Abstract
Previously we showed that galanin, a neuropeptide, is secreted by human squamous cell carcinoma of the head and neck (SCCHN) in which it exhibits an autocrine mitogenic effect. We also showed that rap1, a ras-like signaling protein, is a critical mediator of SCCHN progression. Given the emerging importance of the galanin cascade in regulating proliferation and survival, we investigated the effect of GAL on SCCHN progression via induction of galanin receptor 2 (GALR2)-mediated rap1 activation. Studies were performed in multiple SCCHN cell lines by inducing endogenous GALR2, by stably overexpressing GALR2 and by downregulating endogenous GALR2 with siGALR2. Cell proliferation and survival, mediated by the ERK and AKT signaling cascades, respectively, were evaluated by functional and immunoblot analysis. The role of rap1 in GALR2-mediated proliferation and survival was evaluated by modulating expression. Finally, the effect of GALR2 on tumor growth was determined. GALR2 stimulated proliferation and survival via ERK and AKT activation, respectively. Knockdown or inactivation of rap1 inhibited GALR2-induced, AKT and ERK-mediated survival and proliferation. Overexpression of GALR2 promoted tumor growth in vivo. GALR2 promotes proliferation and survival in vitro, and promotes tumor growth in vivo, consistent with an oncogenic role for GALR2 in SCCHN.
Keywords: Galanin, GALR2, proliferation, apoptosis, rap1
1. Introduction
Rap1 is a tightly regulated, critical mediator of cell proliferation in aggressive cancers, including human squamous cell carcinoma of the head and neck (SCCHN), but the mechanism of rap1 activation by membrane bound receptors, like GALR2, has not been elucidated. Rap1 is a ubiquitously expressed, ras-like protein that executes its functions, by shuttling between an inactive GDP and an active GTP bound form [1–3]. We previously reported that rap1 has a significant role in growth regulatory pathways in normal keratinocytes and SCCHN [4–6]. Consistent with a role for active rap1 in tumor progression, rap1GAP, a protein that inactivates rap1, has a tumor suppressive role in SCCHN via ERK inhibition [6] Subsequent studies have supported these findings in other cancers such as melanoma, pancreatic cancer and thyroid tumors [6–9]. However, the mechanism of rap1 activation in SCCHN is relatively unknown.
Given the emerging importance of the galanin (GAL) cascade in regulating proliferation and survival in keratinocytes, we investigated the effect of GAL on SCCHN progression via induction of galanin receptor 2 (GALR2)-mediated rap1 activation. In previous studies, we showed that GAL, secreted by SCCHN, exhibits an autocrine proliferative effect [10] but the signaling cascade that regulates this effect has not been explored. Human GAL is a 30 amino acid neuropeptide that has mitogenic, neurotrophic and neuroprotective roles [11–12]. Consistent with a mitogenic function, mice overexpressing GAL develop pituitary adenomas [11–12]. GAL induces its functional effects via three different G-protein coupled receptors, galanin receptor 1 (GALR1), GALR2 and GALR3 [13]. Since antagonistic roles have been reported for individual galanin receptors, it is necessary to characterize the signaling cascade of each GALR. In SCCHN, GALR1 inhibits proliferation and its expression is reduced relative to normal tissue [10, 14]. While it is logical to infer that GALR2 and/ or GALR3 regulate proliferation in SCCHN, this role in tumor progression has not been investigated.
GALR2 has a significant role in proliferation and apoptosis in the neuronal system but its signaling mechanisms and functions in non neuronal malignancies are relatively unknown [15–17]. Furthermore, GALR2 may have opposing effects on growth and survival in different tumors, highlighting the importance of establishing its function within the context of SCCHN. In the hippocampus, GALR2 has a neuroprotective effect [18–20] whereas in PC12 cells derived from a tumor of neural crest origin, GALR2 induction inhibits proliferation and promotes apoptosis [21]. In small cell lung cancer, GALR2 induces ERK, which is a central component of mitogenic pathways [15]. Reduced function of ERK prevents cell proliferation in response to many growth factors whereas constitutive activation of upstream molecules is sufficient for ERK-mediated tumorigenesis [22].
Squamous cell carcinoma is a common cancer of the uterine cervix, skin, lung, esophagus and head and neck. SCCHN is one of the ten most common cancers globally with more than 850,000 cases diagnosed worldwide [23]. At ~50%, the 5-year survival rate is poorer than breast cancer or melanoma [24]. Unfortunately, there have been no effective novel therapeutic agents for SCCHN in over 40 years. Identification of the signaling cascade and functional effects of GALR2 will elucidate how GAL promotes SCCHN progression, and is of importance in investigating GALR2’s role as a potential therapeutic target. In the current study, we investigated the role of GALR2 in rap1-mediated proliferation and apoptosis in SCCHN.
2. Materials and Methods
2.1 Cell Culture
Human SCCHN cells and primary human oral keratinocytes (HOK) were cultured as described previously [5, 10]. UM-SCC-1 and UM-SCC-22B cells were obtained from T. Carey (University of Michigan) and were validated by genotyping in his laboratory. OSCC3 cells were obtained from P. Polverini (University of Michigan) and were genotyped in the Sequencing Core at the University of Michigan during this study.
2.2 Immunoblot Analysis
Whole cell lysates, were prepared, quantified, electrophoresed and immunoblotted, as described [5]. For immunoblot with GALR2 antibody lysates were treated with PNGase F (New England Biolabs) according to the manufacturer’s instructions. Primary antibodies for this study were: rabbit anti- GALR2 polyclonal antibody (1:1000, Alpha Diagnostics), mouse anti-rap1 (1:250; BD Transduction Laboratories), mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 1:5000; Chemicon International), anti-phospho MAPK (ERK1/2) (1:500), anti-ERK (1:1000), anti-phospho AKT (1:500), anti-AKT (1:1000) (all from Cell Signaling).
2.3 Induction of GALR2
Endogenous GALR2 was stimulated by two GALR2-specific agonists, [D-Trp2]galanin-(1–29) [25] and Galanin-Like Peptide (GALP) 1–60 (Sigma-Aldrich). Cells were serum starved for 6h prior to treatment with 300 nM [D-Trp2]galanin-(1–29) or 500nM GALP [26]. GAL (5nM to 300nM) (Sigma-Aldrich) was used to induce GALR2 in SCCHN control cells and cells stably overexpressing GALR2 protein.
2.4 Inhibition of ERK and AKT pathway
For ERK and AKT inhibition experiments, cells were pretreated with inhibitors prior to stimulation with GALR2 agonist or GAL. For ERK inhibition experiments, cells were pre-incubated with 10 μM U0126 (Promega). For PI3-kinase inhibition studies, cells were pre-incubated with 25 μM LY294002 (Sigma).
2.5 Proliferation Assay
Cells (1.5–3.0 × 104, in triplicate) were seeded in a 24-well plate. Total cell number was determined by the trypan blue enumeration assay [5].
2.6 Measurement of Apoptosis
Apoptosis was determined using FITC AnnexinV Apoptosis Detection Kit I (BD Biosciences). UM-SCC-1 cells were serum starved for 6h and pre-treated with 25μM Ly294002 for 1h followed by stimulation with 300 nM of [D-Trp2]galanin-(1–29) for 24h. UM-SCC-1 and OSCC3 cells, stably overexpressing GALR2 or control cells, were serum starved for 6h and treated with either 10μM U0126 or 25μM Ly294002 for 24h. Samples were analyzed at the University of Michigan Flow Cytometry Core.
2.7 Rap1 Activation
Active rap1 was assayed with ral-GDS protein (generous gift from Dr. J Bos, University Medical Centre Utrecht), as described [5–6, 27].
2.8 Transfections
UM-SCC-1 and OSCC3 cells were transfected with pcDNA-GALR2 (Missouri S & T cDNA resource center) and pcDNA (Invitrogen) using the Nucleofector-II system (Amaxa). A mixed clonal population was selected in the presence of Geneticin (G418). For rap1GAP stable cell lines, UM-SCC-1 cells were transfected with Flag-tagged rap1GAP in pcDNA vector (generous gift from Dr. P. Stork, Oregon Health Sciences University), as described [28].
2.9 cDNA synthesis and Quantitative Real Time PCR
Cells at 70% confluence were lysed with QIAzol and total RNA was extracted with the RNeasy Assay system (Qiagen). Eluted RNA was treated with RNase-free amplification grade DNaseI. cDNA was synthesized with Cloned AMV First-Strand cDNA Synthesis Kit (Invitrogen). Primers used for this study are: GALR2 Forward 5′-CTCTTCTGCCTCTGCTGGAT-3′, Reverse 5′-GTAGGAGACCAGGTGCGAGA-3′; GAPDH Forward 5′-GAAGGTGAAGGTCGGAGTC-3′, Reverse 5′-GAAGATGGTGATGGGATTTC-3′. Semi quantitative PCR was performed in a multiplex PCR reaction with GAPDH as an internal control. Quantitative Real Time PCR was performed in a ABI-7500 Real Time PCR machine with FastStart SYBR Green Master mix (Roche). The final products were verified by dissociation curves and data were analyzed by the relative quantification method normalized to GAPDH.
2.10 siRNA-mediated knockdown of GALR2 and rap1
Endogeneous GALR2 were knocked down by two GALR2 specific siRNAs (si7 and si10) (Dharmacon) using Oligofectamine (Invitrogen) for transfection. UM-SCC-1 cells overexpressing GALR2 and their corresponding control cells were transfected with on-target-plus rap1 smartpool siRNA (Dharmacon) at a concentration of 2 μM to downregulate rap1.
2.11 In Vivo Studies
Tumor growth was assessed in athymic mice (Ncr nu/nu strain NCI, Frederick), age 4–6 weeks (18–25g). After anesthesia, five mice were injected subcutaneously with 1×105 OSCC3 cells stably transfected with pcDNA-GALR2 or vector control in a suspension of 100μl of DMEM-fetal bovine serum/Matrigel (Becton Dickinson) in an equal ratio. Animals were euthanized after 14 days and tumor weight and volume were quantified.
2.12 Statistical Analysis
All statistical comparisons were performed using Student’s t-Test and all experiments were done in triplicate. A p-value of ≤0.05 was considered significant. For in vivo studies, mice served as their own controls as the same mice were injected with pcDNA and GALR2 cells.
3. Results
3.1 GALR2 is expressed in SCCHN
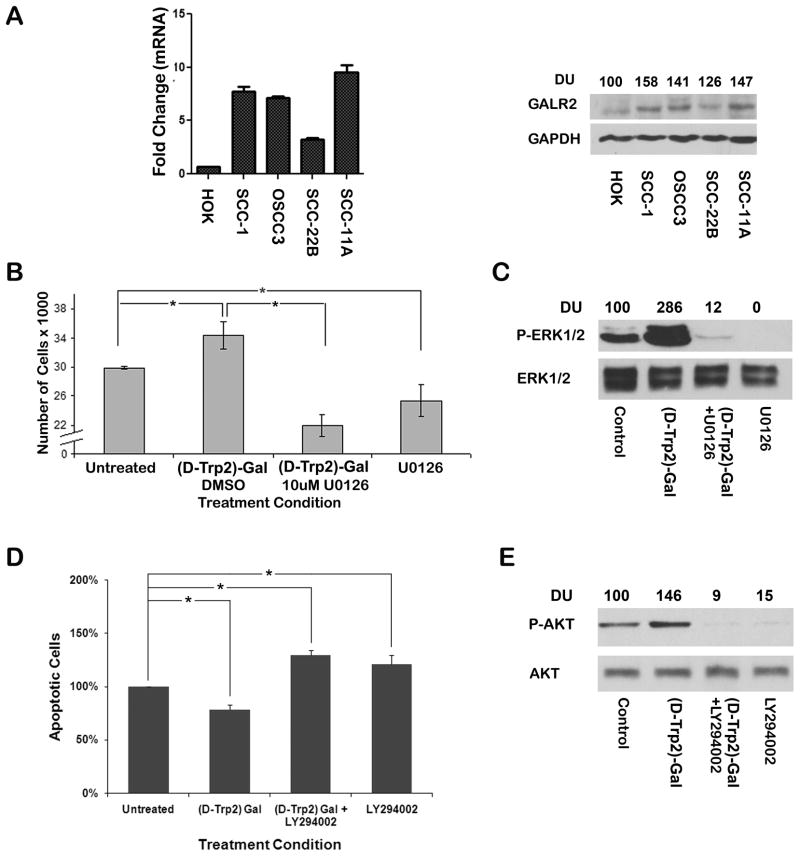
To evaluate GALR2 expression, cDNA was synthesized from normal human oral keratinocyte (HOK) and SCCHN cell lines. An upregulation of GALR2 transcript was observed in SCCHN cells compared to normal keratinocytes (HOK), as determined by qPCR (Fig 1A, left panel). Similarly, GALR2 expression was upregulated in all SCCHN cell lines, UM-SCC-(1, 22B and 11A) and OSCC3 (Fig. 1A, right panel).
Figure 1. Endogenous GALR2 promotes proliferation and survival in SCCHN.
(A) qPCR (left panel) shows the mRNA level of GALR2 in HOK, UM-SCC-1, OSCC3, UM-SCC-(22B and 11A) cells. Data were analyzed by relative quantification and expressed as fold change relative to HOK. Whole cell lysates from these cells were blotted with GALR2 and GAPDH antibodies (right panel). (B and C) GALR2 promotes proliferation via ERK activation. UM-SCC-1 cells were pre-treated with 10μM of U0126 or vehicle control for 1h followed by 300nM of [D-Trp2]galanin-(1–29) for 24h. Total cell number was determined (B; p≤0.05). ERK activation was evaluated by immunoblot analysis of whole cell lysates with phospho-ERK and total ERK (C). (D and E) GALR2 enhances cell survival via PI3K/AKT pathway. UM-SCC-1 cells were pre-incubated for 1h with 25μM of LY294002 followed by 300nM of [D-Trp2]galanin-(1–29) stimulation for 2h prior to etoposide treatment. The percentage of apoptotic cells was determined by staining with AnnexinV-FITC and propidium iodide followed by flow cytometry (D). Data are expressed as percent of the corresponding DMSO-treated control. AKT activation was evaluated by immunoblot analysis of whole cell lysates with phospho-AKT and total AKT (E).
3.2 Stimulation of endogenous GALR2 promotes proliferation and cell survival in SCCHN
Our previous studies showed that GAL, secreted by SCCHN exhibits an autocrine mitogenic effect [10]. To investigate the role of GALR2 in proliferation in SCCHN, we stimulated endogenous GALR2 in UM-SCC-1 with [D-Trp2]galanin-(1–29), a GALR2-specific agonist [25]. Twenty-four hours after treatment with 300 nM [D-Trp2]galanin-(1–29), a significant increase in cell number (p<0.05) was observed in UM-SCC-1 cells (Fig. 1B left), which was abrogated when cells were pre-treated with 10 μM U0126 (p≤0.05 Fig. 1B). U0126 is a MEK inhibitor [29].
To determine whether GALR2-induces ERK activation, UM-SCC-1 cells were stimulated with [D-Trp2]galanin-(1–29) in the presence or absence of U0126 and whole cell lysates were immunoblotted with phospho-ERK antibodies [30]. In UM-SCC-1 cells treated with 300nM [D-Trp2]galanin-(1–29), a prominent increase in phospho-ERK was observed when compared to the corresponding control cells (Fig. 1C). [D-Trp2]galanin-(1–29)-induced ERK activation was inhibited in the presence of U0126 (Fig. 1C). Thus, GALR2 upregulates proliferation via ERK activation in SCCHN.
Since cell survival has a significant role in tumor progression, we investigated whether GALR2 regulates apoptosis in SCCHN. UM-SCC-1 cells treated with [D-Trp2]galanin-(1–29) were protected against apoptosis when compared to corresponding control cells (Fig. 1D; p≤0.05). The GALR2-mediated protective effect was blocked when cells were pre-treated with 25μM LY294002 prior to stimulation with [D-Trp2]galanin-(1–29) (Fig. 1D). LY294002 is a chemical inhibitor of the PI3K/ AKT pathway [31].
To determine whether GALR2-mediated survival is regulated via AKT activation, AKT phosphorylation was investigated in whole cell lysates by immunoblot analysis with phospho-AKT (Ser473). [D-Trp2]galanin-(1–29) induced an increase in phospho-AKT compared to control cells, which was abrogated in the presence of LY294002 (Fig. 1E). Total AKT was used as a loading control.
3.3 Knockdown of endogenous GALR2 inhibits proliferation in SCCHN
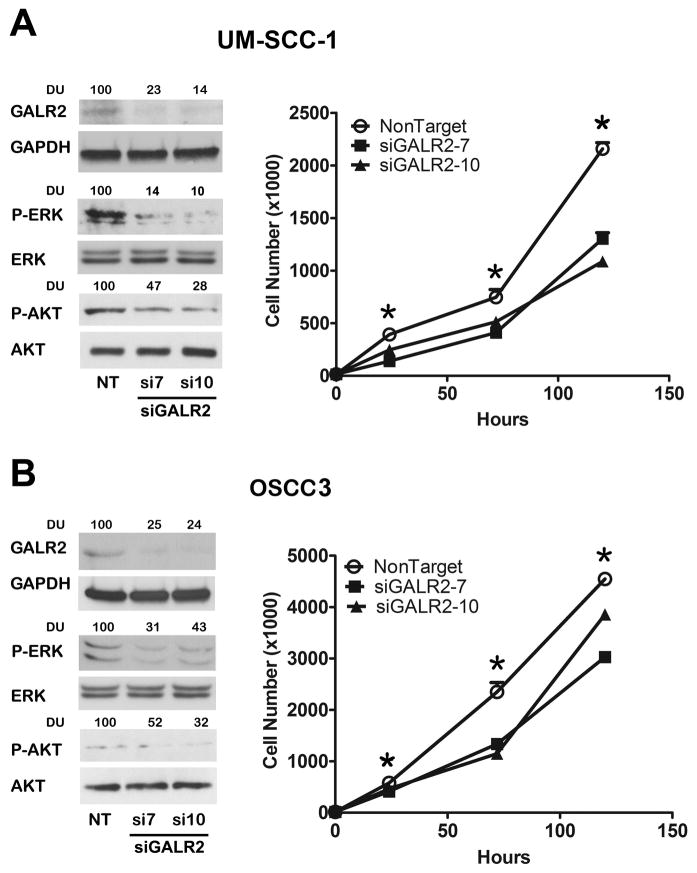
In complementary studies, GALR2 was downregulated by two different siRNAs in UM-SCC-1 (Fig. 2A) and in OSCC3 (Fig. 2B) cells. Downregulation of GALR2 was accompanied by a decrease in ERK and AKT activation when normalized to corresponding total ERK and AKT. Both siRNAs to GALR2 significantly inhibited proliferation in UM-SCC-1 and OSCC3 (Fig 2A & B right panels, respectively) compared to the corresponding cells transfected with non target siRNA.
Figure 2. Downregulation of endogeneous GALR2 by siGALR2 downregulates ERK1/2 and Akt signaling pathway and inhibits proliferation.
(A & B) GALR2 was downregulated in UM-SCC-1 (A) and OSCC3 (B) cells by two siRNAs, siGALR2-7, and siGALR2-10 or with non target siRNA(NT). GALR2 knockdown was verified by immunoblot analysis. ERK and AKT downregulation was also verified by phospho-ERK and phospho-AKT antibodies (Fig 2A left). Total ERK and total AKT was used as loading controls. GALR2 was downregulated in UM-SCC-1 (Fig. 2A right) and OSCC3 (Fig. 2B right) cells with two GALR2 specific siRNA, siGALR2-7 and siGALR2-10. Viable cells were counted on days 1, 3 and 5th post-seeding. Data are representative of three independent experiments, each with three replicates (*p<0.001).
3.4 Overexpression of GALR2 promotes proliferation and cell survival in SCCHN
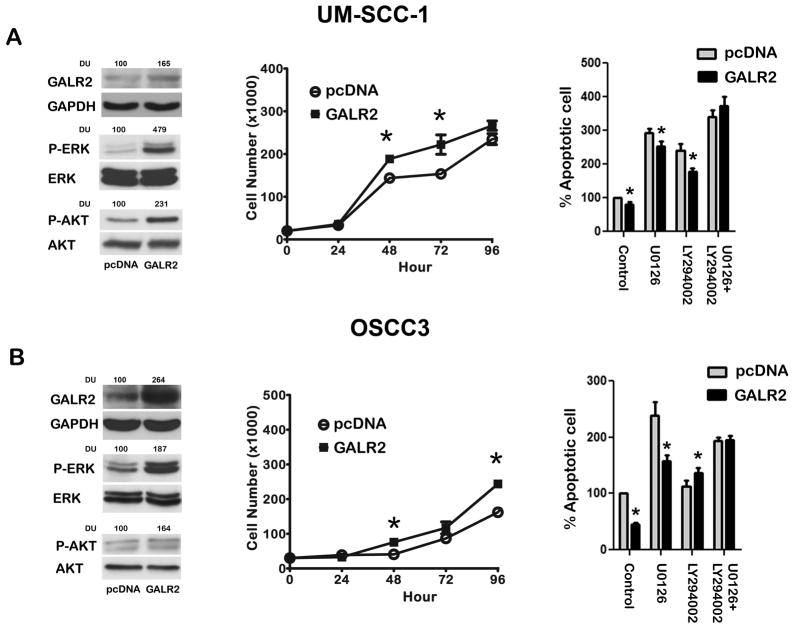
Although [D-Trp2]galanin-(1–29) is highly selective for GALR2 over GALR1 [25], we also verified whether GALR2 induces ERK and AKT activation in cells stably overexpressing GALR2. UM-SCC-1 and OSCC3 (Fig 3A & B) cells, which have different levels of endogenous GALR2 expression, were stably transfected with pcDNA-GALR2 and pcDNA vector alone. Overexpression of GALR2 was verified by Semiquantitative PCR and Quantitative Real Time PCR (Supplementary Fig S1) as well as immunoblot analysis (Fig. 3A & B left panels). Consistent with our observations with [D-Trp2]galanin-(1–29), overexpression of GALR2 induced prominent ERK and AKT activation in UM-SCC-1 and OSCC3 cells (Fig. 3A & B left panels) as compared to the corresponding control cells. Taken together these results show that GALR2 induces the AKT and ERK pathways.
Figure 3. GALR2 induces proliferation and survival.
(A & B) UM-SCC-1 (A, left panel) and OSCC3 (B, left panel) cells were stably transfected with pcDNA or pcDNA-GALR2. GALR2 expression was verified by immunoblot analysis. ERK and AKT activation were detected with phospho-ERK and phospho-AKT antibodies, respectively. Total ERK and total AKT were used to verify total protein. GAPDH was used as a loading control. GALR2 promotes proliferation. UM-SCC-1 (A, middle panel) and OSCC3 (B, middle panel) cells stably transfected with control vector and GALR2 were seeded in triplicate at 20,000 cells/well. Cell number was determined at days 1 through 4 after seeding. Values represent mean of cell number (*p<0.004). GALR2 promotes survival. UM-SCC-1 (A, right panel) and OSCC3 (B, right panel) cells stably transfected with control vector or GALR2 were treated with U0126 or LY294002 or combined were stained with AnnexinV-FITC and propidium iodide. Apoptosis was measured by flow cytometry. Data represent total apoptotic cells and are expressed as percentage of the corresponding vehicle-treated control cells. Data represent mean and SEM of three independent experiments (p≤0.05).
UM-SCC-1 and OSCC3 cells stably overexpressing GALR2 were used for subsequent functional assays. In UM-SCC-1 and OSCC3 (Fig. 3A & 3B, middle panels) GALR2 induced a significant increase in cell number as early as 48h after seeding compared to control cells (p<0.05).
Given that [D-Trp2]galanin-(1–29) protects cells against apoptosis (Fig. 1D), we investigated whether overexpressed GALR2 also protects against apoptosis in UM-SCC-1 and OSCC3. In both cell lines, GALR2 promoted proliferation and was significantly protective against apoptosis compared to pcDNA (p < 0.05) (Fig. 3A & 3B, right panels). In UM-SCC-1 and OSCC3 cells, inhibition of the AKT pathway increased apoptosis in cells stably expressing GALR2 and control vector. When compared to the corresponding pcDNA control cells, the protective effect of GALR2 on apoptosis was abrogated only in the presence of both inhibitors in UM-SCC-1 (Fig. 3A, right panel), suggesting that both the ERK and AKT pathways play a role in survival. In OSCC3, the protective effect of GALR2 on cell survival was abrogated in the presence of LY294002, but not in the presence of U0126 (Fig. 3B, right panel), consistent with a role for the AKT pathway in GALR2-mediated cell survival. These findings support that GALR2-mediated cell survival may be mediated by the AKT or both the AKT and ERK signaling pathways [30]. Thus, in SCCHN, GALR2 regulates cell proliferation and survival.
3.5 GALR2 induces ERK and AKT activation via rap1
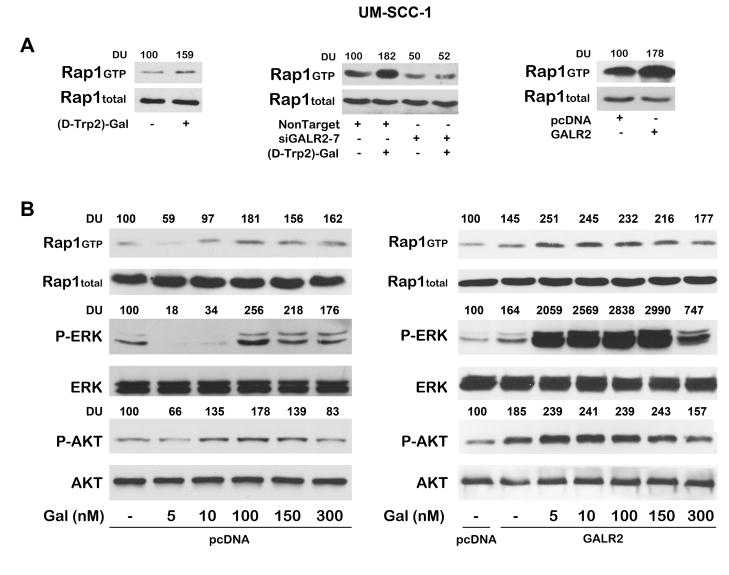
Active, GTP-bound rap1, has a significant role in growth regulatory pathways in normal and malignant keratinocytes and regulates ERK and AKT activation [6]. Therefore, we investigated whether GALR2-induced ERK and AKT activation occurs via rap1. In UM-SCC-1 cells, endogeneous GALR2 was stimulated with 300 nM of [D-Trp2]galanin-(1–29) for 24h. A prominent increase in active, GTP-bound rap1 was observed (Fig. 4A, left panel) whereas total rap1 was unchanged. In a complementary experiment, rap1 activation was significantly decreased when endogeneous GALR2 was downregulated with siGALR2-7, in the presence or absence of [D-Trp2]galanin-(1–29) (Fig. 4A, middle panel). GALR2-mediated rap1 activation was also verified in GALR2 overexpressing cells. UM-SCC-1-GALR2 cells showed an increase in rap1 activation compared to cells transfected with empty vector (Fig. 4A, right panel). Taken together, these findings with endogenous and exogenously expressed GALR2 show that GALR2 mediates rap1 activation.
Figure 4. GALR2 induces rap1, ERK and AKT activation.
(A) UM-SCC-1 cells were stimulated with 300nM of [D-Trp2]galanin-(1–29) for 24h and rap1 activation was evaluated (left panel). UM-SCC-1 cells were transfected with non target siRNA or siGALR2-7 followed by stimulation with 300nM of [D-Trp2]galanin-(1–29) for 24h and rap1 activation was evaluated (middle panel). Rap1 activation was also determined in UM-SCC-1cells overexpressing GALR2 compared to control cells (right panel). Total rap1 was used as loading controls. Active, GTP-bound rap1 was quantified by densitometry and normalized to total rap1. Data are representative of two independent experiments with similar results. (B) Dose response of GAL-mediated activation of rap1, ERK and AKT. UM-SCC-1 control cells (left panel) and GALR2 overexpressing cells (right panel) were stimulated with varying concentrations of GAL, for 1h. Whole cell lysates were blotted with phospho-ERK and phospho-AKT antibodies followed by total ERK and total AKT antibodies respectively. Rap1 activation was evaluated by the rap1 pull down assay and total rap1 was used as loading control. Active rap1, Phospho-ERK and phospho-AKT signals were normalized to corresponding total rap1, ERK and AKT, respectively and then to pcDNA.
Furthermore we investigated whether GAL mediates GALR2-induced ERK and AKT activation via rap1. UM-SCC-1 stably transfected cells with pcDNA-GALR2 and pcDNA were serum starved for 6h and were stimulated with increasing concentrations (5nM to 300nM) of GAL for 1h. SCCHN cells stably transfected with GALR2 showed an increase in rap1 activity in control cells and also upon stimulation with GAL (Fig 4B right panel). Rap1 activation was correlated with ERK and AKT activation. The increase in rap1 activity was maximal with 5 and 10 nM GAL. Active rap1, phospho-ERK and phospho-AKT were normalized to total rap1, ERK, and AKT, respectively. However UM-SCC-1 cells transfected with pcDNA showed lower induction of these pathways with GAL than GALR2 overexpressing cells (Fig 4B left panel).
3.6 GALR2-induced rap1 mediates cell survival by both ERK and AKT-dependent mechanisms
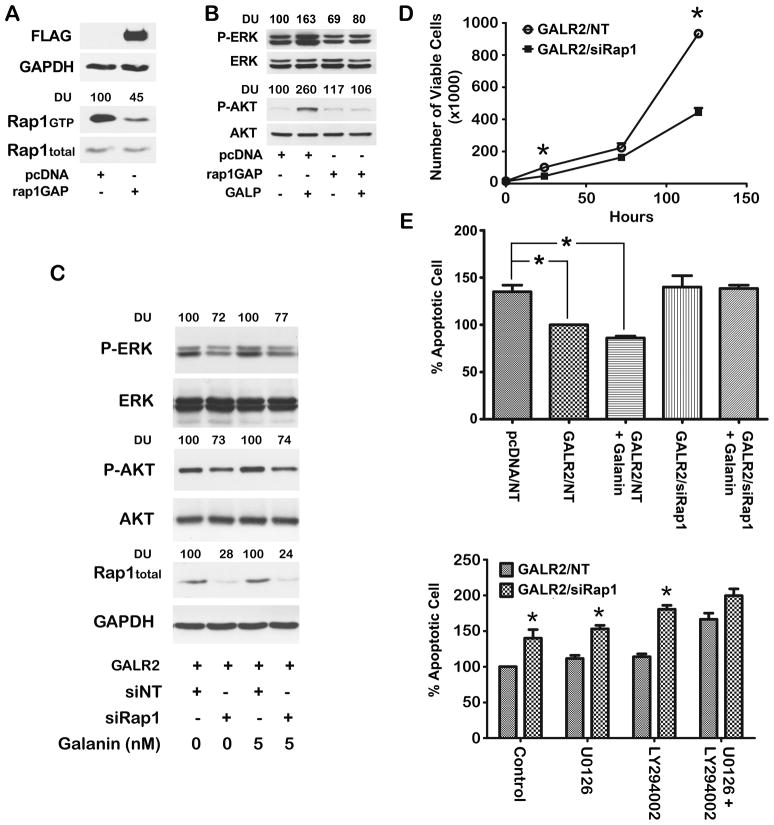
In a complementary approach, UM-SCC-1 cells stably transfected with rap1GAP were stimulated with a GALR2-specific agonist [26]. Rap1GAP is a regulatory protein that specifically inactivates GTP-bound rap1 [6]. As expected, a stronger signal for GTP-bound rap1 was observed in UM-SCC-1 cells transfected with empty vector than in cells expressing rap1GAP (Fig. 5A, left panel). Total rap1 was similar between the two groups. Rap1GAP expression was confirmed using an anti-FLAG antibody. In order to investigate whether GALR2-induced, ERK and AKT activation are mediated by rap1, UM-SCC-1 cells overexpressing rap1GAP and control cells were stimulated with 500 nM of GALP, a GALR2 specific agonist [26]. GALR2-induced ERK and AKT activation were abrogated in the presence of rap1GAP (Fig. 5B, lanes 3 and 4 compared to lanes 1 and 2). These findings are consistent with a role for rap1 in GALR2-mediated ERK and AKT activation.
Figure 5. Rap1 mediates GALR2-induced cell proliferation and survival by ERK and AKT signalling pathways.
(A) Rap1GAP inactivates rap1. UM-SCC-1 cells were stably transfected with empty vector (pcDNA) or FLAG-tagged rap1GAP and whole cell lysates were blotted with FLAG, rap1, and GAPDH antibodies. Rap1 inactivation in rap1GAP transfected cells was confirmed by pull down assay. (B) Rap1 mediates ERK and AKT activation. UM-SCC-1 cells stably transfected with pcDNA and FLAG-tagged-rap1GAP were stimulated with 500 nM GALP for 24h. Active ERK and AKT were evaluated with respect to total ERK and AKT. (C) UM-SCC-1 cells overexpressing GALR2 were transfected with rap1 siRNA and were stimulated with 5 nM GAL for 10 minutes. Active and total ERK, AKT, rap1 and GAPDH were determined. (D) GALR2 promotes proliferation via rap1. UM-SCC-1-GALR2 cells transfected with rap1 siRNA or non target siRNA were seeded in triplicate. Viable cells were counted on days 1, 3 and 5 (*p<.001). (E) GALR2 mediates cell survival via rap1. Upper panel: UM-SCC-1-GALR2 cells transfected with siRap1 or non target siRNA were stimulated with 10 nM GAL for 24h and were stained with AnnexinV-FITC and propidium iodide. The percentage of apoptotic cells was determined by flow cytometry and compared to UM-SCC-1-pcDNA with UM-SCC-1-GALR2 as 100% (*p<0.038). Lower panel: UM-SCC-1-GALR2 cells transfected with siRap1 or non target siRNA were incubated with U0126, LY294002 or both inhibitors and the percentage apoptotic cells was determined by flow cytometry and compared to their corresponding controls (*p< 0.04).
Similar findings were observed with siRNA-mediated downregulation of rap1 expression in UM-SCC-1-GALR2 cell (Fig. 5C). In these GALR2 overexpressing cells, knockdown of rap1 was verified by immunoblot analysis (Fig. 5C, lanes 2, and 4). A decrease in both ERK and AKT activation was observed with knockdown of rap1 (Fig 5C, lane 2 compared to lane 1). Knockdown of rap1 also inhibited GAL-mediated activation of ERK and AKT (Fig 5C, lane 4 compared to lane 3). Taken together the rap1GAP and siRap1 data show that GALR2-induced AKT and ERK activation are mediated by rap1.
To determine whether rap1 mediates GALR2-induced proliferation in SCCHN, rap1 was selectively knocked down. In UM-SCC-1 cells stably overexpressing GALR2, siRap1 inhibited proliferation significantly compared to cells transfected with non target siRNA (Fig. 5D), consistent with a role for rap1 in proliferation in SCCHN.
To investigate the role of rap1 in GALR2-mediated cell survival, rap1 was downregulated in UM-SCC-1 cells stably overexpressing GALR2. Compared to cells transfected with non target siRNA, knockdown of rap1 abrogated the protective effects of GALR2 on cell survival, regardless of the presence of GAL (Fig 5E, upper panel). 10 nM was selected for GAL stimulation in survival assays because of sustained activation of the ERK and AKT pathways at 24h whereas 5 nM did not induce sustained activation of either pathway at 24h.
Furthermore, in UM-SCC-1-GALR2 cells, downregulation of rap1 promoted apoptosis compared to cells transfected with non target siRNA (Fig. 5E, lower panel). Importantly, inhibition of either the ERK or AKT pathway in these cells did not abrogate the rap1-mediated protective effects on cell survival. However, when both ERK and AKT pathways were inhibited, apoptosis was similar in non target and siRap1-transfected cells. Taken together, these studies support a role for rap1 in GALR2-mediated proliferation and cell survival and emphasize that rap1-mediated ERK and AKT activation promote cell survival.
3.7. GALR2 Promotes tumor growth
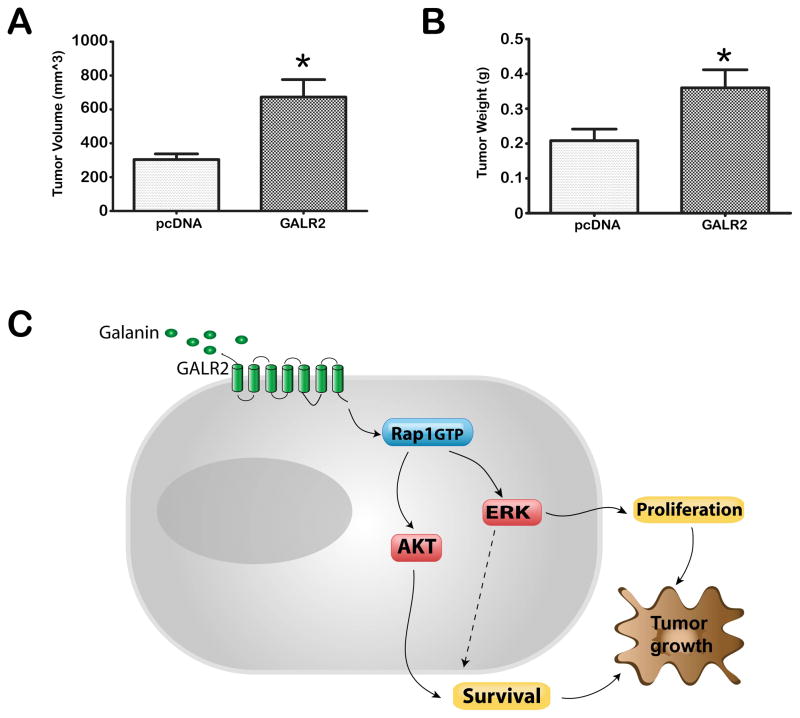
To determine the effects of GALR2 on tumor growth in vivo, SCCHN cells stably expressing GALR2 and control cells were injected subcutaneously into athymic nude mice. OSCC3 cells were used due to ease of generating tumors in mice. Consistent with the in vitro studies, OSCC3-GALR2 cells induced significantly larger tumors than the corresponding control (empty vector) cells (Fig. 6A volume, Fig. 6B weight, p<0.05). We propose a tumor progression model by oncogenic effect of GALR2 mediated through signaling cascade of rap1, ERK and AKT.
Figure 6. GALR2 promotes tumor growth.
(A & B) OSCC3 cells stably transfected with pcDNA or pcDNA-GALR2 were injected subcutaneously in mice. After 2 weeks, the tumors were harvested and tumor volume (A) (*p<0.02) and weight (B) (**p<0.01) were determined. (C) Proposed model: GALR2 mediates tumor growth via rap1-induced, ERK- and AKT-mediated proliferation and cell survival. ERK also regulates cell survival.
4. Discussion
In this study, we show that GALR2 is oncogenic in SCCHN. Stimulation of endogenous GALR2 with specific pharmacologic agonists, and overexpression of GALR2 in SCCHN, activated the ERK signaling cascade to induce tumor cell proliferation and the PI3K/AKT pathway to promote cell survival. The effects of GALR2 on cell proliferation and survival are mediated via rap1-induced ERK and AKT activation. In vivo, GALR2 promoted progression of SCCHN, supporting an oncogenic role for this G-protein coupled receptor.
GAL is a neuropeptide that has a central role in neuropathic pain and memory [32]. GAL knockout mice lose one-third of the cholinergic neurons in the basal forebrain by seven days of age and have impaired memory [11]. In neurons, induction of GALR2 stimulates trophic growth and neuroprotection [32–34]. In small cell lung cancer, GALR2 promotes ERK activation [15]. Little is known about GALR2-mediated survival in non-neuronal cells or in malignancies. Our study showing that GALR2 stimulates the ERK and AKT pathways supports that GALR2 promotes SCCHN tumor growth by enhancing both proliferation and survival. We further investigated whether GALR2 induces cell proliferation and survival via induction of rap1. Rap1 is an effector signaling molecule of G-protein coupled receptors such as the serotonin receptor and the cannabinoid receptor [35–36], that shuttles between an inactive GDP and an active GTP-bound form. In our study, we observed that in SCCHN, rap1 is activated by a GALR2 agonist as well as by overexpressed GALR2. Furthermore, GALR2-mediated ERK and AKT activation is mediated by rap1. Rap1 activates the ERK pathway in several epithelial and mesenchymal cells [3, 30]. Rap1 also activates the PI3K/AKT pathway to promote cell survival in hepatocytes [37].
Rap1 activity is regulated by GTP-GDP exchange factors and GTPase activating proteins or rapGAP [38]. Rap1GAP induces endogenous GTPase activity thereby inactivating rap1. In the present study, rap1GAP inhibited GALR2-mediated ERK and AKT phosphorylation. We previously reported that in serum-treated SCCHN cells, rap1 mediates ERK activation and proliferation but not AKT activation [4]. Since serum contains multiple growth factors, these findings suggest that rap1-mediated ERK and AKT activation in SCCHN are differentially regulated by different growth factors. Consistent with these observations, differential ERK-activation via rap1 may be mediated by different rap1 guanine nucleotide exchange factors, such as C3G and Epac, by specific second messengers [39]. C3G, which activates ERK is stimulated by protein kinase A whereas Epac, which does not activate ERK, is stimulated via a PKA-independent mechanism [40–41].
Several studies by our group and others have shown that GALR2 is expressed in SCCHN [10, 14, 42]. However, a study based on one SCCHN cell line showed that GALR2 is downregulated in SCCHN [43]. Furthermore, these anti-proliferative and pro-apoptotic effects were observed at concentrations of 1uM or higher [43]. In the present study, analyses performed in multiple SCCHN cell lines, using multiple approaches, showed that GALR2 promotes proliferation and survival in SCCHN. Endogenous GALR2 was induced by a GALR2-specific agonist. In complementary studies, overexpressed GALR2 was stimulated by GAL. This induction was at nanomolar concentrations, close to the physiologic range [44], because at very high concentrations, even mitogenic and pro-survival drugs may have toxic effects on cells.
Elucidation of the critical signaling cascades that regulate growth and survival in SCCHN, will facilitate the development of novel treatments. Previously we showed that GAL, which stimulates all three GAL receptors, is pro-proliferative in SCCHN [10]. GALR1 inhibits adenylyl cyclase via a pertusis toxin sensitive G-protein of the Gi/Go family [45–46]. In keratinocytes, GALR1 has an inhibitory effect on proliferation [10]. Consistent with this tumor suppressor role, in ~40% of SCCHN patients, the GALR1 gene, which is located on the D18S70 region of chromosome 18q, exhibits loss of heterozygosity (LOH) [47–48]. Furthermore, SCCHN patients having tumors with 18q LOH have lower survival rates than patients without 18q LOH [48]. A subsequent study showed that GALR1 is reduced in clinical samples of SCCHN relative to normal tissues, GALR2 is unchanged and GALR3 varies [14]. Together these studies suggest that disruption of the balance between expression of the galanin receptors or increased expression of GALR2, may promote tumor growth in SCCHN. In a recent study, an effective positive allosteric modulator of GALR2 was identified for epileptic seizure control [49]. The availability of GALR2 antagonists emphasizes the feasibility of targeting this receptor with currently available agents. However, the role of GALR3 in proliferation of SCCHN remains to be characterized. Subsequent studies will focus on these areas of investigation.
5. Conclusion
This study demonstrates that GALR2 induces the ERK and PI3K/AKT signaling pathways via rap1 to promote growth and survival in SCCHN cells and tumor progression in vivo.
Supplementary Material
Acknowledgments
Grant support: This work was supported by NIH/NIDCR grants R01-DE018512; K02-DE019513, and R21-DE017977and project and developmental grants from NCI P50-CA97248 (University of Michigan Head and Neck SPORE) (NJ D’Silva).
Footnotes
Conflict of Interest. The authors declare no conflict of interest.
Author contribution
RB, BSH, NR, NJD performed experiments and analyzed data. AT performed all statistical analysis. RB, BSH and NJD wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kawata M, Matsui Y, Kondo J, Hishida T, Teranishi Y, Takai Y. J Biol Chem. 1988;263(35):18965–18971. [PubMed] [Google Scholar]
- 2.Takakura A, Miyoshi J, Ishizaki H, Tanaka M, Togawa A, Nishizawa Y, Yoshida H, Nishikawa S, Takai Y. Mol Biol Cell. 2000;11(5):1875–1886. doi: 10.1091/mbc.11.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stork PJ. Trends Biochem Sci. 2003;28(5):267–275. doi: 10.1016/S0968-0004(03)00087-2. [DOI] [PubMed] [Google Scholar]
- 4.D’Silva NJ, Mitra RS, Zhang Z, Kurnit DM, Babcock CR, Polverini PJ, Carey TE. J Cell Physiol. 2003;196(3):532–540. doi: 10.1002/jcp.10331. [DOI] [PubMed] [Google Scholar]
- 5.Mitra RS, Zhang Z, Henson BS, Kurnit DM, Carey TE, D’Silva NJ. Oncogene. 2003;22(40):6243–6256. doi: 10.1038/sj.onc.1206534. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Z, Mitra RS, Henson BS, Datta NS, McCauley LK, Kumar P, Lee JS, Carey TE, D’Silva NJ. Am J Pathol. 2006;168(2):585–596. doi: 10.2353/ajpath.2006.050132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng H, Gao L, Feng Y, Yuan L, Zhao H, Cornelius LA. Cancer Res. 2009;69(2):449–457. doi: 10.1158/0008-5472.CAN-08-2399. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Chenwei L, Mahmood R, van Golen K, Greenson J, Li G, D’Silva NJ, Li X, Burant CF, Logsdon CD, Simeone DM. Cancer Res. 2006;66(2):898–906. doi: 10.1158/0008-5472.CAN-05-3025. [DOI] [PubMed] [Google Scholar]
- 9.Zuo H, Gandhi M, Edreira MM, Hochbaum D, Nimgaonkar VL, Zhang P, Dipaola J, Evdokimova V, Altschuler DL, Nikiforov YE. Cancer Res. 2010;70(4):1389–1397. doi: 10.1158/0008-5472.CAN-09-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henson BS, Neubig RR, Jang I, Ogawa T, Zhang Z, Carey TE, D’Silva NJ. J Biol Chem. 2005;280(24):22564–22571. doi: 10.1074/jbc.M414589200. [DOI] [PubMed] [Google Scholar]
- 11.O’Meara G, Coumis U, Ma SY, Kehr J, Mahoney S, Bacon A, Allen SJ, Holmes F, Kahl U, Wang FH, Kearns IR, Ove-Ogren S, Dawbarn D, Mufson EJ, Davies C, Dawson G, Wynick D. Proc Natl Acad Sci U S A. 2000;97(21):11569–11574. doi: 10.1073/pnas.210254597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perumal P, Vrontakis ME. J Endocrinol. 2003;179(2):145–154. doi: 10.1677/joe.0.1790145. [DOI] [PubMed] [Google Scholar]
- 13.Corwin RL, Rowe PM, Crawley JN. Am J Physiol. 1995;269(3 Pt 2):R511–518. doi: 10.1152/ajpregu.1995.269.3.R511. [DOI] [PubMed] [Google Scholar]
- 14.Sugimoto T, Seki N, Shimizu S, Kikkawa N, Tsukada J, Shimada H, Sasaki K, Hanazawa T, Okamoto Y, Hata A. Genes Chromosomes Cancer. 2009;48(2):132–142. doi: 10.1002/gcc.20626. [DOI] [PubMed] [Google Scholar]
- 15.Wittau N, Grosse R, Kalkbrenner F, Gohla A, Schultz G, Gudermann T. Oncogene. 2000;19(37):4199–4209. doi: 10.1038/sj.onc.1203777. [DOI] [PubMed] [Google Scholar]
- 16.Sethi T, Rozengurt E. Cancer Res. 1991;51(13):3621–3623. [PubMed] [Google Scholar]
- 17.Seufferlein T, Rozengurt E. Cancer Res. 1996;56(24):5758–5764. [PubMed] [Google Scholar]
- 18.Mazarati A, Lu X. Neuropeptides. 2005;39(3):277–280. doi: 10.1016/j.npep.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Mitsukawa K, Lu X, Bartfai T. Cell Mol Life Sci. 2008;65(12):1796–1805. doi: 10.1007/s00018-008-8153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elliott-Hunt CR, Marsh B, Bacon A, Pope R, Vanderplank P, Wynick D. Proc Natl Acad Sci U S A. 2004;101(14):5105–5110. doi: 10.1073/pnas.0304823101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tofighi R, Joseph B, Xia S, Xu ZQ, Hamberger B, Hokfelt T, Ceccatelli S. Proc Natl Acad Sci U S A. 2008;105(7):2717–2722. doi: 10.1073/pnas.0712300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutkind JS. Sci STKE 2000. 2000;(40):RE1. doi: 10.1126/stke.2000.40.re1. [DOI] [PubMed] [Google Scholar]
- 23.Chin D, Boyle GM, Porceddu S, Theile DR, Parsons PG, Coman WB. Expert Rev Anticancer Ther. 2006;6(7):1111–1118. doi: 10.1586/14737140.6.7.1111. [DOI] [PubMed] [Google Scholar]
- 24.Todd R, Donoff RB, Wong DT. J Oral Maxillofac Surg. 1997;55(6):613–623. doi: 10.1016/s0278-2391(97)90495-x. discussion 623–615. [DOI] [PubMed] [Google Scholar]
- 25.Smith KE, Forray C, Walker MW, Jones KA, Tamm JA, Bard J, Branchek TA, Linemeyer DL, Gerald C. J Biol Chem. 1997;272(39):24612–24616. doi: 10.1074/jbc.272.39.24612. [DOI] [PubMed] [Google Scholar]
- 26.Ohtaki T, Kumano S, Ishibashi Y, Ogi K, Matsui H, Harada M, Kitada C, Kurokawa T, Onda H, Fujino M. J Biol Chem. 1999;274(52):37041–37045. doi: 10.1074/jbc.274.52.37041. [DOI] [PubMed] [Google Scholar]
- 27.Franke B, Akkerman JW, Bos JL. Embo J. 1997;16(2):252–259. doi: 10.1093/emboj/16.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitra RS, Goto M, Lee JS, Maldonado D, Taylor JM, Pan Q, Carey TE, Bradford CR, Prince ME, Cordell KG, Kirkwood KL, D’Silva NJ. Cancer Res. 2008;68(10):3959–3969. doi: 10.1158/0008-5472.CAN-07-2755. [DOI] [PubMed] [Google Scholar]
- 29.Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM. J Biol Chem. 1998;273(29):18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 30.Roux PP, Blenis J. Microbiol Mol Biol Rev. 2004;68(2):320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vlahos CJ, Matter WF, Hui KY, Brown RF. J Biol Chem. 1994;269(7):5241–5248. [PubMed] [Google Scholar]
- 32.Hobson SA, Bacon A, Elliot-Hunt CR, Holmes FE, Kerr NC, Pope R, Vanderplank P, Wynick D. Cell Mol Life Sci. 2008;65(12):1806–1812. doi: 10.1007/s00018-008-8154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crawley JN. Regul Pept. 1995;59(1):1–16. doi: 10.1016/0167-0115(95)00083-n. [DOI] [PubMed] [Google Scholar]
- 34.Wraith DC, Pope R, Butzkueven H, Holder H, Vanderplank P, Lowrey P, Day MJ, Gundlach AL, Kilpatrick TJ, Scolding N, Wynick D. Proc Natl Acad Sci U S A. 2009;106(36):15466–15471. doi: 10.1073/pnas.0903360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He JC, Gomes I, Nguyen T, Jayaram G, Ram PT, Devi LA, Iyengar R. J Biol Chem. 2005;280(39):33426–33434. doi: 10.1074/jbc.M502812200. [DOI] [PubMed] [Google Scholar]
- 36.Fricker AD, Rios C, Devi LA, Gomes I. Brain Res Mol Brain Res. 2005;138(2):228–235. doi: 10.1016/j.molbrainres.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 37.Cullen KA, McCool J, Anwer MS, Webster CR. Am J Physiol Gastrointest Liver Physiol. 2004;287(2):G334–343. doi: 10.1152/ajpgi.00517.2003. [DOI] [PubMed] [Google Scholar]
- 38.Christian SL, Lee RL, McLeod SJ, Burgess AE, Li AH, Dang-Lawson M, Lin KB, Gold MR. J Biol Chem. 2003;278(43):41756–41767. doi: 10.1074/jbc.M303180200. [DOI] [PubMed] [Google Scholar]
- 39.Zwartkruis FJ, Bos JL. Exp Cell Res. 1999;253(1):157–165. doi: 10.1006/excr.1999.4695. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Dillon TJ, Pokala V, Mishra S, Labudda K, Hunter B, Stork PJ. Mol Cell Biol. 2006;26(6):2130–2145. doi: 10.1128/MCB.26.6.2130-2145.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stork PJ, Dillon TJ. Blood. 2005;106(9):2952–2961. doi: 10.1182/blood-2005-03-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanazawa T, Iwashita T, Kommareddi P, Nair T, Misawa K, Misawa Y, Ueda Y, Tono T, Carey TE. Oncogene. 2007;26(39):5762–5771. doi: 10.1038/sj.onc.1210384. [DOI] [PubMed] [Google Scholar]
- 43.Kanazawa T, Kommareddi PK, Iwashita T, Kumar B, Misawa K, Misawa Y, Jang I, Nair TS, Iino Y, Carey TE. Clin Cancer Res. 2009;15(7):2222–2230. doi: 10.1158/1078-0432.CCR-08-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grenback E, Bjellerup P, Wallerman E, Lundblad L, Anggard A, Ericson K, Aman K, Landry M, Schmidt WE, Hokfelt T, Hulting AL. Regul Pept. 2004;117(2):127–139. doi: 10.1016/j.regpep.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 45.Wang S, Hashemi T, Fried S, Clemmons AL, Hawes BE. Biochemistry. 1998;37(19):6711–6717. doi: 10.1021/bi9728405. [DOI] [PubMed] [Google Scholar]
- 46.Smith KE, Walker MW, Artymyshyn R, Bard J, Borowsky B, Tamm JA, Yao WJ, Vaysse PJ, Branchek TA, Gerald C, Jones KA. J Biol Chem. 1998;273(36):23321–23326. doi: 10.1074/jbc.273.36.23321. [DOI] [PubMed] [Google Scholar]
- 47.Carey TE, Frank CJ, Raval JR, Jones JW, McClatchey KD, Beals TF, Worsham MJ, Van Dyke DL. Acta Otolaryngol Suppl. 1997;529:229–232. doi: 10.3109/00016489709124130. [DOI] [PubMed] [Google Scholar]
- 48.Pearlstein RP, Benninger MS, Carey TE, Zarbo RJ, Torres FX, Rybicki BA, Dyke DL. Genes Chromosomes Cancer. 1998;21(4):333–339. doi: 10.1002/(sici)1098-2264(199804)21:4<333::aid-gcc7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 49.Lu X, Roberts E, Xia F, Sanchez-Alavez M, Liu T, Baldwin R, Wu S, Chang J, Wasterlain CG, Bartfai T. Proc Natl Acad Sci U S A. 2010;107(34):15229–15234. doi: 10.1073/pnas.1008986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.