Abstract
It has been over 50 years since Sir James Black developed the first beta adrenergic receptor (βAR) blocker to treat heart disease. At that time, the concept of cell-surface receptors was relatively new and not widely accepted, and most of the tools now used to characterize plasma membrane receptors had not been developed. There has been remarkable progress in receptor biology since then, including the development of radioligand binding assays, the biochemical characterization of receptors as discrete membrane proteins, and the cloning of the first G protein-coupled receptors (GPCRs), which led to the identification of other members of the large family of GPCRs. More recently, progress in GPCR structural biology has led to insights into the three-dimensional structures of βARs receptors in both active and inactive states. Despite all of this progress, the process of developing a drug for a particular GPCR target has become more complex, time-consuming, and expensive.
Adrenergic receptor subtypes and complex signaling behavior
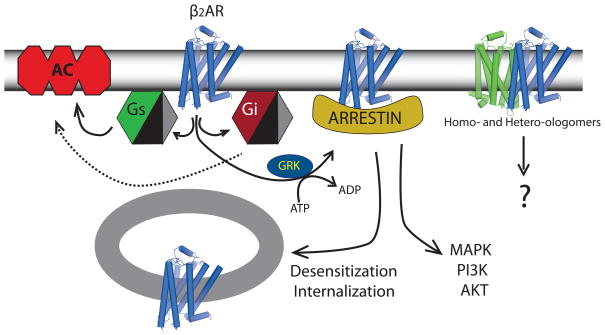
When Sir James Black and colleagues developed propranolol, the scope of adrenergic receptor pharmacology was limited to two subtypes: alpha (α) and beta (β) adrenergic receptors. Catecholamines induced vasoconstriction through αARs and increased heart rate and contraction through βARs. Since then, nine adrenergic receptor genes have been identified [IUPHAR database (http://www.iuphar-db.org/DATABASE/GPCRListForward] : α1AAR, α1BAR, α1DAR, α2AAR, α2BAR, α2CAR, β1AR, β2AR, and β3AR. The β2AR has been one of the principal model systems for hormone-activated G protein-coupled receptors (GPCRs). One of the first radioligands was developed for βARs allowing identification and characterization of β2AR in cells and tissues. Detailed characterization of agonist and antagonist binding behavior provided insight into the allosteric effect of G proteins on receptor structure and agonist binding affinity [1]. Ligand binding assays also played a central role in the purification of β2AR from lung tissue. This purified receptor was used to obtain the amino acid sequence that led to the cloning of the β2AR [2], providing the first insights into the structure of a hormone-activated GPCR and enabling the identification of domains involved in ligand binding and G protein coupling through mutagenesis [3] and chimeric receptor studies [4]. More recently, the β2AR was the first hormone activated GPCR to yield to structure determination by X-ray crystallography [5,6]. Understanding of the cellular mechanisms by which adrenergic receptors mediate the effects of catecholamines has also evolved considerably since the development of propranolol. Figure 1 is a simplified diagram of the diverse signaling pathways that have been identified for the β2AR. Early studies revealed the predominant signaling pathway to be from agonist-activated β2AR to Gs, the stimulatory G protein for adenylyl cyclase. However, it is now known that the β2AR also couples to Gi proteins [7, 8] and activates G protein-independent pathways through arrestins [9–11].
Figure 1.
Signaling pathways regulated by the β2AR. Solid lines indicate activation while dashed lines indicate inhibition. Abbreviations: AC - adenylyl cyclase, GRK -G protein coupled receptors kinase, Gs – the stimulatory G protein for adenylyl cyclase, Gi – pertussis toxin sensitive G proteins, MAPK – mitogen activated protein kinase, PI3K -phosphoinositide 3 kinase, AKT – a serionine/threonine protein kinase initially identified in the AKT virus.
Up until the mid 1990’s, it was generally accepted that GPCRs existed as monomers in the plasma membrane. However, in 1993, Maggio and colleagues observed functional complementation when two inactive chimeric receptors, composed of sequences from the α2AAR and M3 muscarinic receptors, were co-expressed in cells [12]. This result could best be explained by receptor dimerization. Then in 1996, Hébert and Bouvier demonstrated differentially tagged β2ARs could be co-immunoprecipitated from insect cell membranes [13], suggesting that they exist as dimers or oligomers in the plasma membrane. These early reports led to a flood of studies by investigators employing a variety of innovative techniques to characterize both homo-and heterooligomeric GPCRs in living cells. Although GPCR oligomers are now widely accepted, several studies have shown that they are not required for activation of G proteins [14–18], and their physiologic role in GPCR function is not completely understood. Oligomerization has been proposed to play roles in posttranslational processing and trafficking to the plasma membrane, and in generating functional diversity through heterooligomerization [19].
More than just beta blockers
The idea of ligand efficacy was proposed by in 1956 [20]. At the time Sir James was developing propranolol, ligands fell into three categories: full agonists, partial agonists, and antagonists or blockers. Full agonists are drugs that maximally activate the receptor in a signaling assay (either direct G protein activation or activation of an effector protein regulated by a G protein), whereas partial agonists produce submaximal activity even at saturating concentrations. The concept of inverse agonism emerged with the observation that many GPCRs exhibit basal, agonist-independent activity and some ligands, previously classified as blockers or antagonists with “zero” efficacy, could inhibit this basal activity. Propranolol has been classified as an inverse agonist because it inhibits basal activity of both β1AR and β2AR [21, 22].
More recently, pluridimensional efficacy was proposed to describe the observed effect of ligands on specific signaling pathways [11, 23]. As discussed above, for the β2AR, many GPCRs activate more than one G protein. The efficacy profile and rank order of potency of ligands can be G protein specific [24–26]. Moreover, some GPCRs signal through G protein-independent pathways. As an example, the β2AR can regulate mitogen-activated protein kinase (MAPK) signaling pathways through arrestins. The efficacy of ligands for activating these arrestin pathways can differ from those that activate Gs. Carvedilol is an inverse agonist for β2AR activation of Gs but a partial agonist for β2AR activation of arrestin [22]. The complexity of GPCR signaling pathways and ligand efficacy profiles complicate the process of drug discovery. Moreover, specific receptors might exhibit cell-type specific signaling as a consequence of the cell-specific complement of signaling, regulatory and scaffold proteins. As such, it is very difficult to develop a high throughput assay that accurately reflects the physiological environment of the target GPCR.
GPCR structural biology
Two- and three-dimensional crystal structures of rhodopsin purified from bovine retina provided the first three-dimensional images of GPCRs [27–29]. However, hormone-activated GPCRs proved to be more challenging subjects for structural studies due to the lack of a naturally abundant source of protein, as well as their inherent structural instability, and the relatively small amount of structured polar surface available for forming crystal lattice contacts. Three approaches have been used to facilitate crystallogenesis of GPCRs: i) the use of antibody fragments [5]; ii) protein engineering to create T4 lysozyme fusion proteins [6]; and iii) thermostabilization by alanine scanning mutagenesis [30]. These approaches, along with lipid mediated in meso- [31] and bicelle-based [32] screens, and recently developed microfocus diffraction techniques [33, 34] contributed to the first GPCR structures. The first crystal structure of the β2AR was obtained using an antibody fragment that recognized a three-dimensional epitope consisting of the cytoplasmic ends of TM5 and TM6. These crystals were grown in bicelles (a mixture of lipid and detergent) and yielded a partial structure to 3.4 Å [5]. The β2AR was also the first GPCR to be crystallized as a T4Lysozyme (T4L) fusion protein. In this approach, the unstructured third intracellular loop of the β2AR was replaced by T4L, a small well-folded protein known to crystallize under many different conditions [6]. Like the antibody fragment, the T4L component provides a surface for crystal lattice contacts. Crystals of the β2AR-T4L fusion protein were grown in lipidic cubic phase [31] and led to a 2.4Å structure. This T4L approach, initially developed for the β2AR, has been used to obtain structures of three other GPCRs including the A2A adenosine receptor [35], the CXCR4 receptor [36] and the D3 dopamine receptor [37]. Finally, the turkey β1AR was obtained by creating a thremostabilized mutant that was also stable in detergents with short alkyl chains enabling crystallization in small detergent micelles [30].
Structural insights into subtype selectivity
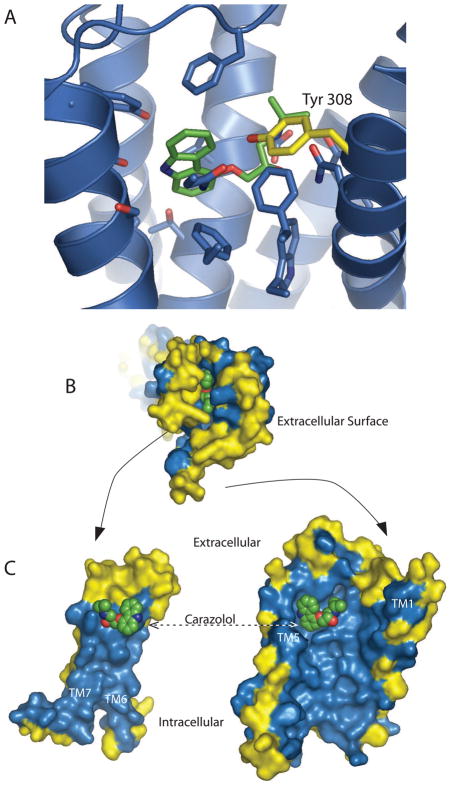
The β1AR and β2AR structures provided the first high-resolution picture of the ligand-binding pocket of an adrenergic receptor, and the first insights into the challenges involved in developing subtype-selective ligands. Figure 2A shows the binding pocket of the human β2AR. Only one of the 15 amino acids that constitute the antagonist binding pocket (defined as being within 4Å of the inverse agonist carazol) differs between β1AR and β2AR. Tyr308 at the top of TM7 in the β2AR is Phe in the β1AR. This high degree of binding pocket homology extends to the more distantly related αARs. For example, there are only four differences between the ligand binding pockets of the α2AAR and β2AR. Of interest, one of these differences (Asn 312 in β2AR is Phe in homologous position 412 the α2AAR) was previously identified in chimeric receptor and mutagenesis studies to contribute to βAR antagonist binding specificity. The single mutation of Phe 412 to Asn in the human α2AR receptor resulted 3,000-fold increase in binding affinity for the β2AR antagonist dihydroalprenolol [38].
Figure 2.
Subtype specific ligand binding in the β2AR. Amino acids that differ between β1AR and β2AR are shown in yellow. The inverse agonist carazolol is shown with green carbons. A, The binding pocket of the human β2AR. Only one of the 15 amino acids that constitute the antagonist binding pocket (defined as being within 4A of the inverse agonist carazol) differs between β1AR and β2AR. Tyr 308 at the top of TM7 in the β2AR is Phe in the β1AR. B, The extracellular surface of the β2AR. C, The interior surface of the β2AR that has been split along the plane of the binding pocket, TMs1 through 5 on the right and TMs 6 and 7 on the left.
Figure 2B shows the extracellular surface of the β2AR, where residues that differ with the β1AR are colored in yellow. Figure 2C shows the interior surface of a β2AR that has been split along the binding pocket, TMs1 through 5 on the right and TMs 6 and 7 on the left. All of the residues that line the interior of the receptor from the binding pocket to the cytoplasm are identical between β1AR and β2AR. It can be seen that structural diversity is much greater for amino acids lining the pathway into the binding pocket. Although these aminoacids do not make direct contact with a bound ligand the size of carazolol, they could contribute to initial transient interactions between the ligand and receptor that contribute to the ligand association rate. These residues might also play a role in binding larger ligands that extend into the vestibule of the binding pocket, such as the long-acting beta agonist salmeterol that is used in the treatment of asthma.
Structural changes upon activation
Thinking about activation of GPCRs has evolved considerably over the past two decades. Early models consisted of two receptor states: an inactive state (R) in equilibrium with an active state (R*) [39]. In these models, the level of receptor activity depends on the effect of ligands on this equilibrium (R DR*), with agonists and partial agonists shifting the equilibrium to the right, and inverse agonists shifting the equilibrium to the left. Although many aspects of GPCR function can be explained by a simple two-state model, evidence from biophysical and functional studies support a multistate model in which ligands stabilize a specific conformational state or set of states [40].
This conformational heterogeneity contributes to the challenges in obtaining structures of GPCRs, particularly active structures. Although most GPCRs exhibit some basal activity, the inactive conformation predominates and can be further stabilized by inverse agonists. The majority of GPCR crystal structures to date represent inverse agonist stabilized inactive states.
For the purpose of this discussion, I will define the active state of the β2AR as the state in which the receptor is bound to a full agonist and forms a complex with nucleotide-free Gs, the G protein that activates adenylyl cyclase. However, it should be noted that it is possible that the Gs activating conformation of the β2AR might differ from the conformations that activate Gi proteins and arrestins. The active state of the β2AR is a relatively unstable, high-energy state that is not adequately stabilized by agonists alone. Evidence for this instability comes from ligand binding studies in which agonist binding affinity is 100-fold lower for β2AR in the absence of Gs [15].
Furthermore, biophysical experiments show that full agonists are not capable of shifting the conformational equilibrium to a single active state [41, 42], and crystallography experiments show that even when bound to a covalent agonist, the β2AR crystallizes in an inactive conformation [43]. The active state can be stabilized through interactions with the G protein Gs, but forming a stable β2AR-Gs complex in detergents used for crystallography is challenging.
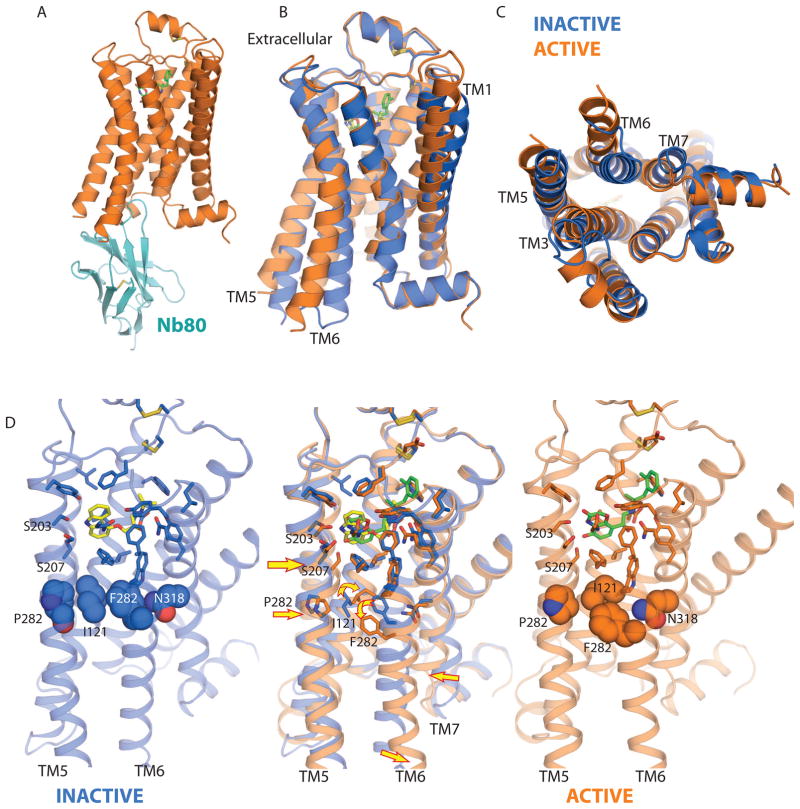
As an alternative approach, we generated a camelid antibody (nanobody) that exhibits G protein-like behavior. This nanobody (Nb80) preferentially binds to agonist occupied β2AR, and when bound to Nb80, the affinity of the β2AR for agonists increases 100-fold, similar to what is observed in a β2AR-Gs complex. We were able to crystallize the β2AR-Nb80 complex with the receptor bound to an ultra high affinity agonist (80 pM) [44]. As shown in Figure 3A, CDR3 of Nb80 projects into the cytoplasmic core of the protein, occupying a position similar to that of the transducin peptide in opsin [45]. Moreover, the structural changes that are observed in the cytoplasmic domains are similar to those observed in opsin. The largest change involves an 11Å outward movement of TM6, and inward movement of TMs 3 and 7 (Fig. 3B, C).
Figure 3.
Active and inactive states of the β2AR. A, β2AR-Nb80 complex. CDR3 of Nb80 projects into the cytoplasmic core of the protein. B, Side view of superimposed active (orange) and inactive (blue) conformations of the β2AR. C. Cytoplasmic view of superimposed active and inactive states. D, Binding pocket of active and inactive states of the β2AR are shown separately (outer panels) and overlapped (center). Small changes in the ligand binding pocket lead to rearrangement of the conserved amino acids (Pro 211, Ile 121, Phe 282, and Asn 318). These changes are indicated by arrows in the center panel. Carazolol is shown with yellow carbons and the agonist BI-167107 is shown with green carbons.
Differences in the ligand binding pocket are more subtle. Figure 3D shows a comparison of the ligand binding pockets for the inactive carazolol-bound β2AR with the active-state agonist-bound receptor. The agonist BI and the inverse agonist carazolol share many interactions with amino acids within the binding pocket. The major difference involves interactions between Ser 203, Ser 207, and Asn 293 with the heterocycle of the agonist. These interactions have been shown to be important for agonist binding and activation in mutagenesis studies [46,47]. There are 4 potential polar interactions between the receptor and the agonist heterocycle; in contrast, in the inactive structure there is only one polar interaction between Ser 203 and the carazolol heterocycle. As a result of the more extensive interactions with the agonist, there is a 2Å inward movement of TM5 around Ser207 that is associated with a disruption of intramolecular interactions around the highly conserved Pro 211. The packing interactions involving Pro211 (TM5), Ile121 (TM3), Phe 282 (TM6) and Asn 318(TM7) contribute to the network of interactions that stabilize the inactive conformation (Fig. 3D left panel). Agonist binding alters these packing interactions resulting in a rotation of TM6 around Phe282, with a consequent outward movement of the cytoplasmic end of TM6 along with the other changes shown in Fig. 3D. Of interest, a similar mechanism has recently been proposed for the H1 histamine receptor based on mutagenesis and modeling studies [48].
The impact of structural biology on GPCR drug discovery
Structural biology has been an integral part of drug discovery for soluble proteins such as kinases and proteases [49], and will likely play a role in the development of drugs for GPCRs. However, there are important differences in the time required to obtain crystal structures and the ability to manipulate crystals. The value of a crystal structure in drug development is greatest when it is available early in the discovery process so that crystals might be used for fragment-based screens and crystal structures might aid in optimizing an early lead from high throughput screens. In contrast, based on the initial efforts, crystallization of GPCRs is likely to take longer than crystallization of soluble protein targets. Each new GPCR target requires the generation of modified forms of the receptor to facilitate expression and purification, and some of these modifications will be receptor specific, necessitating trial and error mutagenesis. Identifying conditions for solubilization of functional receptor protein from cell membranes and purifying monodisperse receptor protein for crystallogenesis usually takes more time than for more tractable soluble protein targets that are readily expressed in E. coli. Moreover, the structures will likely be of lower resolution, and the crystals will be more fragile and radiation sensitive.
Nevertheless, the crystal structure of the inactive state β2AR has enabled the identification of new β2AR ligands through in silico screening [50, 51]. Of interest, all of the ligands identified in one of the screens were characterized in a functional assay as inverse agonists [51], suggesting that the compounds identified through in silico screens will have an efficacy profile similar to that of the ligand used to obtain the crystal structure. Finally, crystals of orphan GPCRs for which no ligand is available pose special problems, because ligands are often needed to stabilize the receptor during expression, purification and crystallogenesis, and for monitoring the function of the protein during protein engineering and stabilization experiments. Nevertheless, although structural biology might never be as versatile a tool for GPCR drug discovery as it is for soluble protein targets, it will likely play an increasingly important role for challenging GPCRs for which a structure of the bound ligand can help chemists improve affinity and selectivity profiles. Moreover, we can expect technological and methodological advances to reduce the time required, and improve the quality GPCR crystals. Finally, as we accumulate more structures from different families and subfamilies of GPCRs, molecular models of the remaining GPCRs will become more accurate and therefore more valuable in drug discovery.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lefkowitz RJ. Seven transmembrane receptors: something old, something new. Acta Physiol (Oxf) 2007;190(1):9–19. doi: 10.1111/j.1365-201X.2007.01693.x. [DOI] [PubMed] [Google Scholar]
- 2.Dixon RA, et al. Cloning of the gene and cDNA for mammalian beta-adrenergic receptor and homology with rhodopsin. Nature. 1986;321:75–79. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- 3.Dixon RA, Sigal IS, et al. Structural features required for ligand binding to the beta-adrenergic receptor. Embo J. 1987;6(11):3269–75. doi: 10.1002/j.1460-2075.1987.tb02645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobilka BK, Kobilka TS, et al. Chimeric alpha 2-,beta 2-adrenergic receptors: delineation of domains involved in effector coupling and ligand binding specificity. Science. 1988;240(4857):1310–6. doi: 10.1126/science.2836950. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen SG, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 6.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 7.Daaka Y, et al. Switching of the coupling of the beta2-adrenergic receptor to different G proteins by protein kinase A. Nature. 1997;390:88–91. doi: 10.1038/36362. [DOI] [PubMed] [Google Scholar]
- 8.Devic E, et al. Beta-adrenergic receptor subtype-specific signaling in cardiac myocytes from beta(1) and beta(2) adrenoceptor knockout mice. Mol Pharmacol. 2001;60:577–583. [PubMed] [Google Scholar]
- 9.Azzi M, et al. Beta-arrestin-mediated activation of MAPK by inverse agonists reveals distinct active conformations for G protein-coupled receptors. Proc Natl Acad Sci U S A. 2003;100:11406–11411. doi: 10.1073/pnas.1936664100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shenoy SK, et al. beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor. J Biol Chem. 2006;281:1261–1273. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 11.Rajagopal S, et al. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maggio R, et al. Coexpression studies with mutant muscarinic/adrenergic receptors provide evidence for intermolecular “cross-talk” between G-protein-linked receptors. Proc Natl Acad Sci U S A. 1993;90:3103–3107. doi: 10.1073/pnas.90.7.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebert TE, et al. A peptide derived from a beta2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. J Biol Chem. 1996;271:16384–16392. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 14.Bayburt TH, et al. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 15.Whorton MR, et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuszak AJ, et al. Purification and functional reconstitution of monomeric mu-opioid receptors: allosteric modulation of agonist binding by Gi2. J Biol Chem. 2009;284:26732–26741. doi: 10.1074/jbc.M109.026922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whorton MR, et al. Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J Biol Chem. 2008;283:4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjee S, et al. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J Mol Biol. 2008;377:10671081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 19.Terrillon S, Bouvier M. Roles of G-protein-coupled receptor dimerization. EMBO Rep. 2004;5:30–34. doi: 10.1038/sj.embor.7400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephenson RP. A modification of receptor theory. Br J Pharmacol Chemother. 1956;11:379–393. doi: 10.1111/j.1476-5381.1956.tb00006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galandrin S, et al. Conformational rearrangements and signaling cascades involved in ligand-biased mitogen-activated protein kinase signaling through the beta1-adrenergic receptor. Mol Pharmacol. 2008;74:162–172. doi: 10.1124/mol.107.043893. [DOI] [PubMed] [Google Scholar]
- 22.Wisler JW, et al. A unique mechanism of beta-blocker action: Carvedilol stimulates beta-arrestin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16657–16662. doi: 10.1073/pnas.0707936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenakin T. Functional Selectivity and Biased Receptor Signaling. J Pharmacol Exp Ther. 2010 doi: 10.1124/jpet.110.173948. [DOI] [PubMed] [Google Scholar]
- 24.Kenakin T. Ligand-selective receptor conformations revisited: the promise and the problem. Trends Pharmacol Sci. 2003;24:346–354. doi: 10.1016/S0165-6147(03)00167-6. [DOI] [PubMed] [Google Scholar]
- 25.Violin JD, Lefkowitz RJ. Beta-arrestin-biased ligands at seven-transmembrane receptors. Trends Pharmacol Sci. 2007;28:416–422. doi: 10.1016/j.tips.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Kenakin TP. ‘7TM receptor allostery: putting numbers to shapeshifting proteins. Trends Pharmacol Sci. 2009;30:460–469. doi: 10.1016/j.tips.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 28.Unger VM, Schertler GF. Low resolution structure of bovine rhodopsin determined by electron cryo-microscopy. Biophys J. 1995;68:1776–1786. doi: 10.1016/S0006-3495(95)80354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, et al. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 30.Warne T, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caffrey M. Crystallizing membrane proteins for structure determination: use of lipidic mesophases. Annu Rev Biophys. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. [DOI] [PubMed] [Google Scholar]
- 32.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 33.Riekel C, et al. Protein crystallography microdiffraction. Curr Opin Struct Biol. 2005 doi: 10.1016/j.sbi.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Fischetti RF, et al. Mini-beam collimator enables microcrystallography experiments on standard beamlines. J Synchrotron Radiat. 2009;16:217–225. doi: 10.1107/S0909049508040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaakola VP, et al. The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. 2008 doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu B, et al. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010 doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chien EY, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suryanarayana S, et al. Identification of intramolecular interactions in adrenergic receptors. J Biol Chem. 1992;267:21991–21994. [PubMed] [Google Scholar]
- 39.Leff P. The two-state model of receptor activation. Trends Pharmacol Sci. 1995;16:89–97. doi: 10.1016/s0165-6147(00)88989-0. [DOI] [PubMed] [Google Scholar]
- 40.Deupi X, Kobilka BK. Energy landscapes as a tool to integrate GPCR structure, dynamics, and function. Physiology (Bethesda) 2010;25:293–303. doi: 10.1152/physiol.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao XJ, et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci U S A. 2009;106:9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghanouni P, et al. Functionally different agonists induce distinct conformations in the G protein coupling domain of the beta 2 adrenergic receptor. J Biol Chem. 2001;276:24433–24436. doi: 10.1074/jbc.C100162200. [DOI] [PubMed] [Google Scholar]
- 43.Rosenbaum DM, et al. Structure and Function of an Irreversible Agonist-β2 Adrenoceptor complex. Nature. 2011 doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. 2011 doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheerer P, et al. Crystal structure of opsin in its G-proteininteracting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 46.Strader CD, et al. Identification of residues required for ligand binding to the β-adrenergic receptor. Proc Natl Acad Sci U S A. 1987;84:4384–4388. doi: 10.1073/pnas.84.13.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liapakis G, et al. The forgotten serine. A critical role for Ser2035.42 in ligans binding to and activation of the beta 2-adrenergic receptor. J Biol Chem. 2000;275:37779–37788. doi: 10.1074/jbc.M002092200. [DOI] [PubMed] [Google Scholar]
- 48.Sansuk K, et al. A structural insight into the reorientation of transmembrane domains 3 and 5 during family A GPCR activation. Mol Pharmacol. 2010 doi: 10.1124/mol.110.066068. [DOI] [PubMed] [Google Scholar]
- 49.Carvalho AL, et al. X-ray crystallography in drug discovery. Methods Mol Biol. 2009;572:31–56. doi: 10.1007/978-1-60761-244-5_3. [DOI] [PubMed] [Google Scholar]
- 50.Sabio M, et al. Use of the X-ray structure of the beta2-adrenergic receptor for drug discovery. Part 2: Identification of active compounds. Bioorg Med Chem Lett. 2008;18:5391–5395. doi: 10.1016/j.bmcl.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 51.Kolb P, et al. Structure-based discovery of beta2-adrenergic receptor ligands. Proc Natl Acad Sci U S A. 2009;106:6843–6848. doi: 10.1073/pnas.0812657106. [DOI] [PMC free article] [PubMed] [Google Scholar]