakoski K, Eerola H, Easton D, Bartkova J, Lukas J, Heikkila P, Aittomaki K, Holli K, Blomqvist C, et al. CHEK2 variant I157T may be associated with increased breast cancer risk. Int J Cancer. 2004;111:543–547. doi: 10.1002/ijc.20299. [DOI] [PubMed] [Google Scholar]
- 13.Seppala EH, Ikonen T, Mononen N, Autio V, Rokman A, Matikainen MP, Tammela TL, Schleutker J. CHEK2 variants associate with hereditary prostate cancer. Br J Cancer. 2003;89:1966–1970. doi: 10.1038/sj.bjc.6601425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weischer M, Bojesen SE, Tybjaerg-Hansen A, Axelsson CK, Nor-destgaard BG. Increased risk of breast cancer associated with CHEK2*1100delC. J Clin Oncol. 2007;25:57–63. doi: 10.1200/JCO.2005.05.5160. [DOI] [PubMed] [Google Scholar]
- 15.de Bock GH, Schutte M, Krol-Warmerdam EM, Seynaeve C, Blom J, Brekelmans CT, Meijers-Heijboer H, van Asperen CJ, Cornelisse CJ, Devilee P, Tollenaar RA, Klijn JG. Tumour characteristics and prognosis of breast cancer patients carrying the germline CHEK2*1100-delC variant. J Med Genet. 2004;41:731–735. doi: 10.1136/jmg.2004.019737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt MK, Tollenaar RA, de Kemp SR, Broeks A, Cornelisse CJ, Smit VT, Peterse JL, van Leeuwen FE, Van’t Veer LJ. Breast cancer survival and tumor characteristics in premenopausal women carrying the CHEK2*1100delC germline mutation. J Clin Oncol. 2007;25:64–69. doi: 10.1200/JCO.2006.06.3024. [DOI] [PubMed] [Google Scholar]
- 17.Dong X, Wang L, Taniguchi K, Wang X, Cunningham JM, McDonnell SK, Qian C, Marks AF, Slager SL, Peterson BJ, Smith DI, Cheville JC, et al. Mutations in CHEK2 associated with prostate cancer risk. Am J Hum Genet. 2003;72:270–280. doi: 10.1086/346094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dufault MR, Betz B, Wappenschmidt B, Hofmann W, Bandick K, Golla A, Pietschmann A, Nestle-Kramling C, Rhiem K, Huttner C, von Lindern C, Dall P, et al. Limited relevance of the CHEK2 gene in hereditary breast cancer. Int J Cancer. 2004;110:320–325. doi: 10.1002/ijc.20073. [DOI] [PubMed] [Google Scholar]
- 19.Cybulski C, Huzarski T, Gorski B, Masojc B, Mierzejewski M, Debniak T, Gliniewicz B, Matyjasik J, Zlowocka E, Kurzawski G, Sikorski A, Posmyk M, et al. A novel founder CHEK2 mutation is associated with increased prostate cancer risk. Cancer Res. 2004;64:2677–2679. doi: 10.1158/0008-5472.can-04-0341. [DOI] [PubMed] [Google Scholar]
- 20.Lee SB, Kim SH, Bell DW, Wahrer DC, Schiripo TA, Jorczak MM, Sgroi DC, Garber JE, Li FP, Nichols KE, Varley JM, Godwin AK, et al. Destabilization of CHK2 by a missense mutation associated with Li-Fraumeni Syndrome. Cancer Res. 2001;61:8062–8067. [PubMed] [Google Scholar]
- 21.Wu X, Webster SR, Chen J. Characterization of tumor-associated Chk2 mutations. J Biol Chem. 2001;276:2971–2974. doi: 10.1074/jbc.M009727200. [DOI] [PubMed] [Google Scholar]
- 22.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410:842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 23.Falck J, Lukas C, Protopopova M, Lukas J, Selivanova G, Bartek J. Functional impact of concomitant versus alternative defects in the Chk2-p53 tumour suppressor pathway. Oncogene. 2001;20:5503–5510. doi: 10.1038/sj.onc.1204811. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell. 2002;9:1045–1054. doi: 10.1016/s1097-2765(02)00527-0. [DOI] [PubMed] [Google Scholar]
- 25.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 26.Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/ RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 27.Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 28.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 29.Walworth NC, Bernards R. Rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 30.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 31.Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 32.Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- 33.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- 35.Jack MT, Woo RA, Hirao A, Cheung A, Mak TW, Lee PW. Chk2 is dispensable for p53-mediated G1 arrest but is required for a latent p53-mediated apoptotic response. Proc Natl Acad Sci USA. 2002;99:9825–9829. doi: 10.1073/pnas.152053599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takai H, Naka K, Okada Y. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 2002;21:5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirao A, Cheung A, Duncan G, Girard PM, Elia AJ, Wakeham A, Okada H, Sarkissian T, Wong JA, Sakai T, De Stanchina E, Bristow RG, et al. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol Cell Biol. 2002;22:6521–6532. doi: 10.1128/MCB.22.18.6521-6532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bahassi el M, Penner CG, Robbins SB, Tichy E, Feliciano E, Yin M, Liang L, Deng L, Tischfield JA, Stambrook PJ. The breast cancer susceptibility allele CHEK2*1100delC promotes genomic instability in a knock-in mouse model. Mutat Res. 2007;616:201–209. doi: 10.1016/j.mrfmmm.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 39.McPherson JP, Lemmers B, Hirao A. Collaboration of Brca1 and Chk2 in tumorigenesis. Genes Dev. 2004;18:1144–1153. doi: 10.1101/gad.1192704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim SS, Cao L, Li C, Xu X, Huber LJ, Chodosh LA, Deng CX. Uterus hyperplasia and increased carcinogen-induced tumorigenesis in mice carrying a targeted mutation of the Chk2 phosphorylation site in Brca1. Mol Cell Biol. 2004;4:9498–9507. doi: 10.1128/MCB.24.21.9498-9507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Einarsdottir K, Humphreys K, Bonnard C, Palmgren J, Iles MM, Sjolander A, Li Y, Chia KS, Liu ET, Hall P, Liu J, Wedren S. Linkage disequilibrium mapping of CHEK2: common variation and breast cancer risk. PLoS Med. 2006;3:e168. doi: 10.1371/journal.pmed.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolonel LN, Henderson BE, Hankin JH, Nomura AM, Wilkens LR, Pike MC, Stram DO, Monroe KR, Earle ME, Nagamine FS. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller CW, Ikezoe T, Krug U, Hofmann WK, Tavor S, Vegesna V, Tsukasaki K, Takeuchi S, Koeffler HP. Mutations of the CHK2 gene are found in some osteosarcomas, but are rare in breast, lung, and ovarian tumors. Genes Chromosomes Cancer. 2002;33:17–21. doi: 10.1002/gcc.1207. [DOI] [PubMed] [Google Scholar]
- 44.Sodha N, Bullock S, Taylor R, Mitchell G, Guertl-Lackner B, Williams RD, Bevan S, Bishop K, McGuire S, Houlston RS, Eeles RA. CHEK2 variants in susceptibility to breast cancer and evidence of retention of the wild type allele in tumours. Br J Cancer. 2002;87:1445–1448. doi: 10.1038/sj.bjc.6600637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hangaishi A, Ogawa S, Qiao Y, Wang L, Hosoya N, Yuji K, Imai Y, Takeuchi K, Miyawaki S, Hirai H. Mutations of Chk2 in primary hematopoietic neoplasms. Blood. 2002;99:3075–3077. doi: 10.1182/blood.v99.8.3075. [DOI] [PubMed] [Google Scholar]
- 46.Durocher D, Jackson SP. The FHA domain. FEBS Lett. 2002;513:58–66. doi: 10.1016/s0014-5793(01)03294-x. [DOI] [PubMed] [Google Scholar]
- 47.Sun Z, Hsiao J, Fay DS, Stern DF. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 48.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–510. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 49.Offit K, Pierce H, Kirchhoff T. Frequency of CHEK2*1100delC in New York breast cancer cases and controls. BMC Med Genet. 2003;4:1. doi: 10.1186/1471-2350-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaag A, Walsh T, Renbaum P, Kirchhoff T, Nafa K, Shiovitz S, Mandell JB, Welcsh P, Lee MK, Ellis N, Offit K, Levy-Lahad E, et al. Functional and genomic approaches reveal an ancient CHEK2 allele associated with breast cancer in the Ashkenazi Jewish population. Hum Mol Genet. 2005;14:555–563. doi: 10.1093/hmg/ddi052. [DOI] [PubMed] [Google Scholar]
- 51.Huang J, Domchek SM, Brose MS, Rebbeck TR, Nathanson KL, Weber BL. Germline CHEK2*1100delC mutations in breast cancer patients with multiple primary cancers. J Med Genet. 2004;41:e120. doi: 10.1136/jmg.2004.022913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mateus Pereira LH, Sigurdson AJ, Doody MM, Pineda MA, Alexander BH, Greene MH, Struewing JP. CHEK2:1100delC and female breast cancer in the United States. Int J Cancer. 2004;112:541–543. doi: 10.1002/ijc.20439. [DOI] [PubMed] [Google Scholar]