Abstract
We employ single-molecule fluorescence resonance energy transfer (smFRET) to study structural dynamics over the first two elongation cycles of protein synthesis, using ribosomes containing either Cy3-labeled ribosomal protein L11 and A- or P-site Cy5-labeled tRNA or Cy3 and Cy5 labeled tRNAs. Pre-translocation (PRE) complexes demonstrate fluctuations between classical and hybrid forms, with concerted motions of tRNAs away from L11 and from each other when classical complex converts to hybrid complex. EF-G·GTP binding to both hybrid and classical PRE complexes halts these fluctuations prior to catalyzing translocation to form the post-translocation (POST) complex. EF-G dependent translocation from the classical PRE complex proceeds via transient formation of a short-lived hybrid intermediate. A-site binding of either EF-G to the PRE complex or of aminoacyl-tRNA·EF-Tu ternary complex to the POST complex markedly suppresses ribosome conformational lability.
INTRODUCTION
Continued dramatic progress in the elucidation of the structures of the bacterial ribosome and its functional complexes has led to proposals for the detailed mechanisms of ribosomecatalyzed protein synthesis (Agirrezabala and Frank, 2009; Schmeing and Ramakrishnan, 2009; Fischer et al., 2010; Ratje et al., 2010). Ensemble rapid reaction kinetics (Antoun et al., 2006; Daviter et al., 2006; Dorner et al., 2006; Grigoriadou et al., 2007; Hetrick et al., 2009; Pan et al., 2007, 2008; Pape et al., 1998; Phelps and Joseph, 2005; Savelsbergh et al., 2003; Walker et al., 2008; Wintermeyer et al., 2004; Zaher and Green, 2009; Zavialov and Ehrenberg, 2003) and single molecule (Aitken et al. 2010; Blanchard 2009; Frank and Gonzalez 2010; Uemura et al. 2010) studies of the translational machinery in the past several years have resulted in increased understanding of some aspects of ribosome dynamics during the initiation, elongation, and termination phases of protein synthesis, but many essential points remain to be elucidated. Here we present single molecule Fluorescence Resonance Energy Transfer (smFRET) experiments focused on elucidating the structural dynamics and translocation mechanism of the elongation process.
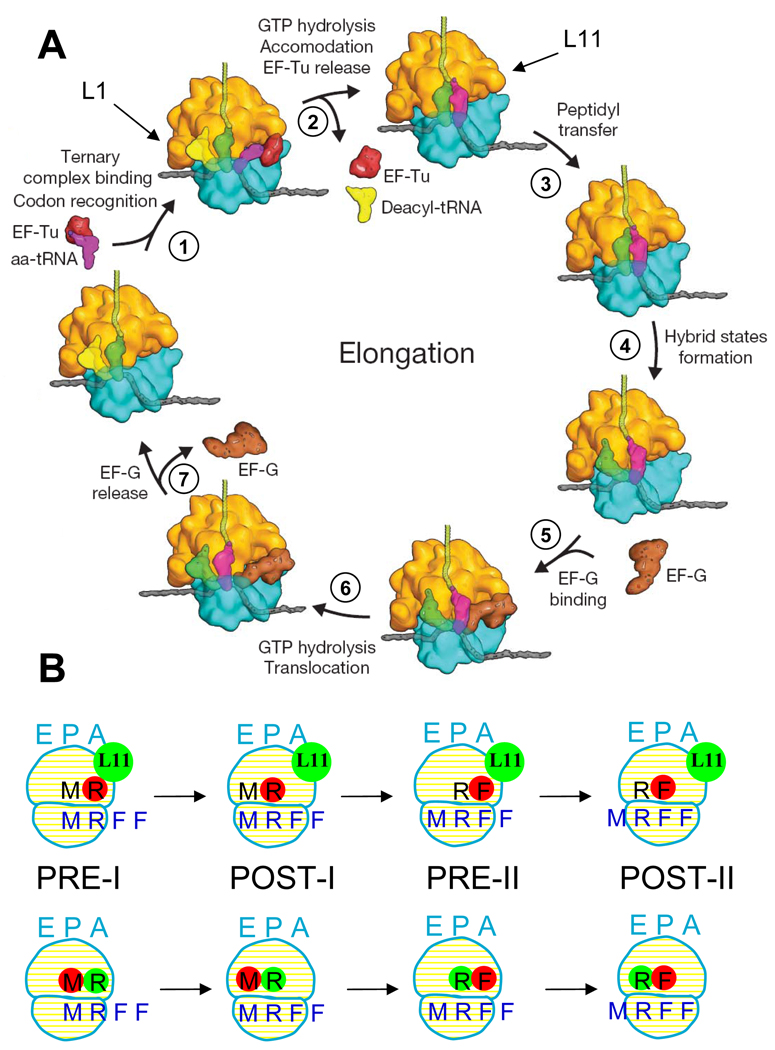
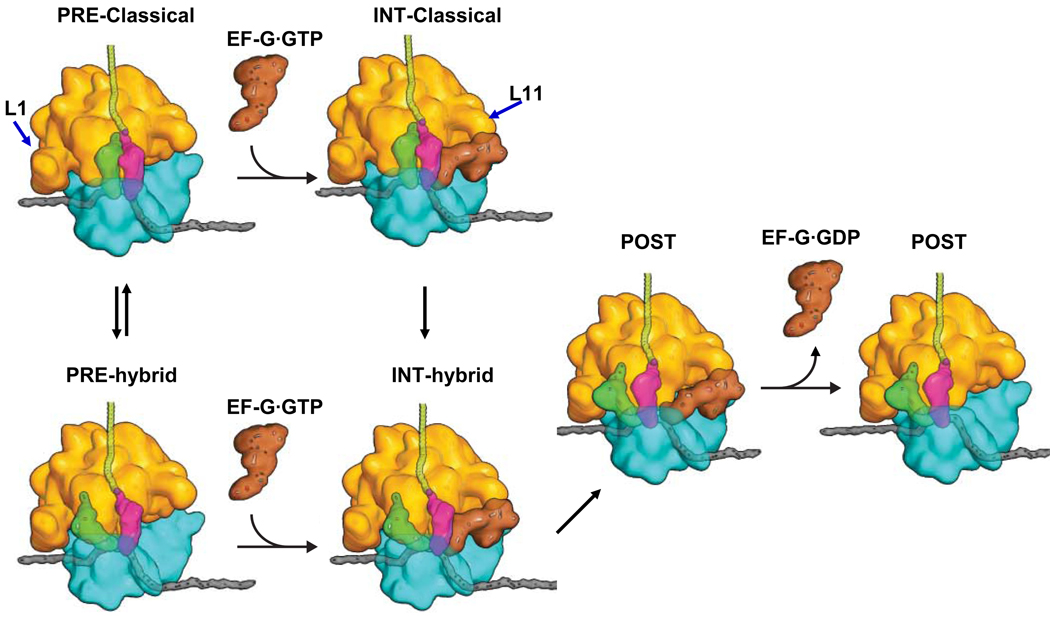
A proposed (Schmeing and Ramakrishnan, 2009) version of this cycle is shown in Figure 1A. In step 1, an incoming aminoacyl-tRNA (aa-tRNA), bound in a ternary complex (TC) with EF-Tu and GTP, binds to the A/T site of the ribosome. The aa-tRNA is next accommodated into the A-site (step 2), with concomitant GTP hydrolysis and dissociation of EF-Tu·GDP and Pi. This is followed rapidly by peptide bond formation (step 3), transferring the nascent peptide to the tRNA in the A-site, and resulting in formation of the pre-translocation (PRE) complex. In the PRE complex, tRNAs fluctuate between classical (A/A and P/P sites) and hybrid (A/P and P/E sites) states (step 4 and its reversal). EF-G·GTP binding to the spontaneously formed hybrid PRE complex (step 5) induces translocation of peptidyl-tRNA and deacylated tRNA into the P- and E-sites, respectively, with movement of the mRNA by one codon and with GTP hydrolysis (step 6). Dissociation of EF-G·GDP and Pi (step 7), leading to formation of the post-translocation (POST) complex, completes the cycle. In this proposed model, EF-G·GTP binds exclusively to the hybrid state of the PRE complex and not to the classical state, thereby imparting directionality to the translocation reaction (Fei et al., 2008), but the available evidence on this point, based mainly on prior smFRET studies, is not definitive (see Discussion).
Figure 1.
A: The elongation cycle ( modified from Schmeing and Ramakrishnan, 2009). The L11 and L1 regions are indicated by arrows. B: Schematic of the complexes studied. Top and bottom rows are for L11-tRNA (Lt) and tRNA-tRNA (tt) FRET complexes, respectively, for elongation cycles I and II. The  colored letters M, R and F indicate mRNA codons for fMet, Arg and Phe. The black letters M, R, and F refer to tRNAfMet, tRNAArg and tRNAPhe.
colored letters M, R and F indicate mRNA codons for fMet, Arg and Phe. The black letters M, R, and F refer to tRNAfMet, tRNAArg and tRNAPhe.  dots indicate Cy5 labeling of the tRNAs.
dots indicate Cy5 labeling of the tRNAs.  dots indicate Cy3 labeling of L11 or tRNA in Lt or tt complexes, respectively.
dots indicate Cy3 labeling of L11 or tRNA in Lt or tt complexes, respectively.
Two mobile domains of the large (50S) ribosomal subunit are strongly involved in the binding and movement of tRNA during the elongation cycle. The GTPase associated center (GAC) (Valle et al., 2003) which includes proteins L10, L11, L7 and L12 as well as several 23S rRNA helices, contributes part of the A/T and A-sites, whereas the L1 stalk interacts with tRNA in the P, E and P/E sites. Fluctuations in the structure of PRE complexes have been examined via smFRET monitoring of various intra-complex distances (reviewed in Aitken et al. 2010; Blanchard 2009; Frank & Gonzalez 2010). Independently, structural studies have demonstrated a rotation (termed ratcheting) of the 30S and 50S subunits relative to each other (Frank and Agrawal, 2000), as well as a swiveling of the head domain of the 30S subunit (Zhang et al., 2009). There is currently debate over the extent to which the localized movements of the L1 stalk, tRNAs, and other ribosomal proteins are coupled to each other and to the more general ratcheting and swiveling movements (Cornish et al., 2008, 2009; Fei et al., 2008, 2009; Marshall et al., 2008, 2009; Munro et al., 2010a, b).
In the present study we examine the first two elongation cycles of protein synthesis using ribosomes programmed with an mRNA coding for the initial tripeptide fMetArgPhe. In these experiments we use smFRET to monitor the distance between tRNA and protein L11 within the GAC (Lt complexes – Figure 1B, top row) and between adjacent tRNAs (tt complexes - Figure 1B, bottom row) both in PRE complexes and during the translocation step resulting in POST complex formation. These measurements allow us to compare fluctuations in PRE complexes monitored by L11-tRNA distance with those monitored by tRNA-tRNA distance, and to determine the effect of EF-G·GTP binding on these fluctuations and subsequent tRNA movements. Our results show that 1) EF-G·GTP binds directly to both classical and hybrid PRE complexes and promotes translocation directly from both PRE complexes with similar probabilities; 2) EF-G binding halts the L11-tRNA and tRNA-tRNA distance fluctuations between classical and hybrid PRE states prior to the translocation step; 3) EF-G dependent translocation from the classical PRE complex proceeds via transient formation of a short-lived hybrid intermediate; and 4) ribosome conformational lability is largely suppressed by binding of the next TC at the A-site. In addition, the generally similar rates of exchange between classical and hybrid PRE states that we measure by tRNA-tRNA FRET and tRNA-L11 FRET suggest that PRE state fluctuations of A-site and P-site tRNAs and L11 are coupled.
RESULTS
Pretranslocation complexes
Four PRE complexes were examined (Figure 1B): PRE-I(Lt) contains L11 labeled with Cy3 at position 87 (L11Cy3) and fMetArg-tRNAArg(Cy5); PRE-I(tt) contains tRNAfMet(Cy5) and fMetArg-tRNAArg(Cy3); PRE-II(Lt) contains L11Cy3 and fMetArgPhe-tRNAPhe(Cy5); PRE-II(tt) contains tRNAArg(Cy3) and fMetArgPhe-tRNAPhe(Cy5). Some percentage of each of these PRE complexes, 23 – 49%, rapidly fluctuates between two states with differing FRET efficiencies, while the remainder is found in two fixed, non-fluctuating states. Distributions between fluctuating and non-fluctuating states have been found in other smFRET studies of ribosomes (Cornish et al., 2009; Fei et al., 2009).
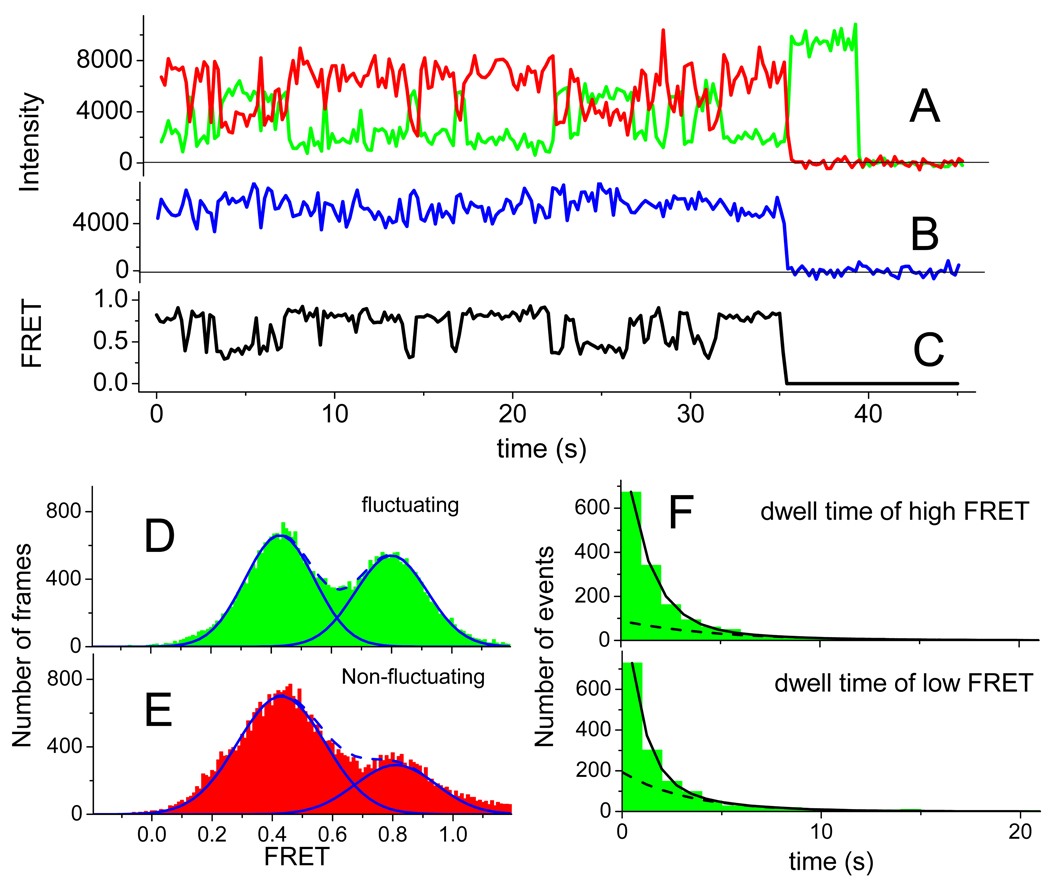
Fluctuating PRE-I(Lt) complexes alternate between two FRET states, present in approximately equal amounts (Figure 2A–C, Table S1), having FRET efficiency values of 0.43 and 0.80 (Figure 2D). Dwell times average ~2 s in both states (Table S1). Other individual complexes are stable in either the high or low FRET state, with the low FRET state predominating (Figure 2E). The zero time points for the fluctuating traces shown in Figures 2, 3, S1a–c, and S2a,b correspond to the moment when fluorescence monitoring of that particular complex began. Since the PRE complexes were assembled and bound to the surface 10 – 15 min beforehand, fluctuations between the two FRET states must continue for at least 10 min. Both the transitions from low FRET to high FRET (low state dwell time, L/H transitions) and from high FRET to low FRET (high FRET dwell time, H/L transitions) are described by at least two exponential rates (Figure 2F).
Figure 2. Time courses of fluorescence intensity and FRET for the PRE-I(Lt) complex.
Time zero designates when the recording was started. A: Cy3 (green) and Cy5 (red) fluorescence intensity traces under excitation of the Cy3 with 532 nm TIRF illumination at 100 ms integration time per frame. Cy5 fluorescence under this condition is sensitized emission due to FRET. Cy5 and Cy3 bleach at 35 s and 40 s, respectively. B: Fluorescence (blue trace) from Cy5 under direct excitation at 640 nm. Alternating laser excitation (ALEX) every other frame was accomplished by synchronizing the AOTF shown in Figure S5 with the camera. C: FRET ratio. D and E: Temporal histograms of FRET values of fluctuating (green) and non-fluctuating (red) complexes. The solid fitted lines are Gaussian distributions and the dashed lines are the sums of two Gaussian distributions. In part D., the distributions are fitted to the intervals of high FRET and low FRET output obtained from hidden Markov (HaMMy) analysis. F: Event histograms of high FRET (upper) and low FRET (lower) and the sum of two exponential decay components fitted to the data. The dashed line within each time course is the slower exponential component. See also Figures S1a–c and Tables S1 and S2.
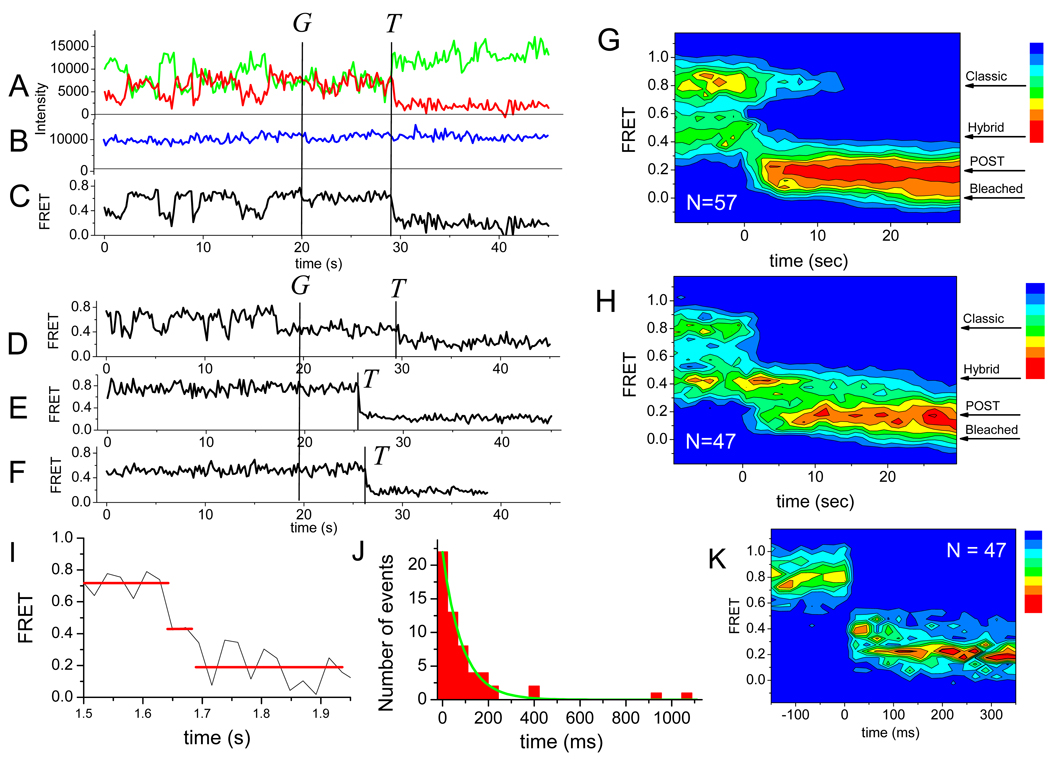
Figure 3. Fluorescence and FRET traces for EF-G·GTP translocation of PRE-I(Lt).
Times of EF-G·GTP injection (G) and of translocation (T) are marked. A: – C: ALEX presentation of data from a fluctuating ribosome that translocated from the high FRET state. Color coding as in Figure 2. D: – F: Sample FRET efficiency traces of translocation from D: a fluctuating ribosome translocating from the low FRET state; E: a stable high FRET state; and F: a stable low FRET state. G: and H: FRET probability density plots for translocation of fluctuating PRE complexes from high and low FRET states, respectively. All traces are aligned to EF-G·GTP addition as t = 0. Noteworthy are the halts in fluctuations that follow EF-G·GTP addition. I: FRET efficiency traces of translocation from a high FRET PRE state through a low FRET intermediate state to a POST state recorded at a higher time resolution (11 ms integration time per frame with ALEX (alternating-laser excitation) between 532 nm and 640 nm lasers.) than other traces shown in this paper – see Experimental J: Dwell time distribution of the low FRET translocation intermediate lifetime. The curve is a single exponential fit to the data. K: FRET probability density plot for translocation from the high FRET state at the higher time resolution shown in part I. All traces are aligned to the translocation event as t = 0.
Quite similar results were also found for the PRE-I(tt) (Figure S1a), PRE-II(Lt) (Figure S1b), and PRE-II(tt) (Figure S1c) complexes. For each, the transition rates between the fluctuating FRET states are biphasic (although the proportions of fast and slow rate processes vary), the rate constants for both phases are similar in magnitude to what is measured for PRE-I(Lt) and the distributions between fluctuating high and low FRET states and mean dwell times in the two states are roughly comparable (Table S1). These similarities suggest that the two FRET states represent two distinct physical states of the PRE(I) and PRE(II) complexes, as monitored either by L11:peptidyl-tRNA(A-site) FRET (the Lt complexes) or peptidyl-tRNA(A-site):deacylated-tRNA(P-site) FRET (the tt complexes). Two fluctuating tRNA-tRNA FRET states were reported in an earlier single molecule FRET study of a PRE-I complex (Blanchard et al., 2004a,b), which only differs from the present PRE-I(tt) complex in that it contained fMetPhe-tRNAPhe rather than fMetArg-tRNAArg in the A site and the positions of fluorophore labeling were nt 47 in tRNAPhe and nt 8 in tRNAfMet rather than in the D-loops of both tRNAs as here. Those workers identified the state having higher tRNA-tRNA FRET efficiency with the classical orientation of the tRNAs in the A/A and P/P positions, and the lower FRET state with tRNAs in the hybrid A/P and P/E positions. In these hybrid positions, structural studies (Agirrezabala et al., 2008) show that the positions of the tRNAs relative to the 30S subunit are similar to their classical positions, whereas their locations in the 50S subunit are displaced in the direction of translocation. These structural results support the assignment of the higher and lower FRET states to the classical and hybrid states, respectively, in all four PRE complexes (Table S2).
Translocation
Translocation of the PRE-I-(Lt) complex
Upon addition of EF-G·GTP to the PRE-I(Lt), transitions to a very low FRET state (0.20) are observed from either the classical, high (0.80) or hybrid, low (0.43) FRET state (Figure 3). These changes are consistent with an increase in distance between L11 and peptidyl-tRNA in either the A/A or A/P positions to the P/P position on translocation, as expected (Table S2). Fluorescence observed on direct excitation of the Cy5 fluorophore in alternating camera frames (Figure 3B) shows that the large decrease of FRET following EF-G injection cannot be ascribed to Cy5 dissociation, photobleaching, or blinking. EF-G·GTP-induced translocation occurs from PRE-I(Lt) complexes that fluctuate between both FRET states (Figure 3C, D) and from complexes that are fixed in either state (Figure 3E, F). The percentages of fluctuating complexes that translocate from the high FRET state (55%) or low FRET state (49%) are similar and are considerably higher than the percentages of complexes that translocate from either the fixed high FRET (35%) or fixed low FRET (21%) state (Table S3). Interestingly, EF-G·GTP addition results in a rapid halt in fluctuations (Figure 3G, H) an indication that EF-G binding locks L11 and peptidyl-tRNA in a fixed conformation, corresponding to either the classical or hybrid state, prior to translocation. Fluctuations are not seen in the POST complexes formed from either fluctuating or fixed PRE complexes, indicating that POST complexes achieve a single steady FRET state within 5 s following translocation.
Measurements of translocation from the classical state at higher time resolution show that it proceeds via formation of a transient intermediate, having a FRET efficiency of 0.42 ± 0.03, indistinguishable from that of the hybrid state of the PRE complex (Figures 3I, K; S2c), and a lifetime of 86 ± 4 ms (Figure 3J). This supports our earlier ensemble stopped-flow kinetic studies of translocation that also indicated formation of a short-lived transient intermediate (Pan et al., 2007), with an apparent lifetime of 180 ± 20 ms. The relatively minor 2-fold difference from the current value can plausibly be ascribed to several differences in the protocols of the two experiments, including buffer composition, the tRNA transferring the second amino acid (tRNAArg vs. tRNAPhe), the specific fluorophores in the labeled tRNAs (Cy5 vs. proflavin), and the ribosome preparation (Cy3-labeled AM77 ribosomes vs. unlabeled MRE600 ribosomes).
Apparent overall rate constants for the translocation process that is initiated by EF-G injection (k´t) were determined by single exponential fitting of histograms of the waiting times between injection of the EF-G and translocation for both fluctuating and fixed PRE-I(Lt) complexes. Translocation generally proceeds more rapidly for non-fluctuating complexes than for fluctuating complexes and more rapidly from high FRET than from low FRET complexes (Table 1), although in each case the difference is modest (~ 2-fold).
Table 1.
Translocation rate constants (s−1)a
| Fluctuating Complexes |
Non-fluctuating complexes |
|||||||
|---|---|---|---|---|---|---|---|---|
| High FRET | Low FRET | High FRET |
Low FRET |
|||||
| Complex | k't | k'b | k't | k'b | kH/Lb | kL/Hb | k't | k't |
| PRE-I- Lt |
0.31±0.06 | 0.37±0.07 | 0.13±0.04 | 0.14±0.02 | 0.59±0.06 | 0.77±0.05 | 0.50±0.10 | 0.28±0.05 |
| PRE-II- Lt |
0.30±0.03 | 0.48±0.11 | 0.36±0.05 | 0.32±0.04 | 0.56±0.05 | 0.73±0.10 | 0.77±0.06 | 0.36±0.06 |
| PRE-II- tt |
0.25±0.06 | 0.31±0.05 | 0.68±0.03 | 0.83±0.13 | 0.59±0.04 | 0.67±0.06 | n. d. | n. d. |
k't, apparent overall translocation constant following EF-G injection; k'b, measured apparent rate constant for approach to the final FRET efficiency before translocation; ± indicates s.d.
kH/L and kL/H are equal to the reciprocals of the average dwell times of the high and low FRET states , respectively, of those fluctuating PRE complexes that translocate.
Current models of the EF-G-dependent translocation process (Pan et al., 2007; Savelsbergh et al., 2003; Walker et al., 2008) suggest that it includes a number of discrete steps that culminate in tRNA and mRNA movement. To determine the effect of EF-G binding on PRE complex fluctuations prior to the major translocation event, kinetic FRET traces from individual fluctuating PRE complexes (Figures 3C, D; S2a – A, B; S2b – A, B) were temporally aligned at the time of the sudden decrease or increase of FRET efficiency that signals POST complex formation and averaged. This was done separately for translocation from the high and low FRET states. Such post-synchronized traces show positive or negative curvatures (Figure 4) for fluctuating complexes that translocated from the high or low FRET state, respectively. These curvatures reflect the increase or decrease in average FRET value as fluctuating complexes become fixed at high or low FRET efficiency subsequent to EF-G binding. Single exponential fitting of the curves from the time of translocation (t = 0), and extending backwards in time to their asymptotes (Figure 4A), following a known procedure (Veigel et al., 2002) produced a rate constant k´b. Determination of the value of k´b is thus experimentally distinct from determination of the value of k´t, the overall translocation rate constant, as described above.
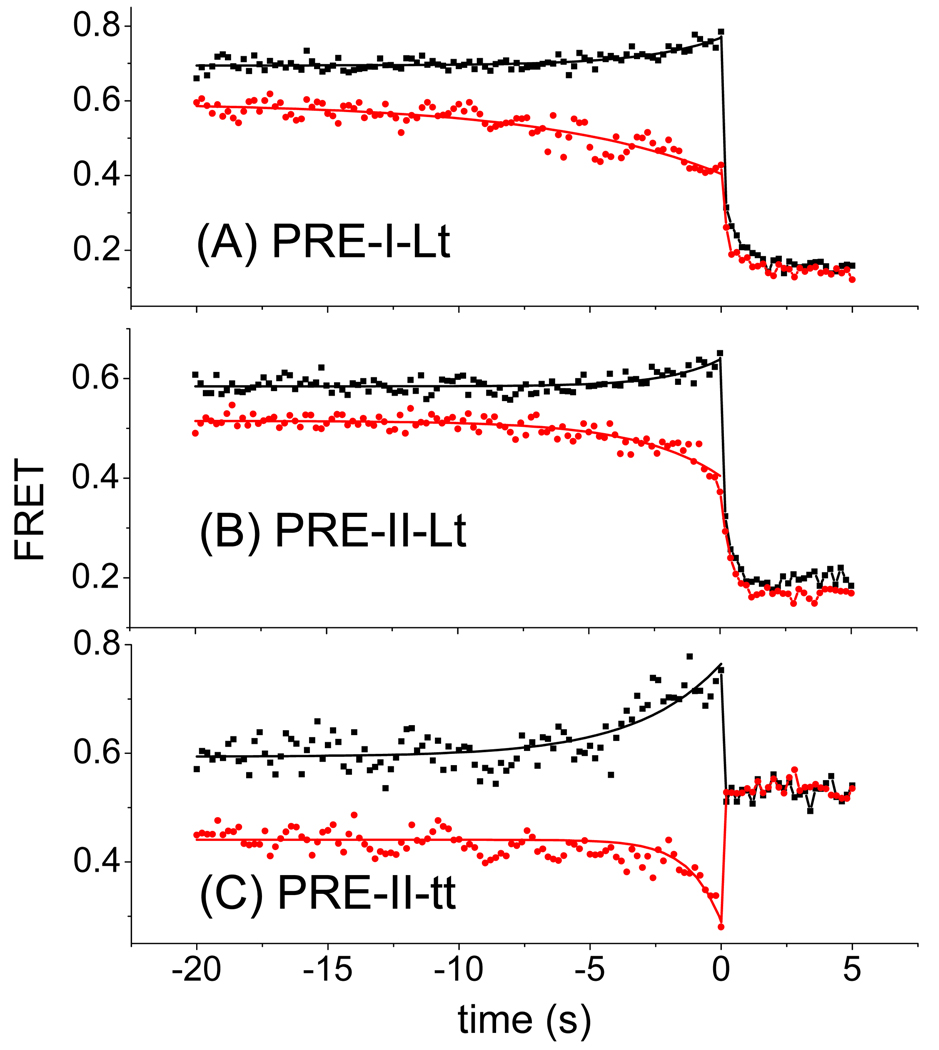
Figure 4. Post-synchronized, averaged translocation traces of fluctuating PRE complexes translocated from high FRET (black squares) and low FRET (red circles) PRE states.
A: PRE-I(Lt) B: PRE-II(Lt) C: PRE(II-tt). Average FRET values as a function of time were calculated by post-synchronizing all translocation events to the time (t = 0) of maximum FRET change. The solid curves are fits of the equation E = Eo + ΔE (1 − ekt) to the results from t = 0 s to t = −20 s, where E is the average FRET value as a function of time, Eo is the FRET value at t = 0, Eo + ΔE is the asymptotic FRET value at large negative time, and k is the apparent rate constant k’b.
If the binding of EF-G to PRE complex had no effect on the transition rates between classical and hybrid PRE states, k´b would be equal to the sum of the transition rate constants between these states of fluctuating PRE complexes that translocate (kH/L + kL/H, 1.4 s−1, Table 1) and would be the same for ribosomes that translocate from the high or low FRET states. In contrast, k´b values are not only differ significantly from each other [0.37 s−1 and 0.14 s−1 for ribosomes that translocate from the high and low FRET states, respectively (Table 1)], but also are considerably lower than 1.4 s−1. These differences between measured and ‘expected’ values may be attributed to the marked EF-G suppression of fluctuations between FRET states, resulting in pausing of the ribosome in either the high or low (classical or hybrid) state of the PRE-I(Lt) complex, as shown in Figures 3G and 3H. Furthermore, the similarities in the determined values of k´t and k´b, and the fact that k´t is less than (kH/L + kL/H) (Table 1) suggest that the transition to POST complex from the EF-G bound classical or hybrid PRE complex, either not fluctuating or fluctuating slowly, largely determines the rate of the overall process induced by addition of EF-G (see Supplemental – “Kinetic Modeling of Translocation”).
Interestingly, the sum of the transition rate constants between these states of fluctuating PRE complexes that translocate (1.4 ± 0.1 s−1) is significantly larger than the corresponding sum for all fluctuating PRE complexes (1.00 ± 0.08 s−1, Table S1), suggesting that translocation proceeds with a higher probability for fast-fluctuating vs. slow-fluctuating complexes, extending our observation that fluctuating complexes translocate with higher probability than non-fluctuating complexes (Table S3). Taken together, these differences lead us to speculate that PRE complexes are bound to the surface in a range of conformational energy wells, due, at least in part, to heterogeneity in ribosome preparation and/or ribosome:surface interaction, with those bound in the deepest wells being most resistant to conformational changes that accompany classical to hybrid state interconversion and translocation.
Translocation of the PRE-II complexes
Real-time monitoring of translocation of the PRE-II(Lt) and PRE-II(tt) complexes gives results largely paralleling those found for the PRE-I(Lt) complex. For both PRE-II complexes, translocations are seen from the high FRET (0.62, PRE-II(Lt); 0.69, PRE-II(tt)) and low FRET (0.34, PRE-II(Lt); 0.38, PRE-II(tt)) states, both fluctuating and non-fluctuating, to a POST state characterized by a single FRET efficiency (0.15, PRE-II(Lt), Figures 4B, S2a; 0.55, PRE-II(tt), Figures 4C, S2b). The POST state FRET efficiency values (0.15, 0.55) are consistent with reported distances between the D-loop of P-site bound tRNA and position L87 of L11, and between D-loops of E-site and P-site bound tRNAs, respectively (Table S2). As is the case with the PRE-I(Lt) complex, the fractions of fluctuating complexes that translocate from the high or low FRET state are similar. Translocation from fixed high and low FRET PRE-II complexes proceed with considerably lower frequency than for the corresponding fluctuating complexes (Table S3).
Values of k´t, k´b, kH/L and kL/H for the PRE-II(Lt) and PRE-II(tt) complexes were determined as described above for the PRE-I(Lt) complex (Table 1). For each of the complexes, k´t is similar in value to k´b and less than (kH/L + kL/H), leading to conclusions paralleling those described above for translocation of the PRE-I(Lt) complex. EF-G·GTP directly binds to both classical and hybrid PRE complexes, leading to the suppression of fluctuations between L11 and tRNA and between tRNAs. The transition from EF-G bound PRE complex, either not fluctuating or fluctuating slowly, to POST complex largely determines the rate of the overall translocation process. However, there is an as yet unexplained difference between the small variation in the k´t values determined for translocation of the three fluctuating PRE complexes from the high FRET state (0.25 – 0.31 s−1) and the considerably larger differences observed for the k´t values of the fluctuating PRE complexes from the low FRET state (0.1 – 0.7 s−1).
The k´t values collected in Table 1 are somewhat lower than the value of ~2 – 3 s−1 which can be calculated from prior ensemble measurements (Pan, 2008; Pan et al., 2007) of translocation of a PRE(I) complex containing tRNAfMet and fMetPhe-tRNAPhe at 2 µM EF-G and at ~21 – 23 °C, as in the present experiments. The lower rate constants may be attributable to some combination of the following differences: higher Mg2+ concentration (15 mM here vs. 7 mM in the ensemble experiment); a different tRNA (tRNAArg, Figure S3); bulkier fluorescent labels; and the possible slower binding of EF-G·GTP to immobilized PRE complexes than to a PRE complex in solution.
From the curves presented in Figure 4 it is clear that average FRET values approach asymptotic values 15 – 20 s before translocation. If the PRE complexes were kinetically homogenous, the asymptotes from post-synchronized traces translocated from high and low states would be expected to converge and be equal to the weighted mean of the high and low FRET efficiencies, based on the populations given in Table S1. However, this expectation is not met. Rather, the traces for each of the three PRE complexes examined indicate that PRE complexes that eventually translocate from the low or high FRET states show a preference for the low or high FRET states, respectively, even prior to EF-G addition, with the preference being more marked for PRE complexes translocating from the high FRET state. Such preferences provide additional evidence that PRE complexes are kinetically heterogeneous (see above), providing a plausible rationale for the biphasic behavior observed for the dwell times of PRE states (Figures 2F, S1a–F, S1b–F, S1c–F).
The effect of A-site occupancy on ribosome conformational lability during translocation
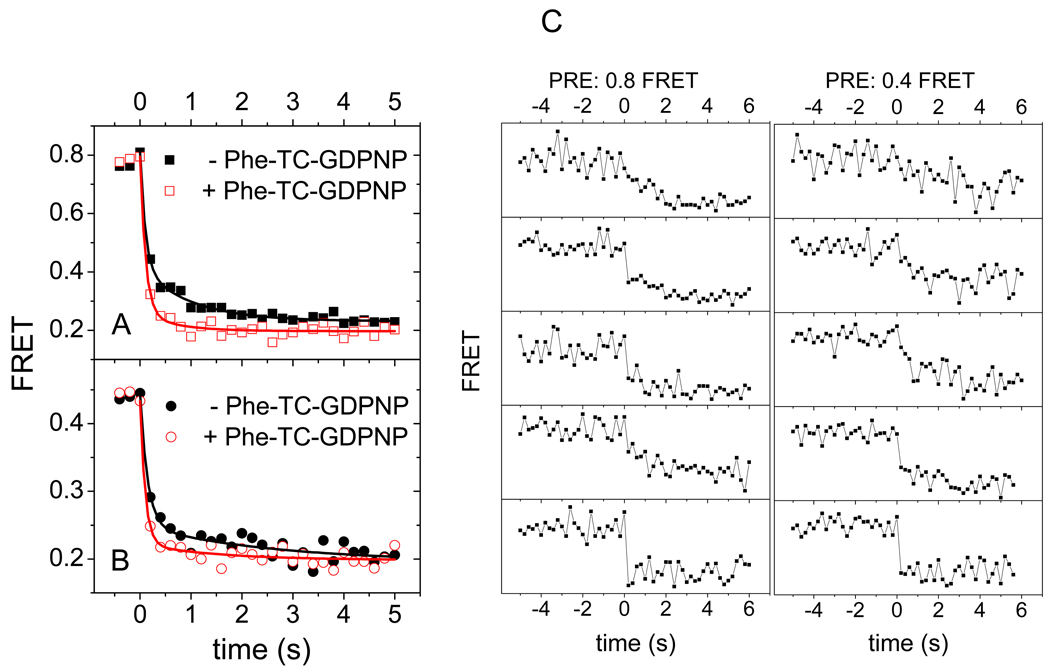
Averaged postsynchronized traces for the PRE-I(Lt) complex are biphasic after translocation begins from both the high and low FRET states (Figure 5A,B). The initial phase, accounting for ~ 70% of the decrease, occurs in a single frame (of duration 200 ms). The second decay phase occurs over several subsequent frames, and is evidenced by the FRET components seen within 0.5 s following the initial phase that have efficiencies intermediate between those of the POST complex (0.2) and either the classical PRE complex (0.8, Figure S4A) or the hybrid PRE complex (0.43, Figure S4B). This ‘tail’ on the FRET efficiency after translocation (Figure 5 A, B) is a property of the individual traces, not just the average (Figure 5C). Such gradual transitions are unusual in single molecule studies, because the elementary motions of macromolecules are generally fast (sub-microsecond), discrete, and rate-limited by briefly populated transient intermediates, but may be interpreted as signaling a sequence of small conformational changes (Yao et al., 2003) as the ribosome completes the translocation process (Munro et al., 2009). Similar biphasic transitions are seen for the PRE-II(Lt) (Figure 4B), but translocation apparently proceeds in a single rapid phase for PRE-II(tt) (Figure 4C). When the translocation experiment is repeated in the presence of the GDPNP form of the Phe-tRNAPhe TC, the next cognate tRNA, the late phase of the averaged PRE-I(Lt) FRET trace is very much decreased, contributing only ~10% of the total FRET decrease on translocation from either the high or low FRET PRE state (Figure 5A, B; Figure S4C, D).
Figure 5. Effect of added Phe-tRNAPhe·EF-Tu·GDPNP (F-TC) on Pre-I(Lt) translocation.
Post-synchronized, averaged translocation traces from A: high FRET state and B: low FRET state of PRE-I(Lt) complexes in the absence (solid dots) or presence (hollow dots) of the next cognate TC, added as Phe-tRNAPhe·EF-Tu·GDPNP. Average FRET values as a function of time were calculated by post-synchronizing all translocation events to the time (t = 0) of maximum FRET change. The solid curves are fits of the equation E = Eo + ΔEf exp(−kft) + ΔEs exp(−kst) to the results from t = 0 s to t = 5 s, where E is the average FRET value as a function of time, E∞ is the FRET value at t = ∞, kf > ks, ΔEf is the amplitude of fast phase, and ΔEs is the amplitude of slow phase. ΔE values are: A: (−TC) ΔEf 0.39 ± 0.04; ΔEs 0.19 ± 0.03: (+TC) ΔEf 0.53 ± 0.04; ΔEs 0.06 ± 0.02. B: (−TC) ΔEf 0.20 ± 0.02; ΔEs 0.06 ± 0.02: (+TC) ΔEf 0.24 ± 0.02; ΔEs 0.022 ± 0.015. FRET probability density plots corresponding to parts A and B are presented in Figure S4. C: Selected FRET trajectories of EF-G catalyzed translocation of PRE-I(Lt) in the absence of the next cognate TC from classical (FRET ~0.8) (left) or hybrid (FRET ~0.4) (right) pretranslocation states.
A plausible interpretation of these results is that the slower phase, observed for the PRE-I(Lt) and PRE-II(Lt) complexes, but not for the PRE-II(tt) complex, reflect conformational changes occurring within the highly mobile L11 region of the ribosome (Schuwirth et al., 2005) that is fluorescently-labeled in both PRE-I(Lt) and PRE-II(Lt) but not in PRE-II(tt). According to this interpretation, immediate occupancy of the A-site following translocation and EF-G·GDP dissociation, mimicking what occurs during protein synthesis in vivo, reduces the conformational lability of the ribosome, as would be expected from the known interactions between the TC and the L11 region (Schmeing et al., 2009). The apparent rate constant for the slow phase of translocation from the high FRET state (Figure 5A) increases from 1.0 ± 0.2 s−1 to 2.3 ± 0.9 s−1 in the presence of TC (Table S4), indicating that this phase is due to residual conformational lability of the L11 region even when TC is bound, rather than to translocated complexes which fail to bind TC. By analogy, the same interpretation is likely to apply to the slow phase of translocation from the low FRET state (Figure 5B), although here the poor signal-to-noise ratio resulting from the low amplitude of change in the presence of TC does not permit reliable estimation of the apparent rate constant.
DISCUSSION
Previously we showed that ribosomes containing L11 fluorescently-labeled at position 38 could be used in both ensemble FRET and smFRET experiments designed to monitor EF-G (Seo et al., 2006; Wang et al., 2007) and IF2 (Qin et al., 2009) interactions with the ribosome. Here we extend this approach to monitoring tRNA–ribosome interactions. Position 87 in L11, which was chosen for Cy3 labeling, falls within the C-terminal domain of L11 that interacts strongly with 23S rRNA (Conn et al., 1999; Wimberly et al., 1999), and is well positioned to provide a sensitive monitor of the movement of the Cy5-labeled peptidyl-tRNA D-loop both within the PRE complex and during translocation (Table S2). In this work we complement smFRET measurements of L11-tRNA interaction with parallel smFRET measurements of tRNA-tRNA interaction.
PRE complexes
Each of the four PRE complexes (Lt-I, Lt-II, tt-I, tt-II) fluctuates between two principal FRET states. As described above, for each PRE complex it is reasonable to assign the higher and lower FRET states to the classical (A/A, P/P) and hybrid (A/P, P/E) states of tRNA binding, respectively. A similar assignment was made in an earlier smFRET study of a fluctuating PRE(I)-tt complex (Blanchard et al., 2004a, b), which for ease of reference we denote PRE(I)-tt-B. As mentioned above, PRE(I)-tt-B contains labeled tRNAs that differ from those used in the present study with respect both to amino acid specificity and positions of labeling. Both our PRE(I)-tt complex and the PRE(I)-tt-B complex show a slightly higher occupancy in the classical state, about 60% of the time in fluctuating PRE complexes. However, there are two noteworthy differences. First, a subsequent more complete analysis of the PRE(I)-tt-B provided evidence for an additional hybrid state, assigned as (A/A, P/E) (Munro et al., 2007). We used hidden Markov analysis (McKinney et al., 2006) to test specifically for the presence of a third distinct FRET state in PRE(I)-tt, but none was detected. It is possible that observation of the putative (A/A, P/E) intermediate depends on the precise placement of fluorescent probes within the two bound tRNAs especially since interconversion of the two hybrids involves only a minor displacement of peptidyl-tRNA. Such small displacement might also make putative additional hybrid intermediates undetectable by L11-tRNA FRET. Second, the rates of classical to hybrid interconversion are 2 – 5 times faster for PRE(I)-tt-B than for PRE(I)-tt. This may reflect effects of different tRNAs on rates of conformational change in the ribosome and of fluorophore placement at different positions within tRNA, as well as differences in experimental protocols employed for the two PRE complexes.
In the present work, the general similarities between the Lt complexes and the tt complexes, evident from the results summarized in Table S1 and taking into account the differences in labeling positions among the Lt and tt complexes studied, strongly suggest that the reversible classical to hybrid transition is monitored by changes in L11-tRNA as well as tRNA-tRNA distances. Although these results suggest coupling of the movements of tRNAs and of the L11 region, it has been observed previously that not all motions within the PRE complex are simultaneous. For example, fluctuations monitored by smFRET between deacylated tRNA and either the N-terminal domain of L1 or peptidyl-tRNA, are largely uncoupled from one another (Munro et al. 2010a), and cryoelectron microscopy studies of the PRE complex (Fischer et al., 2010) indicate significant movements of the L1 stalk that are only loosely coupled with tRNA movement.
Translocation
The results presented in Figures 3 and S2a–c clarify two significant ambiguities that currently exist in the literature concerning the mechanism of EF-G·GTP catalyzed translocation, as revealed by previous smFRET studies. The first concerns whether EF-G·GTP can bind directly to the classical PRE state and translocate directly from that state. The present results demonstrate that it can, and that translocation then proceeds via transient formation of a species having a FRET efficiency indistinguishable from that of the hybrid complex. Our results thus stand in marked contrast to the model put forward by Fei et al (2008) that EF-G selectively binds to the hybrid state in catalyzing translocation. This model was based on measurements of the real-time evolution of a FRET signal between protein L1 and deacylated tRNA during translocation when cognate TC and EF-G·GTP were added simultaneously to a POST-I complex (using our terminology), leading to POST-I to PRE-II to POST-II conversion. However, because the POST-I and POST-II complexes had FRET efficiencies indistinguishable from those of classical PRE-II and hybrid PRE-II complexes, respectively, the results obtained do not exclude alternative models. Indeed, the Fei et al. (2008) results can be interpreted in a manner fully consistent with the results presented in this work. In subsequent papers Fei et al. (2009) and others (Munro et al., 2010b) have speculated that EF-G could catalyze conversion of classical PRE state to a hybrid PRE state during translocation, a speculation for which the present results (Figures 3I–K, S2c) provide the first direct demonstration.
The second ambiguity concerns the effect of EF-G·GTP binding on L11-tRNA and tRNA-tRNA distance fluctuations in the PRE complex prior to translocation. We show by direct measurement that EF-G·GTP addition suppresses such fluctuations, in agreement with the results of both Fei et al. (2008) and Cornish et al. (2008) who also measured real-time evolution of FRET signals during translocation. Conformational fluctuations between L1 and tRNA are also reduced by RF1 binding to the A-site containing a termination codon (Sternberg et al., 2009), paralleling the effects of EF-G binding measured here (Figures 3 and S2a,b). However, other studies have shown that addition of EF-G·GDPNP to ribosomes containing a single tRNA actually stimulates tRNA fluctuations between a hybrid P/E and a classical P/P position (Fei et al. 2009; Munro et al., 2010b), results that could be interpreted as evidence that EF-G increases the dynamic behavior of the ribosome during translocation. The disparity between such an interpretation and our present results suggests that the ribosome complexes containing EF-G·GDPNP and a single tRNA bound to the P site, examined in these latter references, are not good models for the PRE complex immediately prior to translocation, which contains EF-G·GDP·Pi (see below) and two tRNAs bound to the A- and P-sites. Alternatively it might be argued that EF-G·GTP binding to PRE complex could have contrasting effects on fluctuations near the A-site, where L11 is located, from those near the E-site and L1. While the results of Fei et al. (2008), Cornish et al. (2008) and Sternberg et al. (2009) do not provide support for this interpretation, it would be of interest to examine L1 movement during an authentic translocation reaction to definitively select between these possibilities.
Earlier ensemble studies of translocation kinetics (Pan et al. 2007; Rodnina et al. 1997; Savelsbergh et al. 2003; Walker et al. 2008) led to a model of translocation in which EF-G·GTP binding to a PRE complex very rapidly results in GTP hydrolysis (rate constant ~ 250 s−1 at 37 °C), which is followed by slower conformational changes (~35 s−1 at 37 °C) including subunit rotation (“ratcheting”), specific motions within the GAC (Diaconu et al. 2005; Frank and Agrawal, 2000; Seo et al. 2006) and conversion of the PRE complex to a hybrid PRE complex, denoted the INT complex (Pan et al. 2007). Thus, following EF-G·GTP addition to PRE complex, GTP hydrolysis precedes all tRNA and mRNA movements. During formation of the INT complex, the 3’ terminus of peptidyl-tRNA moves from a position in which it has negligible puromycin reactivity (<10−3 relative to a POST complex) to a position in which it has appreciable puromycin reactivity (~5% of that of a POST complex) (Pan et al. 2007). Translocation is completed by further movement of the tRNAs into the P/P and E/E positions to form the POST complex, with dissociation of EF-G·GDP and Pi and reversion of the ribosome to the unratcheted conformation.
Incorporation of the present results into this kinetic scheme leads to a simplified model of the translocation process, focused on EF-G binding and tRNA movements, illustrated in Figure 6. According to this model, the ensemble results showing movement of both ribosome-bound tRNAs on EF-G·GTP-induced conversion of PRE complex to INT complex (Pan et al., 2007) should principally reflect movement of that fraction of tRNAs which are bound in the high FRET (classical) state of the PRE complex on formation of the transient hybrid complex, as shown in Figure 3I, K. Translocation from the hybrid PRE complex may proceed via the same or a very similar EF-G-bound intermediate complex, although one that would be difficult to detect given that the L11-tRNA FRET efficiencies of the two complexes were not distinguishable. The very short lifetime of the transient complex (86 ms, Figure 3J) is compatible with it being an obligatory intermediate during translocation in vivo, given an average elongation rate of ~20 s−1 at 37 °C (Liang et al, 2000), and allowing for ~ 5-fold slowing in experiments performed at 21° C.
Figure 6. EF-G binding and tRNA movement during translocation.
Earlier ensemble and smFRET results demonstrated that PRE complex, which fluctuates between classical and hybrid states, is converted to POST complex via an INT intermediate (Pan et al., 2007), such as that depicted. A major new result from this work is the demonstration that EF-G·GTP can bind directly to the classical state of the PRE complex before forming a transient hybrid complex as a short-lived, kinetically competent intermediate in POST complex formation. The hybrid PRE-complex may proceed to POST complex via a similar intermediate. The L11 and L1 regions are indicated by the blue arrows. Additional intermediates, not observed in the present work, would be required for a full description of the translocation mechanism (see text).
A more complete view of translocation dynamics would also account for how such tRNA movements are coordinated with larger structural changes within the ribosome (Fischer et al. 2010, Ratje et al. 2010), involving ratcheting and unratcheting of the ribosomal subunits with respect to one another and the swiveling of the 30S subunit head with respect to the body (Zhang et al. 2009). SmFRET experiments, in particular those involving movements of the highly mobile L1 protein (Munro et al. 2010a,b), along with reaction coordinate calculations (Munro et al., 2009), suggest that the ratcheting step that follows ribosome-dependent EF-G·GTP hydrolysis is not well described as a two-state process, but rather requires a series of loosely coupled conformational changes that converge into the overall conformational change associated with ratcheting. The translocation results presented in Figure 5 provide evidence that the unratcheting step associated with the completion of translocation may also be a multi-step process. Most notably, the gradual component of FRET decrease that follows the larger sharp decrease on translocation of the Lt complexes signals a series of conformational changes within the L11 region that follow major tRNA movement.
In summary, FRET distance measurements between adjacent tRNAs in the A and P sites of the ribosome and between A-site tRNA and protein L11 demonstrates similar fluctuations of both distances, suggesting a concerted motion of the tRNAs away from L11 and away from each other when the classical PRE state spontaneously transitions into the hybrid state. EF-G·GTP binds with similar probabilities to both the classical and hybrid PRE complexes, and, in so doing, rapidly decreases these fluctuations. Translocation from the classical PRE state proceeds via transient formation of a short-lived hybrid complex. The transition from the stabilized, EF-G·GTP-bound, classical or hybrid PRE complex to the POST complex is, in each case, the main rate-limiting step of the translocation process. A surprisingly slow final motion of the tRNAs relative to L11 is observed during translocation that is much diminished in the presence of the next cognate ternary complex. The overall picture that emerges is that ribosome conformational lability is markedly suppressed by the binding of protein factors at or near the A-site that mediate the next reaction step in the elongation cycle. Such decreased lability may be important for optimizing reaction rates.
EXPERIMENTAL METHODS
Materials
(for details see Supplemental):
C38S/S87C-L11Cy3
The Stratagene QuikChange site-directed mutagenesis kit was used to construct the double variant C38S/S87C from the cloned, N-terminal His-tagged L11 employed previously (Qin et al., 2009; Seo et al., 2006; Wang et al., 2007). In addition, mutation of the genetic sequence encoding the N-terminal region MGSSHHHHHHSSGLVPRGSHMAKK…, (the beginning of the encoded native sequence is underlined) to replace the mRNA sequence GGCAGCAGC, encoding GlySerSer, by UAUUAUUAU, encoding TyrTyrTyr, led to a 2.8-fold increase in L11 expression. Initial codons with high AU content increase expression levels (de Valdivia and Isaksson, 2005). Purification and labeling with Cy3-maleimide was carried out as described (Qin et al., 2009; Wang et al., 2007). Typical Cy3/L11 ratios were 0.9 ± 0.3.
70SCy3 ribosomes
A 2-fold molar excess of C38S/S87C-L11Cy3 was incubated with 2 µM AM77 70S ribosomes, which lack L11. Reconstituted ribosomes typically containing 0.8 Cy3/ribosome were isolated by ultracentrifugation as described (Qin et al., 2009; Wang et al., 2007).
Cy3- and Cy5-labeled tRNAs
Labeled tRNAs were prepared using the reduction, charging, and labeling protocol as described (Pan et al., 2009), starting with E. coli tRNAfMet, E. coli tRNAArg and yeast tRNAPhe purchased from Chemical Block (Moscow). Separations of fMet-tRNAfMet from tRNAfMet and Phe-tRNAPhe from tRNAPhe were achieved by reversed-phase HPLC using a LiChrospher WP-300 RP-18 (5µm bead) column (250–4mm) (Merck KGaA-Darmstadt). Partial resolution of Arg-tRNAArg from tRNAArg was achieved by FPLC (MonoQ) (Pan et al., 2009), which also removed polyA. Stoichiometries of labeling varied from 0.7 – 0.9 probe per tRNA. Arg-tRNAArg(Cy3), Arg-tRNAArg(Cy5), and Phe-tRNAPhe(Cy5) showed substantial activity in ensemble assays of translation (Figure S3).
tRNAfMet has a single isoacceptor and a single dihydroU at nt position 20. tRNAArg2, which is cognate to the CGU codon in the mRNAs we employ [E. coli tRNAArg has four isoacceptors (Dittmar et al., 2005)] and tRNAPhe, which does not have alternate isoacceptors, contain two dihydroUs, at positions 17 and 20. We have recently determined that Cy3 labeling is spread approximately equally among these two positions (J.K. & B.S.C., in preparation). The Cy5 distribution is likely to be similar. The distribution of fluorescent labels among the two potential positions of labeling (16 and 17) in yeast tRNAPhe is presently unknown.
mRNA
5’-biotin-labeled mRNA (Dharmacon, Inc.) had the following sequence: GGG AAU UCA AAA AUU UAA AAG UUA AUA AGG AUA CAU ACU AUG CGU UUC UUC CGU UUC UAU CGU UUC. The underlined sequence is a strong Shine-Dalgarno region. The italized sequence codes for MRFFRFYRF.
smFRET protocols
General conditions
All smFRET studies were carried out at 21 °C. All complex formations and single molecule detections were carried out in TAM15 buffer (20 mM Tris-HCl (pH 7.5), 15 mM Mg(OAc)2, 30 mM NH4Cl, 70 mM KCl, 0.75 mM EDTA, 1 mM DTT, and 0.2% (w/v) Tween 20). An enzymatic deoxygenation system of 3 mg/mL glucose, 100 µg/mL glucose oxidase (Sigma-Aldrich), 40 µg/mL catalase (Roche), and 1.5 mM 6-hydroxy-2,5,7,8-tetramethyl-chromane-2-carboxylic acid (Trolox, Sigma-Aldrich – by dilution from a DMSO solution) was present in the final single-molecule imaging solutions to diminish fluorophore photobleaching and blinking.
Immobilization of ribosomes
The sample flow chamber (10 µL) was formed on a PEG-coated slide (Roy et al., 2008) and coverslip and held together by double-sided adhesive tape that served as spacers and borders of the flow chamber. Immobilized ribosome complexes were formed by treating the PEGylated surface with streptavidin and biotinylated initiation complex followed by various reagents (see Supplemental). Except as described below for real-time translocation, unbound or unreacted reagents were always washed away before single molecule data was collected.
Real time translocation
Collection of real-time translocation traces began 20 s prior to injecting 2 µM EF-G and 3 mM GTP onto surfaces containing PRE complexes, and was carried out without further washing. 10 nM pre-formed Phe TC (Phe-tRNAPhe·EF-Tu·GDPNP with 10 nM extra EF-Tu and 15 µM GDPNP) was mixed with EF-G and GTP 20–30 s prior to injection.
TIRF measurements on immobilized ribosomes
A custom-built objective-type total internal reflection fluorescence (TIRF) microscope was based on a commercial inverted microscope (Eclipse Ti, Nikon) with a 1.49 N.A. 100× oil immersion objective (Apo TIRF; Nikon, Tokyo, Japan) (Figure S5). Alternating-laser excitation (Kapanidis et al., 2004) (Alex) was performed as described (see Supplemental). Only spots showing anti-correlation behavior between 532-nm-illuminated Cy3 and Cy5 channels, FRET changes in 532 nm laser-illuminated frames and no significant changes in the 640 nm laser-illuminated frames (to eliminate photo-bleaching and blinking), were considered as displaying real FRET changes. Actual FRET efficiencies were calculated as described in Supplemental. Except as otherwise indicated (Figure 3I, J, K) fluorescence intensities were recorded under 100 ms integration time per frame with ALEX (alternating-laser excitation) between 532 nm and 640 nm lasers.
All FRET traces were analyzed by a Hidden Markov Model based software [HaMMy (McKinney et al., 2006)]. No more than two FRET states were found in any single traces despite vigorous efforts to identify additional states. Traces with only one FRET state were termed stable traces, whereas traces with two FRET states were termed fluctuating traces. Dwell times of high and low FRET states of fluctuating traces were calculated by HaMMy and fitted to a biexponential curve:
FRET ratios
All FRET ratios are calculated using Eq. S1.
FRET probability density plots
FRET probability density plots are two-dimensional contour maps plotted from time-resolved FRET histograms. For each plot, FRET traces were synchronized at the same specific event, such as EF-G injection or translocation-induced FRET change.
Supplementary Material
ACKNOWLEDGMENTS
Supported by NIH R01 grant GM080376 to BSC and YEG, NIST ATP grant 70NANB7H7011 through Anima Cell Metrology, Inc., and NIH grant GM071014 to BSC. We thank Dr. Knud Nierhaus for helpful comments. C. C., B. S. C. and Y. E. G. designed experiments, analyzed data and wrote the paper; C. C., B. S., and Y. W. performed smFRET experiments; B. S., C. C. and Y. E. G. built the microscope; J. K., D. C., H. L., H. Z., and G. R. prepared and characterized fluorescent-labeled ribosomes and tRNAs; Z. S. custom wrote custom software for analysis of smFRET results.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: B.S.C. and Y.E.G are paid consultants of Anima Cell Metrology, Inc.
REFERENCES
- Agirrezabala X, Frank J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q Rev Biophys. 2009;42:159–200. doi: 10.1017/S0033583509990060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Lei JL, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Visualization of the Hybrid State of tRNA Binding Promoted by Spontaneous Ratcheting of the Ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors tune the rate of initiation of protein synthesis in bacteria. Embo J. 2006;25:2539–2550. doi: 10.1038/sj.emboj.7601140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004a;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. P Natl Acad Sci USA. 2004b;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn GL, Draper DE, Lattman EE, Gittis AG. Crystal structure of a conserved ribosomal protein-RNA complex. Science. 1999;284:1171–1174. doi: 10.1126/science.284.5417.1171. [DOI] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. P Natl Acad Sci USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviter T, Gromadski KB, Rodnina MV. The ribosome's response to codon-anticodon mismatches. Biochimie. 2006;88:1001–1011. doi: 10.1016/j.biochi.2006.04.013. [DOI] [PubMed] [Google Scholar]
- de Valdivia EIG, Isaksson LA. Abortive translation caused by peptidyl-tRNA drop-off at NGG codons in the early coding region of mRNA. Febs J. 2005;272:5306–5316. doi: 10.1111/j.1742-4658.2005.04926.x. [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. Embo Rep. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Kosuri P, MacDougall DD, Gonzalez RL. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Fei JY, Bronson JE, Hofman JM, Srinivas RL, Wiggins CH, Gonzalez RL. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. P Natl Acad Sci USA. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Grigoriadou C, Marzi S, Kirillov S, Gualerzi CO, Cooperman BS. A quantitative kinetic scheme for 70 S translation initiation complex formation. J Mol Biol. 2007;373:562–572. doi: 10.1016/j.jmb.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick B, Lee K, Joseph S. Kinetics of Stop Codon Recognition by Release Factor 1. Biochemistry-Us. 2009;48:11178–11184. doi: 10.1021/bi901577d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E, Weiss S. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. P Natl Acad Sci USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Puglisi JD. GTP Hydrolysis by IF2 Guides Progression of the Ribosome into Elongation. Mol Cell. 2009;35:37–47. doi: 10.1016/j.molcel.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. P Natl Acad Sci USA. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Altman RB, Tung CS, Cate JHD, Sanbonmatsu KY, Blanchard SC. Spontaneous formation of the unlocked state of the ribosome is a multistep process. P Natl Acad Sci USA. 2010a;107:709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. Embo J. 2010b;29:770–781. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Sanbonmatsu KY, Spahn CMT, Blanchard SC. Navigating the ribosome's metastable energy landscape. Trends Biochem Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol. 2010c;17:1470–1477. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Zhang CM, Kirillov S, Hou YM, Cooperman BS. Perturbation of the tRNA tertiary core differentially affects specific steps of the elongation cycle. J Biol Chem. 2008;283:18431–18440. doi: 10.1074/jbc.M801560200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan DL. Department of Chemistry. Philadelphia: University of Pennsylvania; 2008. [Google Scholar]
- Pan DL, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan DL, Qin HO, Cooperman BS. Synthesis and functional activity of tRNAs labeled with fluorescent hydrazides in the D-loop. Rna. 2009;15:346–354. doi: 10.1261/rna.1257509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E-coli ribosome. Embo J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps SS, Joseph S. Non-bridging phosphate oxygen atoms within the tRNA anticodon stem-loop are essential for ribosomal A site binding and translocation. J Mol Biol. 2005;349:288–301. doi: 10.1016/j.jmb.2005.03.079. [DOI] [PubMed] [Google Scholar]
- Qin HO, Grigoriadou C, Cooperman BS. Interaction of IF2 with the Ribosomal GTPase-Associated Center during 70S Initiation Complex Formation. Biochemistry-Us. 2009;48:4699–4706. doi: 10.1021/bi900222e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, Weir JR, Ramakrishnan V. The Crystal Structure of the Ribosome Bound to EF-Tu and Aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3.5 angstrom resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Seo HS, Abedin S, Kamp D, Wilson DN, Nierhaus KH, Cooperman BS. EF-G-dependent GTPase on the ribosome. Conformational change and fusidic acid inhibition. Biochemistry-Us. 2006;45:2504–2514. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, Fei JY, Prywes N, McGrath KA, Gonzalez RL. Translation factors direct intrinsic ribosome dynamics during translation termination and ribosome recycling. Nat Struct Mol Biol. 2009;16:861–868. doi: 10.1038/nsmb.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the processive molecular motor, myosin V. Nat Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. P Natl Acad Sci USA. 2008;105:9192–9197. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qin H, Kudaravalli RD, Kirillov SV, Dempsey GT, Pan D, Cooperman BS, Goldman YE. Single-molecule structural dynamics of EF-G-ribosome interaction during translocation. Biochemistry-Us. 2007;46:10767–10775. doi: 10.1021/bi700657d. [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Guymon R, McCutcheon JP, White SW, Ramakrishnan V. A detailed view of a ribosomal active site: The structure of the L11-RNA complex. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W, Peske F, Beringer M, Gromadski KB, Savelsbergh A, Rodnina MV. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem Soc T. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- Yao G, Fang XH, Yokota H, Yanagida T, Tan WH. Monitoring molecular beacon DNA probe hybridization at the single-molecule level. Chem-Eur J. 2003;9:5686–5692. doi: 10.1002/chem.200304977. [DOI] [PubMed] [Google Scholar]
- Zaher HS, Green R. Fidelity at the Molecular Level: Lessons from Protein Synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- Zhang W, Dunkle JA, Cate JHD. Structures of the Ribosome in Intermediate States of Ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.