Abstract
The transcription regulatory properties of murine B-myb protein were compared to those of c-myb. Whereas c-Myb trans-activated an SV40 early promoter containing multiple copies of an upstream c-Myb DNA-binding site (MBS-1), and similarly the human c-myc promoter, B-Myb was unable to do so. Full-length B-Myb translated in vitro did not bind MBS-1; however, truncation of the B-Myb C-terminus or fusion of the B-Myb DNA-binding domain to the c-Myb C-terminus showed that it was inherently competent to interact with this motif. Further evidence from co-transfection experiments, demonstrating that B-Myb inhibited trans-activation by c-Myb, suggested that failure of B-Myb to trans-activate these promoters did not simply occur through lack of binding to MBS-1. Moreover, using GAL4/B-Myb fusions, it was found that an acidic region of B-Myb, which by comparison to c-Myb was expected to contain a transcription activation domain, actually had no inherent trans-activation activity and indeed appeared to trans-inhibit c-Myb. In contrast to the above findings, both B-Myb and c-Myb were able to weakly trans-activate the DNA polymerase alpha promoter. Results obtained here demonstrate that the activities of B-Myb and c-Myb are clearly distinct and suggest that these related proteins may have different functions in regulation of target gene expression.
Full text
PDF

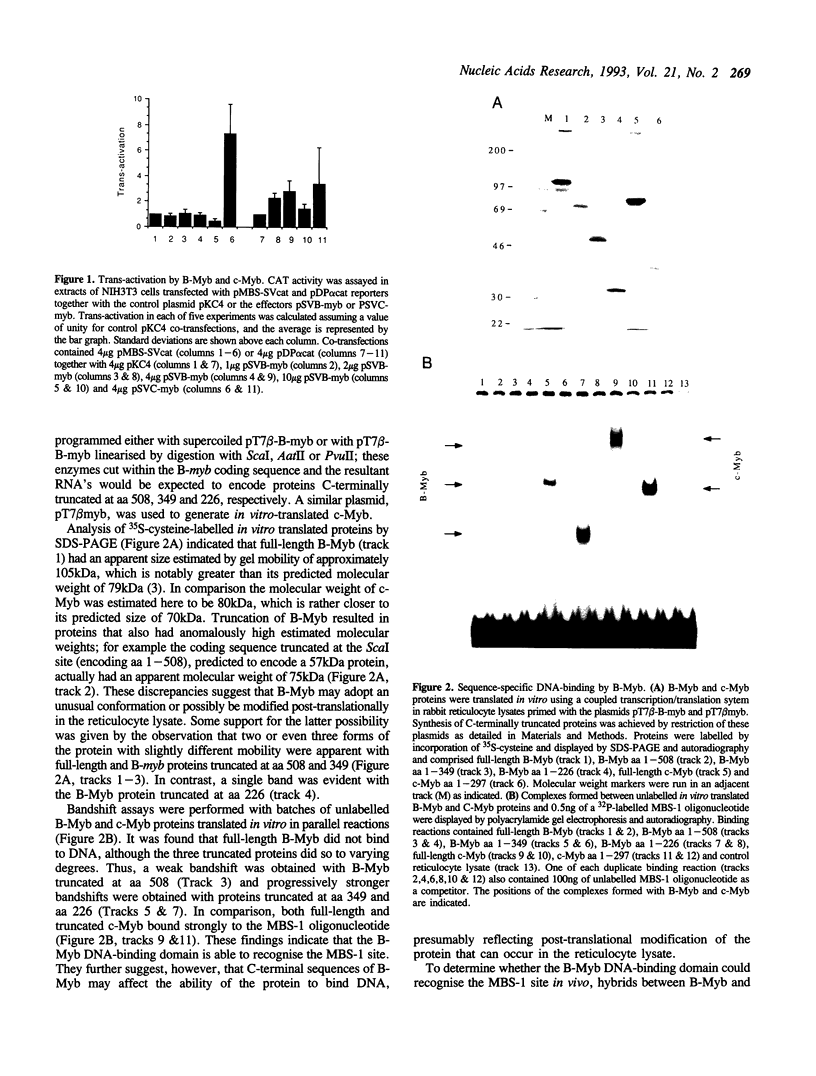
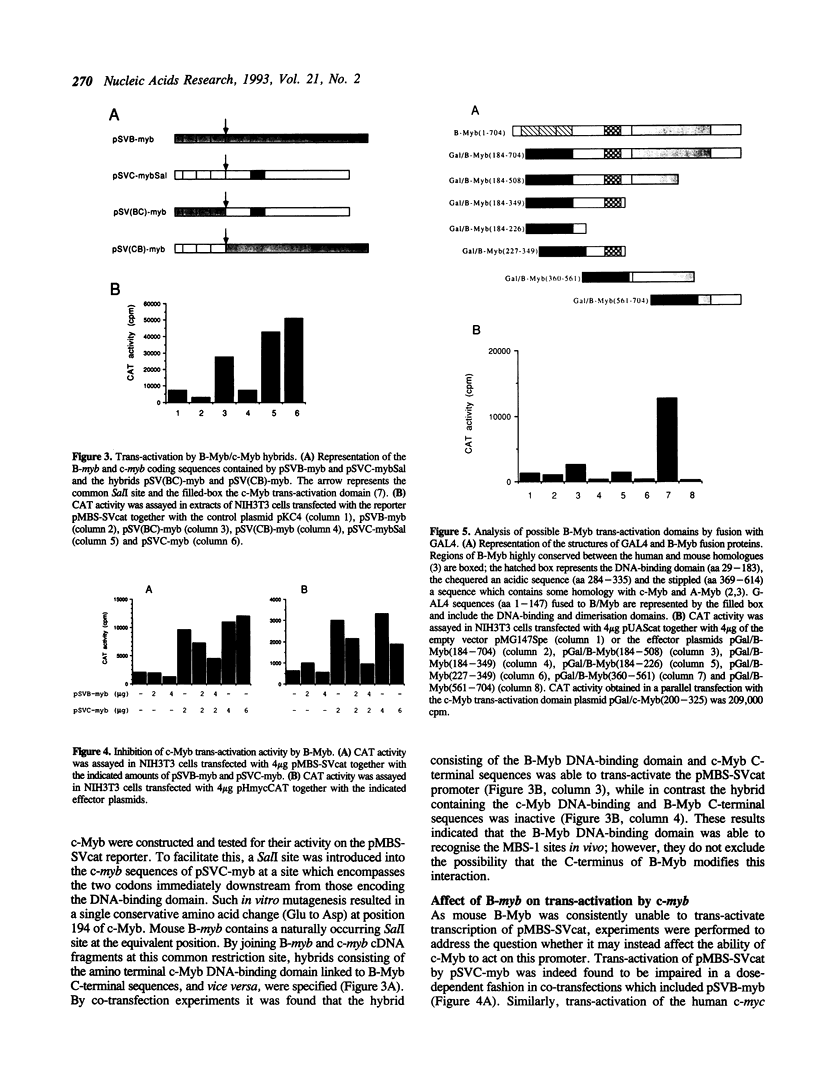
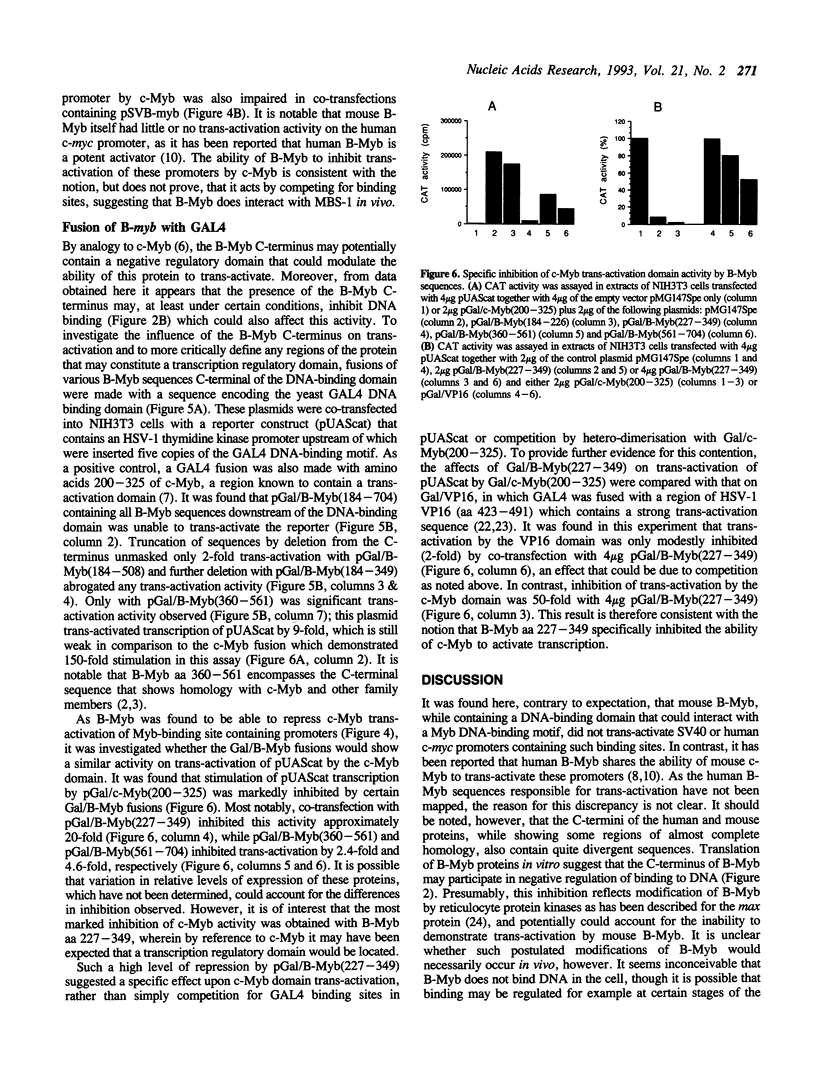

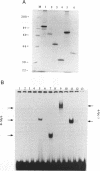
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berberich S. J., Cole M. D. Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers. Genes Dev. 1992 Feb;6(2):166–176. doi: 10.1101/gad.6.2.166. [DOI] [PubMed] [Google Scholar]
- Biedenkapp H., Borgmeyer U., Sippel A. E., Klempnauer K. H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988 Oct 27;335(6193):835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- Cress W. D., Triezenberg S. J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991 Jan 4;251(4989):87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Dalrymple M. A., McGeoch D. J., Davison A. J., Preston C. M. DNA sequence of the herpes simplex virus type 1 gene whose product is responsible for transcriptional activation of immediate early promoters. Nucleic Acids Res. 1985 Nov 11;13(21):7865–7879. doi: 10.1093/nar/13.21.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golay J., Capucci A., Arsura M., Castellano M., Rizzo V., Introna M. Expression of c-myb and B-myb, but not A-myb, correlates with proliferation in human hematopoietic cells. Blood. 1991 Jan 1;77(1):149–158. [PubMed] [Google Scholar]
- Hay N., Bishop J. M., Levens D. Regulatory elements that modulate expression of human c-myc. Genes Dev. 1987 Sep;1(7):659–671. doi: 10.1101/gad.1.7.659. [DOI] [PubMed] [Google Scholar]
- Howe K. M., Reakes C. F., Watson R. J. Characterization of the sequence-specific interaction of mouse c-myb protein with DNA. EMBO J. 1990 Jan;9(1):161–169. doi: 10.1002/j.1460-2075.1990.tb08092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K. M., Watson R. J. Nucleotide preferences in sequence-specific recognition of DNA by c-myb protein. Nucleic Acids Res. 1991 Jul 25;19(14):3913–3919. doi: 10.1093/nar/19.14.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanei-Ishii C., MacMillan E. M., Nomura T., Sarai A., Ramsay R. G., Aimoto S., Ishii S., Gonda T. J. Transactivation and transformation by Myb are negatively regulated by a leucine-zipper structure. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):3088–3092. doi: 10.1073/pnas.89.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempnauer K. H., Arnold H., Biedenkapp H. Activation of transcription by v-myb: evidence for two different mechanisms. Genes Dev. 1989 Oct;3(10):1582–1589. doi: 10.1101/gad.3.10.1582. [DOI] [PubMed] [Google Scholar]
- Lam E. W., Robinson C., Watson R. J. Characterization and cell cycle-regulated expression of mouse B-myb. Oncogene. 1992 Sep;7(9):1885–1890. [PubMed] [Google Scholar]
- Luckow B., Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987 Jul 10;15(13):5490–5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon J., Howe K. M., Watson R. J. The induction of Friend erythroleukaemia differentiation is markedly affected by expression of a transfected c-myb cDNA. Oncogene. 1988 Dec;3(6):717–720. [PubMed] [Google Scholar]
- Mizuguchi G., Nakagoshi H., Nagase T., Nomura N., Date T., Ueno Y., Ishii S. DNA binding activity and transcriptional activator function of the human B-myb protein compared with c-MYB. J Biol Chem. 1990 Jun 5;265(16):9280–9284. [PubMed] [Google Scholar]
- Mucenski M. L., McLain K., Kier A. B., Swerdlow S. H., Schreiner C. M., Miller T. A., Pietryga D. W., Scott W. J., Jr, Potter S. S. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991 May 17;65(4):677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- Nakagoshi H., Kanei-Ishii C., Sawazaki T., Mizuguchi G., Ishii S. Transcriptional activation of the c-myc gene by the c-myb and B-myb gene products. Oncogene. 1992 Jun;7(6):1233–1240. [PubMed] [Google Scholar]
- Nishina Y., Nakagoshi H., Imamoto F., Gonda T. J., Ishii S. Trans-activation by the c-myb proto-oncogene. Nucleic Acids Res. 1989 Jan 11;17(1):107–117. doi: 10.1093/nar/17.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura N., Takahashi M., Matsui M., Ishii S., Date T., Sasamoto S., Ishizaki R. Isolation of human cDNA clones of myb-related genes, A-myb and B-myb. Nucleic Acids Res. 1988 Dec 9;16(23):11075–11089. doi: 10.1093/nar/16.23.11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. E., Nasheuer H. P., Wang T. S. Human DNA polymerase alpha gene: sequences controlling expression in cycling and serum-stimulated cells. Mol Cell Biol. 1991 Apr;11(4):2081–2095. doi: 10.1128/mcb.11.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay R. G., Ishii S., Gonda T. J. Increase in specific DNA binding by carboxyl truncation suggests a mechanism for activation of Myb. Oncogene. 1991 Oct;6(10):1875–1879. [PubMed] [Google Scholar]
- Sakura H., Kanei-Ishii C., Nagase T., Nakagoshi H., Gonda T. J., Ishii S. Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5758–5762. doi: 10.1073/pnas.86.15.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleigh M. J. A nonchromatographic assay for expression of the chloramphenicol acetyltransferase gene in eucaryotic cells. Anal Biochem. 1986 Jul;156(1):251–256. doi: 10.1016/0003-2697(86)90180-6. [DOI] [PubMed] [Google Scholar]
- Venturelli D., Travali S., Calabretta B. Inhibition of T-cell proliferation by a MYB antisense oligomer is accompanied by selective down-regulation of DNA polymerase alpha expression. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5963–5967. doi: 10.1073/pnas.87.15.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston K., Bishop J. M. Transcriptional activation by the v-myb oncogene and its cellular progenitor, c-myb. Cell. 1989 Jul 14;58(1):85–93. doi: 10.1016/0092-8674(89)90405-4. [DOI] [PubMed] [Google Scholar]