Abstract
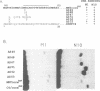
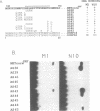
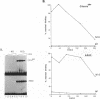
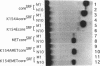
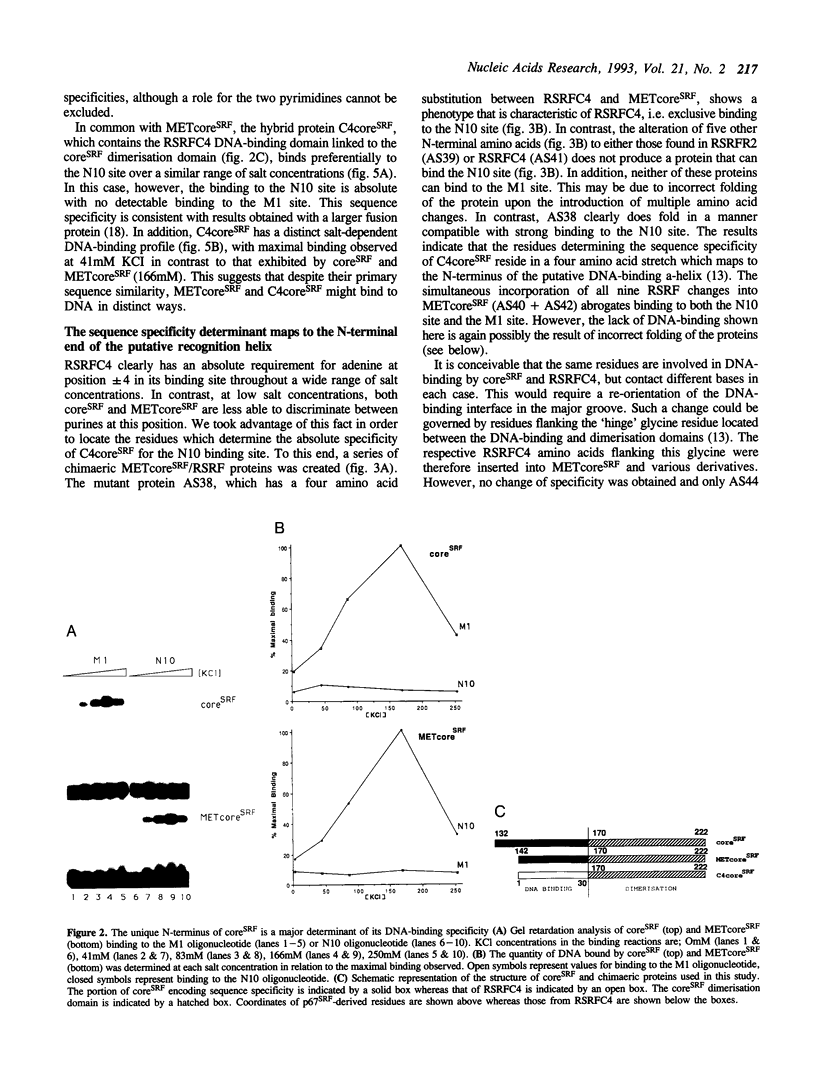
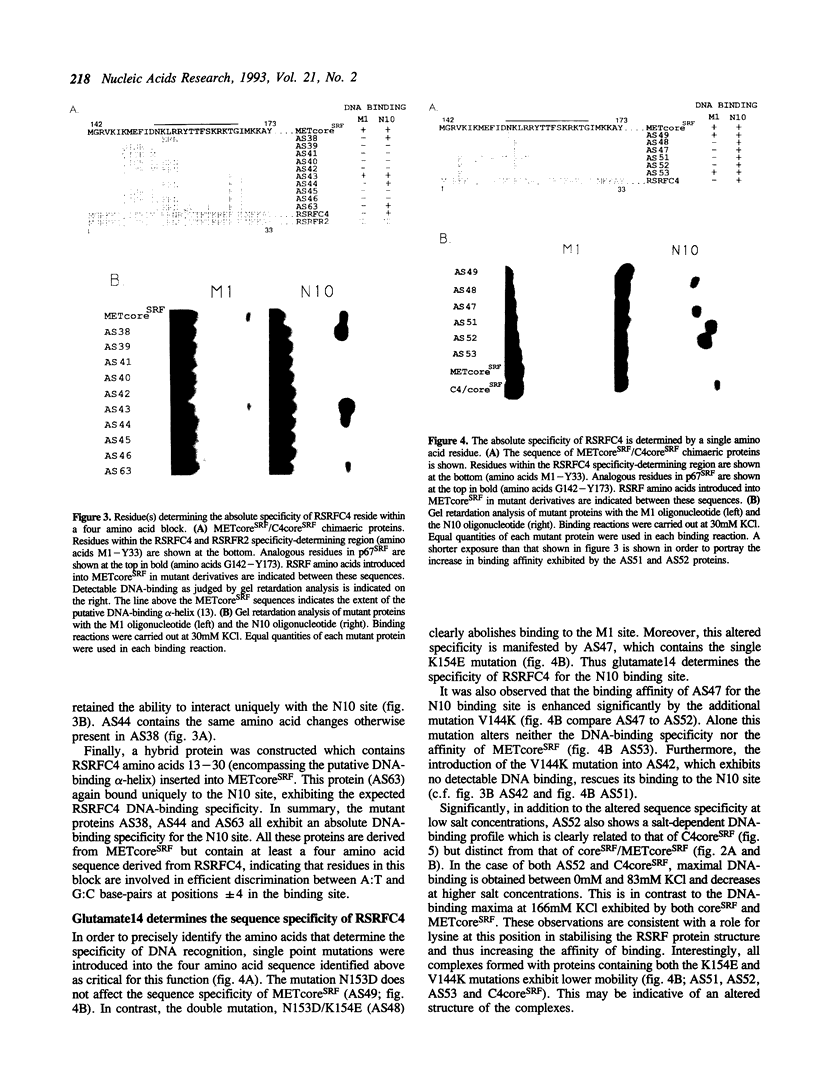
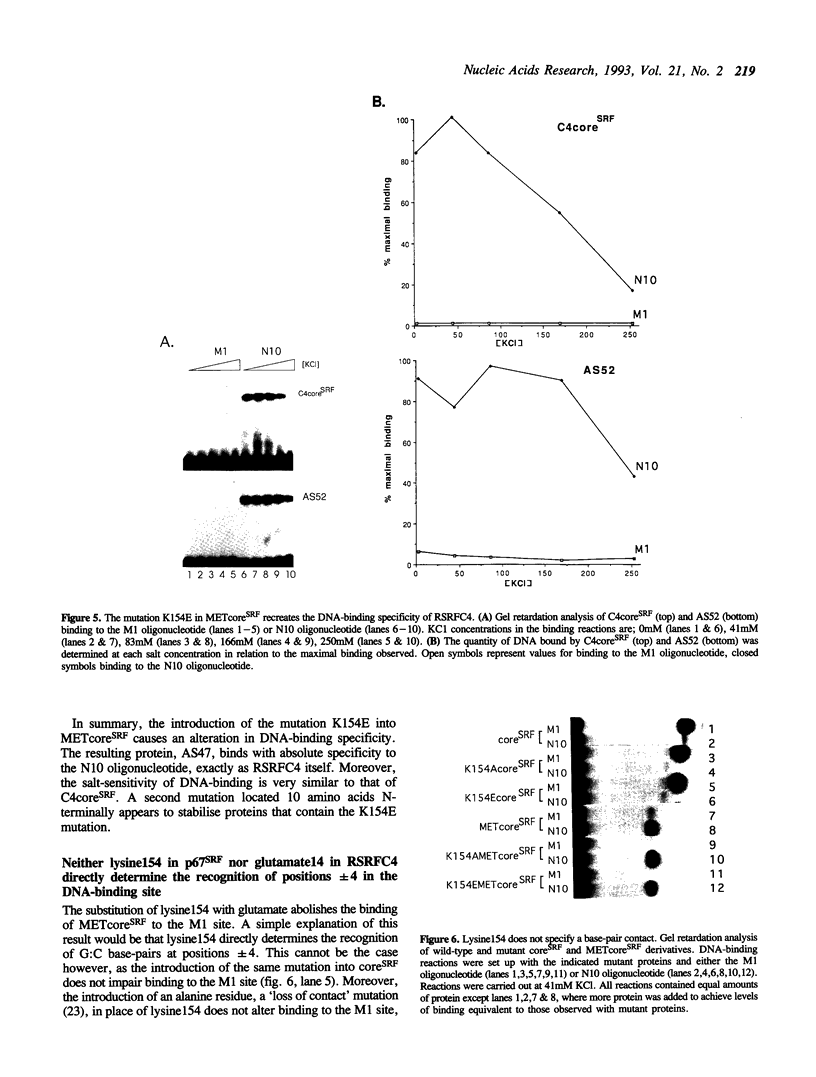
The human transcription factors p67SRF and RSRFC4 recognise similar but distinct binding sites which are found in the promoters of both muscle-specific and 'immediate early' genes. Both proteins share a common basic DNA-binding domain, which is defined by the MADS box homology region. The DNA-binding specificity of a truncated form of p67SRF (coreSRF) can be converted to that of RSRFC4. Removal of residues immediately N-terminal to the MADS box relaxes the specificity of coreSRF for its cognate sequence (CC(A/T)6GG) as it improves binding to the RSRFC4 site (CTA(A/T)4TAG). Moreover, the introduction of a single, additional mutation, K154E, into the N-terminal truncated derivative completes the change in specificity to the RSRFC4 binding site. It also influences the salt dependence of DNA binding and ternary complex formation with p62TCF. However, residues at this position do not appear to be involved in direct basepair recognition. These results indicate that although the DNA binding specificity of p67SRF can be converted to that of RSRFC4, the two proteins may bind DNA in different ways. Furthermore, they suggest that binding site specificity can be determined by an indirect mechanism involving residues which are not directly involved in base recognition.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ebright R. H. Identification of amino acid-base pair contacts by genetic methods. Methods Enzymol. 1991;208:620–640. doi: 10.1016/0076-6879(91)08032-d. [DOI] [PubMed] [Google Scholar]
- Gille H., Sharrocks A. D., Shaw P. E. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992 Jul 30;358(6385):414–417. doi: 10.1038/358414a0. [DOI] [PubMed] [Google Scholar]
- Hayes T. E., Sengupta P., Cochran B. H. The human c-fos serum response factor and the yeast factors GRM/PRTF have related DNA-binding specificities. Genes Dev. 1988 Dec;2(12B):1713–1722. doi: 10.1101/gad.2.12b.1713. [DOI] [PubMed] [Google Scholar]
- Hipskind R. A., Rao V. N., Mueller C. G., Reddy E. S., Nordheim A. Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature. 1991 Dec 19;354(6354):531–534. doi: 10.1038/354531a0. [DOI] [PubMed] [Google Scholar]
- König H., Ponta H., Rahmsdorf U., Büscher M., Schönthal A., Rahmsdorf H. J., Herrlich P. Autoregulation of fos: the dyad symmetry element as the major target of repression. EMBO J. 1989 Sep;8(9):2559–2566. doi: 10.1002/j.1460-2075.1989.tb08394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucibello F. C., Lowag C., Neuberg M., Müller R. trans-repression of the mouse c-fos promoter: a novel mechanism of Fos-mediated trans-regulation. Cell. 1989 Dec 22;59(6):999–1007. doi: 10.1016/0092-8674(89)90756-3. [DOI] [PubMed] [Google Scholar]
- Ma H., Yanofsky M. F., Meyerowitz E. M. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991 Mar;5(3):484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Mueller C. G., Nordheim A. A protein domain conserved between yeast MCM1 and human SRF directs ternary complex formation. EMBO J. 1991 Dec;10(13):4219–4229. doi: 10.1002/j.1460-2075.1991.tb05000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman C., Runswick M., Pollock R., Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988 Dec 23;55(6):989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- Ofir R., Dwarki V. J., Rashid D., Verma I. M. Phosphorylation of the C terminus of Fos protein is required for transcriptional transrepression of the c-fos promoter. Nature. 1990 Nov 1;348(6296):80–82. doi: 10.1038/348080a0. [DOI] [PubMed] [Google Scholar]
- Pollock R., Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990 Nov 11;18(21):6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R., Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991 Dec;5(12A):2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- Prywes R., Roeder R. G. Purification of the c-fos enhancer-binding protein. Mol Cell Biol. 1987 Oct;7(10):3482–3489. doi: 10.1128/mcb.7.10.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryseck R. P., Macdonald-Bravo H., Mattéi M. G., Ruppert S., Bravo R. Structure, mapping and expression of a growth factor inducible gene encoding a putative nuclear hormonal binding receptor. EMBO J. 1989 Nov;8(11):3327–3335. doi: 10.1002/j.1460-2075.1989.tb08494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P., Verma I. M. Modulation of c-fos gene transcription by negative and positive cellular factors. Nature. 1987 Apr 2;326(6112):507–510. doi: 10.1038/326507a0. [DOI] [PubMed] [Google Scholar]
- Schröter H., Mueller C. G., Meese K., Nordheim A. Synergism in ternary complex formation between the dimeric glycoprotein p67SRF, polypeptide p62TCF and the c-fos serum response element. EMBO J. 1990 Apr;9(4):1123–1130. doi: 10.1002/j.1460-2075.1990.tb08218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Huijser P., Nacken W., Saedler H., Sommer H. Genetic Control of Flower Development by Homeotic Genes in Antirrhinum majus. Science. 1990 Nov 16;250(4983):931–936. doi: 10.1126/science.250.4983.931. [DOI] [PubMed] [Google Scholar]
- Schönthal A., Büscher M., Angel P., Rahmsdorf H. J., Ponta H., Hattori K., Chiu R., Karin M., Herrlich P. The Fos and Jun/AP-1 proteins are involved in the downregulation of Fos transcription. Oncogene. 1989 May;4(5):629–636. [PubMed] [Google Scholar]
- Sharrocks A. D., Shaw P. E. Improved primer design for PCR-based, site-directed mutagenesis. Nucleic Acids Res. 1992 Mar 11;20(5):1147–1147. doi: 10.1093/nar/20.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P. E., Frasch S., Nordheim A. Repression of c-fos transcription is mediated through p67SRF bound to the SRE. EMBO J. 1989 Sep;8(9):2567–2574. doi: 10.1002/j.1460-2075.1989.tb08395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw P. E., Schröter H., Nordheim A. The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c-fos promoter. Cell. 1989 Feb 24;56(4):563–572. doi: 10.1016/0092-8674(89)90579-5. [DOI] [PubMed] [Google Scholar]
- Shaw P. E. Ternary complex formation over the c-fos serum response element: p62TCF exhibits dual component specificity with contacts to DNA and an extended structure in the DNA-binding domain of p67SRF. EMBO J. 1992 Aug;11(8):3011–3019. doi: 10.1002/j.1460-2075.1992.tb05371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. The SRE: a growth factor responsive transcriptional regulator. Semin Cancer Biol. 1990 Feb;1(1):47–58. [PubMed] [Google Scholar]
- Tuil D., Clergue N., Montarras D., Pinset C., Kahn A., Phan-Dinh-Tuy F. CC Ar GG boxes, cis-acting elements with a dual specificity. Muscle-specific transcriptional activation and serum responsiveness. J Mol Biol. 1990 Jun 20;213(4):677–686. doi: 10.1016/S0022-2836(05)80255-4. [DOI] [PubMed] [Google Scholar]
- Wilson T., Treisman R. Fos C-terminal mutations block down-regulation of c-fos transcription following serum stimulation. EMBO J. 1988 Dec 20;7(13):4193–4202. doi: 10.1002/j.1460-2075.1988.tb03316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]