Abstract
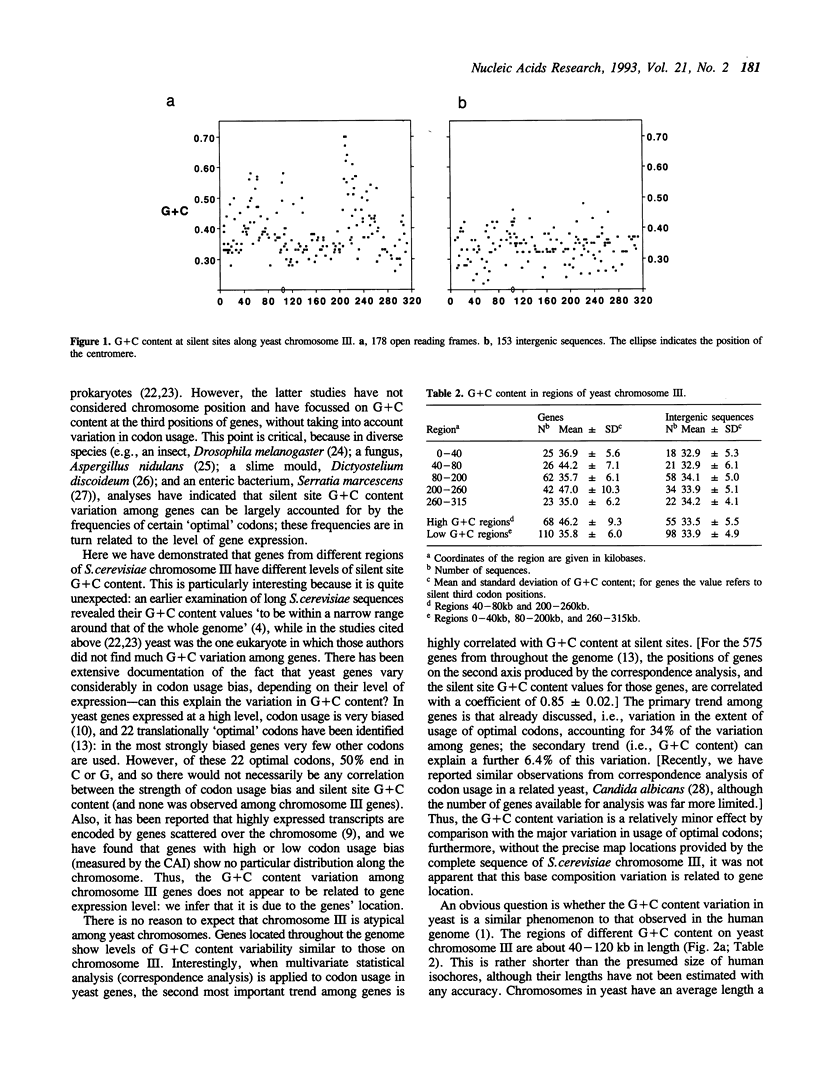
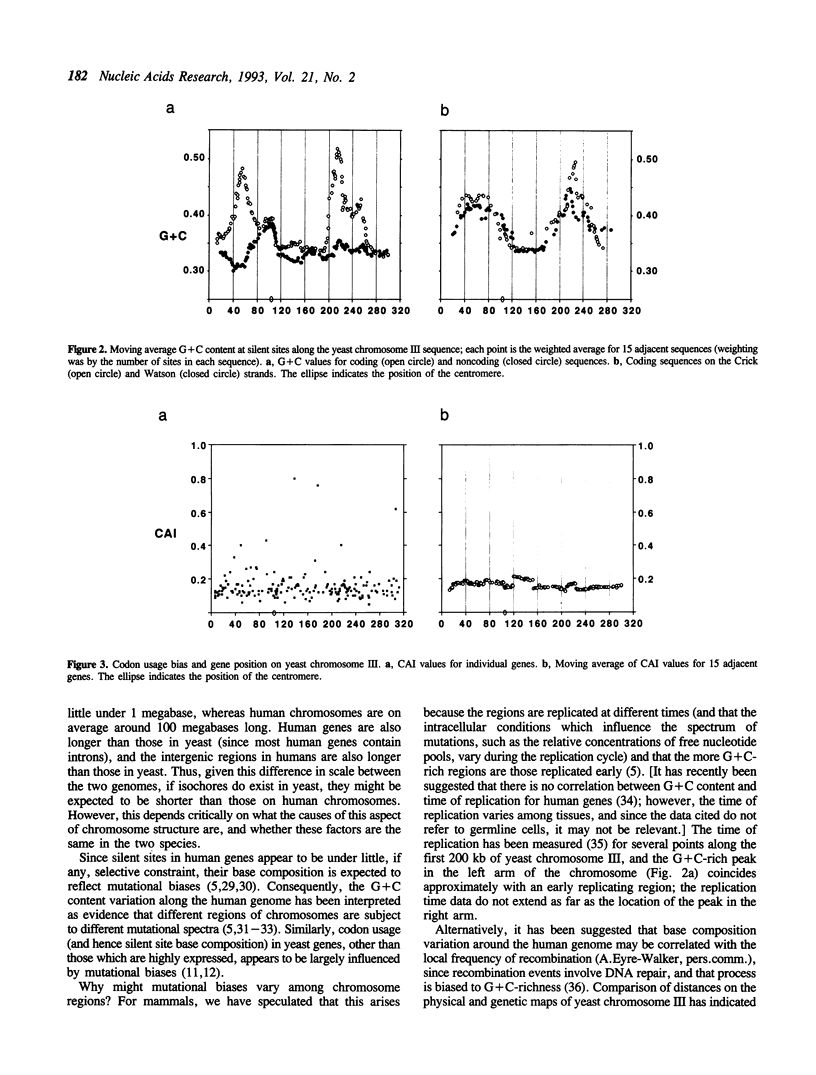
The recent determination of the complete sequence of chromosome III from the yeast Saccharomyces cerevisiae allows, for the first time, the investigation of the long range primary structure of a eukaryotic chromosome. We have found that, against a background G+C level of about 35%, there are two regions (one in each chromosome arm) in which G+C values rise to over 50%. This effect is seen in silent sites within genes, but not in noncoding intergenic sequences. The variation in G+C content is not related to differential selection of synonymous codons, and probably reflects mutational biases. That the intergenic regions do not exhibit the same phenomenon is particularly interesting, and suggests that they are under substantial constraint. The yeast chromosome may be a model of the structure of the human genome, since there is evidence that it is also a mosaic of long regions of different base compositions, reflected in wide variation of G+C content at silent sites among genes. Two possible causes of this regional effect, replication timing, and recombination frequency, are discussed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aota S., Ikemura T. Diversity in G + C content at the third position of codons in vertebrate genes and its cause. Nucleic Acids Res. 1986 Aug 26;14(16):6345–6355. doi: 10.1093/nar/14.16.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985 May 24;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Bernardi G. The isochore organization of the human genome. Annu Rev Genet. 1989;23:637–661. doi: 10.1146/annurev.ge.23.120189.003225. [DOI] [PubMed] [Google Scholar]
- Bork P., Ouzounis C., Sander C., Scharf M., Schneider R., Sonnhammer E. What's in a genome? Nature. 1992 Jul 23;358(6384):287–287. doi: 10.1038/358287a0. [DOI] [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988 Aug 26;54(5):705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- Bulmer M. The effect of context on synonymous codon usage in genes with low codon usage bias. Nucleic Acids Res. 1990 May 25;18(10):2869–2873. doi: 10.1093/nar/18.10.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Onofrio G., Bernardi G. A universal compositional correlation among codon positions. Gene. 1992 Jan 2;110(1):81–88. doi: 10.1016/0378-1119(92)90447-w. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. C. An analysis of codon usage in mammals: selection or mutation bias? J Mol Evol. 1991 Nov;33(5):442–449. doi: 10.1007/BF02103136. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. Evidence that both G + C rich and G + C poor isochores are replicated early and late in the cell cycle. Nucleic Acids Res. 1992 Apr 11;20(7):1497–1501. doi: 10.1093/nar/20.7.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipski J. Correlation between molecular clock ticking, codon usage fidelity of DNA repair, chromosome banding and chromatin compactness in germline cells. FEBS Lett. 1987 Jun 15;217(2):184–186. doi: 10.1016/0014-5793(87)80660-9. [DOI] [PubMed] [Google Scholar]
- Filipski J., Salinas J., Rodier F. Chromosome localization-dependent compositional bias of point mutations in Alu repetitive sequences. J Mol Biol. 1989 Apr 5;206(3):563–566. doi: 10.1016/0022-2836(89)90501-9. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C., Attimonelli M., Lanave C., di Paola G. ACNUC--a portable retrieval system for nucleic acid sequence databases: logical and physical designs and usage. Comput Appl Biosci. 1985 Sep;1(3):167–172. doi: 10.1093/bioinformatics/1.3.167. [DOI] [PubMed] [Google Scholar]
- Ikemura T., Aota S. Global variation in G+C content along vertebrate genome DNA. Possible correlation with chromosome band structures. J Mol Biol. 1988 Sep 5;203(1):1–13. doi: 10.1016/0022-2836(88)90086-1. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985 Jan;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Lloyd A. T., Sharp P. M. CODONS: a microcomputer program for codon usage analysis. J Hered. 1992 May-Jun;83(3):239–240. doi: 10.1093/oxfordjournals.jhered.a111205. [DOI] [PubMed] [Google Scholar]
- Lloyd A. T., Sharp P. M. Codon usage in Aspergillus nidulans. Mol Gen Genet. 1991 Nov;230(1-2):288–294. doi: 10.1007/BF00290679. [DOI] [PubMed] [Google Scholar]
- Lloyd A. T., Sharp P. M. Evolution of codon usage patterns: the extent and nature of divergence between Candida albicans and Saccharomyces cerevisiae. Nucleic Acids Res. 1992 Oct 25;20(20):5289–5295. doi: 10.1093/nar/20.20.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassi G., Montero L. M., Salinas J., Bernardi G. The isochore organization and the compositional distribution of homologous coding sequences in the nuclear genome of plants. Nucleic Acids Res. 1989 Jul 11;17(13):5273–5290. doi: 10.1093/nar/17.13.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Reynolds A. E., McCarroll R. M., Newlon C. S., Fangman W. L. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989 Oct;9(10):4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Cowe E. Synonymous codon usage in Saccharomyces cerevisiae. Yeast. 1991 Oct;7(7):657–678. doi: 10.1002/yea.320070702. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Devine K. M. Codon usage and gene expression level in Dictyostelium discoideum: highly expressed genes do 'prefer' optimal codons. Nucleic Acids Res. 1989 Jul 11;17(13):5029–5039. doi: 10.1093/nar/17.13.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M. Processes of genome evolution reflected by base frequency differences among Serratia marcescens genes. Mol Microbiol. 1990 Jan;4(1):119–122. doi: 10.1111/j.1365-2958.1990.tb02020.x. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields D. C., Sharp P. M. Evidence that mutation patterns vary among Drosophila transposable elements. J Mol Biol. 1989 Jun 20;207(4):843–846. doi: 10.1016/0022-2836(89)90252-0. [DOI] [PubMed] [Google Scholar]
- Shields D. C., Sharp P. M., Higgins D. G., Wright F. "Silent" sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol Biol Evol. 1988 Nov;5(6):704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. Directional mutation pressure, selective constraints, and genetic equilibria. J Mol Evol. 1992 Feb;34(2):95–114. doi: 10.1007/BF00182387. [DOI] [PubMed] [Google Scholar]
- Van Dyck E., Foury F., Stillman B., Brill S. J. A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J. 1992 Sep;11(9):3421–3430. doi: 10.1002/j.1460-2075.1992.tb05421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas D. F., Boeke J. D. Yeast retrotransposon revealed. Nature. 1992 Aug 27;358(6389):717–717. doi: 10.1038/358717a0. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Sharp P. M., Li W. H. Mutation rates differ among regions of the mammalian genome. Nature. 1989 Jan 19;337(6204):283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- Yoshikawa A., Isono K. Chromosome III of Saccharomyces cerevisiae: an ordered clone bank, a detailed restriction map and analysis of transcripts suggest the presence of 160 genes. Yeast. 1990 Sep-Oct;6(5):383–401. doi: 10.1002/yea.320060504. [DOI] [PubMed] [Google Scholar]
- Zenvirth D., Arbel T., Sherman A., Goldway M., Klein S., Simchen G. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J. 1992 Sep;11(9):3441–3447. doi: 10.1002/j.1460-2075.1992.tb05423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



