SUMMARY
Background
Optimal management of clinically localized prostate cancer presents unique challenges because of its highly variable and often indolent natural history. To predict disease aggressiveness, clinicians combine clinical parameters to create prognostic models, but the accuracy of current models is very limited. There is significant clinical need for biomarkers that improve our ability to predict disease outcome.
Methods
Using quantitative RT-PCR on RNA from formalin fixed paraffin-embedded tumour samples, we measured the expression level of 31 genes involved in cell cycle progression (CCP genes), created a predefined score and evaluated its ability to predict disease outcome. The signature was tested in a retrospective cohort of 366 patients from the U.S. who had undergone radical prostatectomy, and in a retrospective cohort of 337 men with clinically localized prostate cancer diagnosed by a transurethral resection (TURP) in the UK and managed conservatively.
Findings
The cell cycle progression signature was a highly significant predictor of outcome in both cohorts. After prostatectomy the CCP score predicted biochemical recurrence in univariate (Hazard ratio (HR) for a one unit change in CCP (doubling) = 1.89; 95% CI (1.54, 2.31) χ2 = 34·0, 1df, p = 5·6 × 10−9) and multivariate analysis (HR = 1.74; 95% CI (1.39, 2.17) χ2 = 21·65, 1df, p = 3·3 ×10−6). The CCP score and PSA were the dominant variables in the best predictive model and were much more significant than any other clinical measure. In the TURP cohort, the CCP score was the dominant variable for predicting death from prostate cancer in both univariate (HR= 2.92; 95% CI (2.38, 3.57) χ2 = 92·7, 1df, p = 6.1 × 10−22) and multivariate analyses (χ2 = 42·2, p = 8·2 × 10−11), where it was much stronger than all other prognostic factors. In no case 4 was there significant evidence for heterogeneity in the hazard ratio for the CCP score across any clinical parameter.
Interpretation
The CCP score provides a substantial amount of independent information about the risk of recurrence after radical prostatectomy and the risk of death in conservatively managed prostate cancer diagnosed by TURP. Taken together, these studies provide strong evidence that the CCP score is a highly robust prognostic marker which, after additional validation, could have a central role in determining appropriate treatment for prostate cancer patients.
Funding
Study funded by Cancer Research UK, the Orchid Appeal, US National Institutes of Health (SPORE CA92629), and the Koch Foundation. Molecular testing performed at Myriad Genetics.
Keywords: Prostate cancer, predictive model, Cell cycle progression genes
INTRODUCTION
Prostate cancer is very common especially where PSA screening is used [0] and its natural history highly variable and difficult to predict. While some men have indolent disease which can be safely watched, others have aggressive cancer and need immediate intervention. Accurately predicting disease behaviour is critical because radical treatment is associated with a high morbidity and indiscriminate over treatment of indolent disease is a serious problem. This fact has led to calls for a more conservative approach to prostate cancer care. However, this approach is not without consequences as prostate cancer is already the second or third commonest form of cancer death in men in most developed countries. Furthermore, conservative management can lead to considerable anxiety, especially when the clinical outcome is so uncertain.
The problems associated with uncertain prognosis have been exacerbated by the introduction of population prostate specific antigen (PSA) screening in some countries, leading to a much higher reported incidence rate, but having a modest influence on mortality rates 2,3,. Autopsy series have confirmed that histologically proven prostate cancer can be identified in approximately 40% of men over 50 years of age who die of other causes 4,5. This is approximately 10 times higher than the lifetime risk of dying of prostate cancer in the Western world 1 indicating that more intensive screening is likely to uncover even more indolent disease.
To help predict prostate cancer outcome, clinical parameters like Gleason score, tumour stage, margin status, and PSA level have been combined into models that aim to predict disease outcome 6–9 in the most favourable of clinical settings (post surgical resection) these prognostic models are about 75–85% accurate 10, but they perform less well in conservatively treated patients11–13 To improve prediction of outcome, many groups have tried to develop tumour-derived RNA expression markers to enhance the predictive power of clinical parameters 15–21 However, these studies have achieved only limited success and have not changed clinical care.
In contrast, tumour-derived RNA expression signatures 22 have proven prognostic value in breast cancer and have significantly influenced clinical care 23–28. These different signatures have very few genes in common, yet they perform similarly in terms of predicting disease recurrence 29. This apparent inconsistency can be reconciled by noting that the predictive power of each signature is generated primarily from genes whose expression is regulated as a function of cell cycle progression 30. These genes, hereafter referred to as cell cycle progression (CCP) genes, were originally identified as having RNA expression levels that oscillated as cells progressed though various stages of the cell cycle 31.
Since the expression levels of CCP genes probably reflect fundamental aspects of tumour biology, we reasoned that CCP genes might also be useful in prostate cancer. To investigate this possibility, we selected a set of 31 CCP genes and tested their ability to predict disease outcome using a predefined score based on their expression levels. First, we tested their utility to predict biochemical recurrence in a large retrospective cohort of patients who were treated with radical prostatectomy. Next, we evaluated the ability of this same CCP signature to predict death from prostate in a cohort of conservatively treated patients diagnosed from a TURP specimen. The primary aim of the study was to evaluate the prognostic value of the predefined CCP score in two very different settings. In both cohorts we tested the CCP score univariately and in multivariate models to determine how much additional prognostic information was provided that was not available from standard clinicopathological variables.
PATIENTS AND METHODS
CCP Gene Selection
To develop a CCP signature that would reliably generate expression data from formalin fixed paraffin embedded (FFPE) tissue, we selected 126 CCP genes from the GEO database and tested their performance on RNA derived from 96 commercially available prostate FFPE tumour sections (Asterand, Detroit MI) that were anonymous and not associated with clinical data. No demographic data were available for these specimens. Previous data 30 indicated that expression levels of different CCP genes were highly correlated, presumably reflecting the fraction of actively dividing cells within the sampled tissue. Therefore, we selected genes for inclusion in the signature based on their individual correlation with the mean expression level of the entire set of CCP genes (Supplement). The final signature consisted of 31 CCP genes (Table 1). Use of a large number of highly correlated genes was designed to provide a robust and highly reproducible measure of cell proliferation and it was not intended to capture information related to other factors (e.g. invasive potential). Derivation of the CCP score was done prospectively and is described below.
Table 1.
List of genes used to generate CCP score
| FOXM1 | ASPM | TK1 | PRC1 |
| CDC20 | BUB1B | PBK | DTL |
| CDKN3 | RRM2 | ASF1B | CEP55 |
| CDC2 | DLGAP5 | C18orf24 | RAD51 |
| KIF11 | BIRC5 | RAD54L | CENPM |
| KIAA0101 | KIF20A | PTTG1 | CDCA8 |
| NUSAP1 | PLK1 | CDCA3 | ORC6L |
| CENPF | TOP2A | MCM10 |
Radical Prostatectomy (RP) Cohort
All patients undergoing radical prostatectomy for prostate cancer from 1985–1995 were identified through the tumour registry at the Scott and White clinic, in Temple, Texas. This list was submitted to the pathology department and matched with the pathology accession number for the radical prostatectomy specimen. An attempt was made to identify tissue blocks for every patient identified for inclusion in this cohort. Pathology blocks were retrieved, and 10 slides were re-cut from the block for analysis. The middle slide was stained with H&E to confirm the presence of cancer. A total of 804 consecutive radical prostatectomy patients were identified, for which 775 had clinical data (Figure 1). They were followed for a median of 9·4 (IQR (6.9, 11.5)) years. The patient characteristics and the treatment outcomes of the entire cohort have been reported32. Median patient age was 68 (63, 72) years. The clinical stage was T1 33%, T2 67% and T3 <1%. The median preoperative PSA was 6·9 (4.3, 12.4) ng/ml and 68% of patients had values less than 10 ng/ml. The specimens were inked and pathological findings were recorded as to positive bladder neck or urethral margin, invasion into the capsule, extension through the capsule, positive margins and the involvement of the seminal vesicles. Biochemical recurrence was defined as a PSA > 0·3 ng/ml. This was based on the management policy used in the 1990s, and was not changed for the present analysis. Death from disease was defined as any patient who had biochemical recurrence and subsequently died with disease progression. We selected at random 442 patients for whom tumour tissue was available for inclusion in this study to create a retrospective cohort with a median follow up time of 9.2 (IQR (6.8, 10.9)) years. A total of 32 patients were excluded on the basis of having been treated with neoadjuvant therapies, because of the influence they have on interpreting Gleason score and the CCP score. Another 44 were excluded because of inadequate sample quality to compute the CCP score leaving 336 for analysis.
Figure 1.
Flow chart of patient selection for the radical prostatectomy cohort. The selection of 442 patients for inclusion in this study was based on tissue availability only, not on any clinical parameters.
TURP Cohort
This was a population-based retrospective cohort in which potential cases were identified from six cancer registries in Great Britain. Within each region, cases from collaborating hospitals were reviewed and full details have been reported 14. National approval was obtained from the Northern Multicentre Research Ethics Committee, followed by local ethics committee approval at each of the collaborating hospitals (Appendix).
Men were included in this study if they had clinically localized prostate cancer diagnosed by transurethral resection of the prostate (TURP) between 1990 and 1996 (inclusively), were under age 76 years at the time of diagnosis and had a baseline PSA measurement. Patients treated by radical prostatectomy or radiation therapy or who died or showed evidence of metastatic disease14 within six months of diagnosis were excluded. Men who had hormone therapy prior to diagnostic biopsy were also excluded, because of the influence of hormone treatment on interpreting Gleason score.
Original histological specimens from the diagnostic procedure were requested, collected and centrally reviewed by a panel of expert urological pathologists to confirm the diagnosis and, where necessary, to reassign Gleason scores for all the prostate cancers using a contemporary interpretation17 of the Gleason scoring system. Follow-up was via the cancer registries and the last review took place in December 2006. Deaths were divided into two categories, death from prostate cancer and death from other causes, according to standardised World Health Organisation criteria. Here we report on a randomly selected subset of 337 men diagnosed by TURP for which we were able to compute the CCP score (Figure 4). Median follow up was 9.8 (IQR (5.0–11.6)) years.
Figure 4.
Flow chart of patient selection for the TURP cohort. The 398 patients were chosen randomly, as specified in the predetermined analysis plan, without regard to clinical or outcome parameters.
Gene Expression
Depending on tumour volume, 5–12 consecutive 5 µM (TURP) or 10 µM (prostatectomy) FFPE tumour sections were used to isolate tumour derived RNA. The tumour region was dissected from the slide using a razor blade according to the pathologist’s instructions into a microfuge tube and the paraffin was removed using xylene and washed with ethanol. Samples were then treated overnight with proteinase K digestion at 55°C.
Total RNA was extracted using either RNeasy FFPE or miRNeasy (Qiagen) as described by the manufacturer (the exception being the extended proteinase K digestion described above). The miRNeasy kit became available after we had isolated RNA from about 1/3 of the RP cohort. We switched from RNeasy FFPE to miRNeasy because the new kit consistently generated better RNA yields. There was no difference in gene expression data between the kits.
Total RNA was treated with DNase I (Sigma) prior to cDNA synthesis. We employed the High-capacity cDNA Archive Kit (Applied Biosystems) to convert total RNA into single strand cDNA as described by the manufacturer. Ideally, at least 200ng RNA was required for the RT reaction, but smaller input amounts were also successful. The quality of the RNA was not ideal, as is expected when isolating nucleic acids from old FFPE biopsies. Careful attention was given as to how to obtain a reliable score from this material in the development of this assay. RNA quality was determined via the amplifiability of the CCP and HK genes. In order to generate a CCP score, essentially all of the house-keeping genes and at least 21 CCP genes needed to amplify. We attempted to generate a CCP score from every sample. For some of the samples some genes failed to amplify indicating that the RNA quality was too poor to create a score. However, most samples (90% of the RP cohort and 85% of the TUPR cohort) generated CCP scores, and therefore, had adequate quality RNA.
Prior to measuring expression levels, the cDNA was pre-amplified with a pooled reaction containing TaqMan assays. Pre-amplification reaction conditions were as follows: 14 cycles of 95°C for 15 seconds and 60°C for 4 minutes. The first cycle also included a ten minute incubation at 95°C. The amplification reaction was diluted 1:20 using the 1XTE buffer prior to loading on Taqman Low Density Arrays (TLDA, Applied Biosystems) to evaluate the amplified genes. Expression data were recorded as a CT value, the PCR cycle at which the fluorescence intensity exceeded a predefined threshold. A total of 31 predefined CCP genes and 15 housekeeper genes were amplified on a single TLDA array.
CCP Score
The CCP score for each individual was calculated as follows: For each of three replicates of each of the 31 CCP genes, CT values were normalised by subtracting the average of up to 15 non-failed housekeeper (HK) genes (centred using a predefined value) to yield . Then, a predefined baseline value was subtracted from to create a quantity labelled . This was then converted to a quantity proportional to copy number, calculated as . For missing values due to low expression, was set equal to 0. The mean was calculated for each CCP gene as the mean of the qualifying replicates, i.e. those with expression of at least 13 HK genes, which was then averaged over the qualifying CCP genes. A CCP gene was considered failed if more than one replicate did not qualify, or if two replicates qualified and one of them had equal to zero, or if the standard deviation between the three replicate values exceeded 0.5. Finally, this was converted back to the CCP score by taking a base 2 logarithm. CCP scores with the number of failing CCP genes in excess of 9 out of the 31, or a high standard deviation between scores calculated from the three replicates, were rejected and excluded from the analysis. The inter-assay variability has been established in our laboratory and the standard deviation of the CCP score for experimental replicates is 0.1.
Statistical Analyses
Survival analysis was performed using Cox proportional hazards models. The primary endpoint for the prostatectomy cohort was time to biochemical recurrence. Follow-up times commenced at date of surgery, and observations were censored at the date of last follow-up. A secondary endpoint was death after progression. Observations were censored at the date of last follow-up or death with no evidence of disease or death with stable disease. The primary endpoint for the TURP cohort was death from prostate cancer. Biochemical progression is not an appropriate outcome for this cohort, since baseline levels remain elevated and some patients will have chosen to start hormones or have radical treatment without evidence of increasing PSA levels, making this unevaluable in many cases.
Observations were censored at the date of last follow-up, or death from other causes. Follow-up times commenced six months after the date of diagnosis.
The following clinical variables were recorded for the prostatectomy cohort: diagnostic Gleason score, most recent pre-biopsy PSA, clinical stage, clinical grade, primary treatment (no pre- or post-operative hormones, orchiectomy, or adjuvant radiation), age at surgery, pathologic tumour stage, pathologic grade, pathologic Gleason score, and invasion features.
The following variables were recorded for the TURP cohort: centrally reviewed Gleason grade and score, baseline PSA value, clinical stage, extent of disease (proportion of TURP chips with disease), age at diagnosis, and initial treatment (no initial treatment or early hormone management). Ki-67 (percent cells positive) was measured by an immunohistochemical assay. Baseline PSA was defined as the last PSA value within six months of diagnosis (including pre diagnostic values), but before initiation of hormone therapy and at least three weeks after any biopsy.
In both cohorts, PSA was modelled as the natural logarithm of (1 + PSA (ng/ml)). Values over 100 were excluded as likely metastatic disease. As indicated above, for the RP cohort, only a single pathological Gleason score was recorded at diagnosis; for the TURP cohort it was previously determined that 3+4 and 4+3 had nearly identical prognosis 13,14. Therefore Gleason score 7 was analyzed as a single group. For simplicity, Gleason scores were grouped into less than 7, Gleason 7, and greater than Gleason 7 (Prostatectomy cohort: less than 7 (56% Gleason 6), greater than Gleason 7 (75% Gleason 8, 19% Gleason 9); TURP cohort: less than 7 (88% Gleason 6 (all 3+3)), Gleason 7 (34% 4+3) and greater than Gleason 7 (51% Gleason 8, 46% Gleason 9)). Model fitting was performed using Gleason score as a 3-level categorical variable. All p-values are 2-sided and 95% confidence intervals and p-values are based on χ2 statistics with 1 degree of freedom obtained from partial likelihood models.
The main analysis consisted of a univariate evaluation of the association between outcome and CCP score. A further predefined evaluation of the added prognostic information after adjustment for the baseline variables was also undertaken. This later effect was measured by the decrease in the likelihood ratio chi-square (χ2) when the CCP score was omitted from a model containing it and the other relevant baseline clinic-pathological variables. In constructing the best prognostic model, the goal was to capture most of the prognostic information from the CCP score and the clinical covariates in a simple linear model without over fitting the data. A multivariate proportional hazards (Cox) model was used to achieve this and to create a combined score based on the major clinical variable plus the CCP score. The CCP score was evaluated as a linear and a quadratic term, and tested for interactions with individual covariates. We employed a forward stepwise regression in which a new variable was added only if it had a p-value less than 0·02 for the prostatectomy study and 0·05 for the TURP study (since fewer variables were tested). Statistical analyses were performed in STATA/IC Version 10.1, S+ Version 8.1.1 for Linux (TIBCO Spotfire) and/or R Version 2.9.0 (http://www.r-project.org).
Role of the funding source
The study sponsor (QMUL) had no role in the design of the study, collection, analysis, or interpretation of the data, or in writing of the paper, or the decision to publish the results. Authors Lanchbury, Gutin, Stone, Wagner, Younus, Flake, Park, Reid and Warren are employees of Myriad Genetics. No other authors report any conflict of interest. Myriad Genetics have provided funding support to Queen Mary University of London to facilitate preparation of tumour blocks. The cell cycle expression profiles were assayed blind to all other data by Myriad Genetics. Analysis of the TURP cohort was conducted at QMUL under the direction of Prof Cuzick, following a predefined Statistical Analysis Plan. Interpretation of the data and the final content of this report were approved by all authors. The corresponding author had full access to the data (as did SS, JR, DM) and took full responsibility to submit for publication,
RESULTS
Radical Prostatectomy Cohort
We tested the utility of the predefined CCP score to predict biochemical recurrence on 410 patients who had undergone radical prostatectomy 32. The clinical characteristics of this cohort have been described previously, and the patient selection for this study is summarized in Figure 1. Thirty-six percent of the cohort had biochemical recurrence by 10 years post surgery. After applying our exclusion rules, there were 366 valid samples (89%) for statistical analysis. The CCP score distribution is shown in Figure 2A. Each unit change in the score corresponds to a 2-fold change in expression level.
Figure 2.
Analysis of the CCP score in the radical prostatectomy cohort. A) Distribution of the CCP scores (N=366). The median value was 0·16 with an interquartile range of −0·30 to 0·64, as indicated by red tick marks on the x-axis. B) Forest plot graphing the CCP score hazard ratio (HR) in different clinical subgroups. The recurrence rate for each subgroup is also given (recurrences/size), and the size of the each box is proportional to the number of recurrences within that patient subgroup. The thin lines indicate the 95% CI for each HR. The diamond at the bottom is the 95% CI of the HR for the entire cohort. C) Kaplan-Meier plot of recurrence versus time for patients grouped by integer values of CCP score. Each bin corresponds to a 2-fold increase in CCP expression. The green line (149 patients) corresponds CCP score • 0, purple line (161 patients) corresponds to 0 < CCP • 1, pink line (50 patients) corresponds to 1 < CCP • 2, and red line (6 patients) corresponds to CCP score > 2. Also indicated are the 10-year recurrence rates for each group. For these four groups, the 10-year biochemical recurrence rates (%) are: 23.7, 44.5, 61.9 and 83.3, respectively.
In a univariate analysis (Table 2), a high CCP gene expression value was predictive of biochemical recurrence (χ2 = 33·96, p = 5·6 ×10−9). Addition of a quadratic term for CCP score was not significant (p = 0·18), indicating that a simple linear model would capture most of the prognostic information from the signature. The increase in hazard ratio for a one-unit change in CCP score was 1·89 (95% CI (1·54, 2·31)). The CCP score was only weakly correlated with other variables, the highest correlation being with Gleason score and PSA (both ρ = 0·22). The CCP score was also higher in patients with positive lymph nodes (p-value 0.003), but this only involved 13 patients and was marginally significant after correcting for multiple tests. CCP was prognostic across different Gleason scores, PSA levels, and pathological stages (Figure 2B). However, there was a non-significant trend toward a reduced CCP hazard ratio in patients with higher clinical risk (i.e. in Gleason score > 6 and PSA levels > 10 ng/ml). A Kaplan-Meier plot of the proportion of patients who developed biochemical recurrence at different follow up times grouped by unit intervals of the CCP score is shown in Figure 2C. Finally, 3·3% (12/366) of our cohort died with disease progression. The CCP score was predictive of death after disease progression (p = 0·0007, HR = 2·99, 95% CI (1·69, 5·28)).
Table 2.
Summary of the statistical analysis of prostatectomy cohort
| Variable | Biochemical recurrence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysisc | Final model | ||||||||||
| + (%) |
N | Hazard Ratio (95% CI) |
χ2 (1 df) | p-value | Hazard Ratio (95% CI) |
χ2 (1 df) | p-value | N | Hazard Ratio (95% CI) |
χ2 (1 df) | p-value | |
| CCP Scorea | 366 | 1.89 (1.54, 2.31) | 33.96 | 5.6 × 10−9 | 1.77 (1.40, 2.22) | 21.13 | 4.3 × 10−6 | 1.74 (1.39, 2.17) | 21.65 | 3.3 × 10−6 | ||
| log(1+baseline PSA) | 395 | 3.07 (2.38, 3.97) | 71.12 | 3.4 × 10−17 | 1.98 (1.47, 2.65) | 20.65 | 5.5 × 10−6 | 2.24 (1.71, 2.93) | 33.47 | 7.2 × 10−9 | ||
| Gleason Scoreb | ||||||||||||
| <7 | 269 | 1 (ref) | 1 (ref) | 240 | 1 (ref) | |||||||
| 7 | 123 | 2.81 (2.01, 3.94) | 54.56 | 1.5 × 10−13 | 1.11 (0.71, 1.71) | 2.58 | 0.11 | 110 | 1.35 (0.92, 2.00) | 7.57 | 5.9 × 10−3 | |
| >7 | 18 | 6.32 (3.65, 10.93) | 2.10 (1.05, 4.18) | 16 | 2.69 (1.43, 5.08) | |||||||
| pathological stage | 410 | 1.66 (1.48, 1.88) | 67.70 | 2.2 × 10−16 | 1.15 (0.87, 1.51) | 1.09 | 0.30 | 1.32 (1.12, 1.56) | 10.30 | 1.3 × 10−3 | ||
| pathological grade | 410 | 2.42 (1.89, 3.11) | 48.45 | 3.4 × 10−12 | 1.24 (0.89, 1.73) | 1.65 | 0.20 | |||||
| surgical margins | 96 (23) | 410 | 3.44 (2.48, 4.76) | 49.53 | 2.0 × 10−12 | 1.88 (0.94, 3.75) | 3.55 | 0.06 | 1.89 (1.23, 2.91) | 8.61 | 3.3 × 10−3 | |
| extracapsular extension | 121 (29) | 410 | 3.34 (2.43, 4.61) | 51.53 | 7.0 × 10−13 | 1.29 (0.58, 2.84) | 0.38 | 0.54 | ||||
| bladder | 19 (5) | 410 | 3.02 (1.67, 5.46) | 10.03 | 1.5 × 10−3 | 2.11 (1.09, 4.08) | 4.17 | 0.041 | ||||
| seminal vesicle | 44 (11) | 410 | 4.53 (3.05, 6.71) | 42.42 | 7.3 × 10−11 | 1.43 (0.67, 3.07) | 0.83 | 0.36 | ||||
| lymph node | 13 (3) | 410 | 4.14 (2.10, 8.18) | 11.53 | 6.9 × 10−4 | 1.54 (0.70, 3.38) | 1.09 | 0.30 | ||||
| age at surgery | 410 | 1.02 (0.99, 1.05) | 2.47 | 0.12 | 1.01 (0.98, 1.05) | 0.76 | 0.38 | |||||
| Diagnostic: | ||||||||||||
| Gleason Score | 410 | 2.42 (1.84, 3.17) | 31.78 | 1.7 × 10−8 | ||||||||
| clinical stage | 410 | 1.48 (1.17, 1.87) | 11.51 | 6.9 × 10−4 | ||||||||
| clinical grade | 410 | 2.21 (1.76, 2.77) | 45.99 | 1.2 × 10−11 | ||||||||
Hazard ratio for a unit increase in CCP score.
χ2 values of 54.93, 4.15, and 8.41 when Gleason score considered as a 3-level categorical variable with 2 df.
353 patients in full multivariate analysis
Next, we evaluated the prognostic utility of CCP signature after accounting for clinical variables. We included in our multivariate analysis all available post-surgical clinical variables and any pre-surgical variable that did not have a post-surgical equivalent. The results of the full multivariate analysis are summarized in Table 2. CCP and PSA were the most significant predictors of biochemical recurrence, and provided much more prognostic information than any other variable. In fact, by comparing χ2 values, it is evident that CCP and PSA are the only variables that retain a substantial portion of their univariate information. The increase in hazard ratio for a one-unit increase in CCP score was 1·77 (95% CI (1·40, 2·22), p = 4·3 × 10−6) in the multivariate model.
To construct the best prognostic multivariate model we employed a forward stepwise regression. As in the full multivariate analysis, CCP and PSA were the most predictive variables and retained a substantial amount of their univariate information.
The best predictor based on these variables was:
Combined Risk Score = 0·55*CCP + 0·81*log(1+PSA) + 0·28*T-stage + 0·64*Margins { + 0·30*(Gleason = 7) + 0·99*(Gleason > 7)}
Finally, we graphed the risk of biochemical recurrence by 10-years as a function of the Combined Risk Score, and given the central prognostic role of Gleason, included a histogram showing the scores distribution in different Gleason categories (Figure 3).
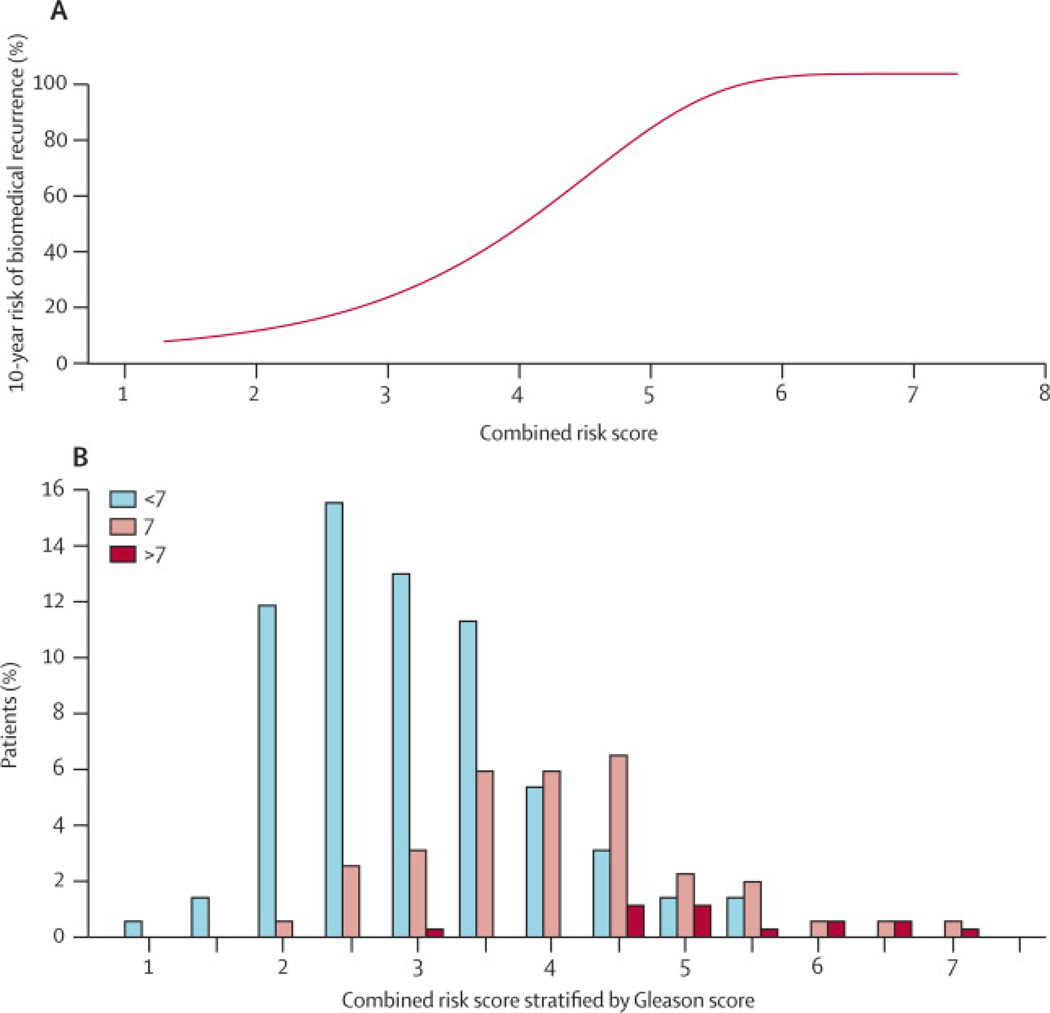
Figure 3.
Ten-year predicted risk of recurrence for different values of Combined Risk Score for the radical prostatectomy cohort, and below, a histogram showing the distribution of the Combined Risk Score for different subgroups based on Gleason score. The probability of recurrence was estimated from a Cox proportional hazard model using the Combined Risk Score. Gleason scores are shown in categories of <7, white bars; =7, grey bars; >7 black bars.
Conservatively Managed TURP Cohort
To determine if prognostic utility of CCP score was robust to patient composition and specific clinical setting, we next evaluated the ability of the CCP score to predict death from prostate cancer in a large cohort of conservatively treated patients who were incidentally diagnosed with prostate cancer after TURP. The details of the full cohort have been presented previously 13. The selection of patients for this study is summarized in Figure 4. After 10 years of follow-up, 61% of the men had died, 35% from prostate cancer and 26% from other causes.
Four hundred and twenty-nine patients were randomly selected for this study. Of these, 31 were later found to be ineligible (see cohort exclusion criteria 14), and a further 61 (15%) were excluded due to a poor CCP quality score leaving 337 in the primary multivariate analysis set. This analysis set and a complete analysis plan was agreed upon, and all CCP scores were assigned, before unblinding of the clinical and outcome data.
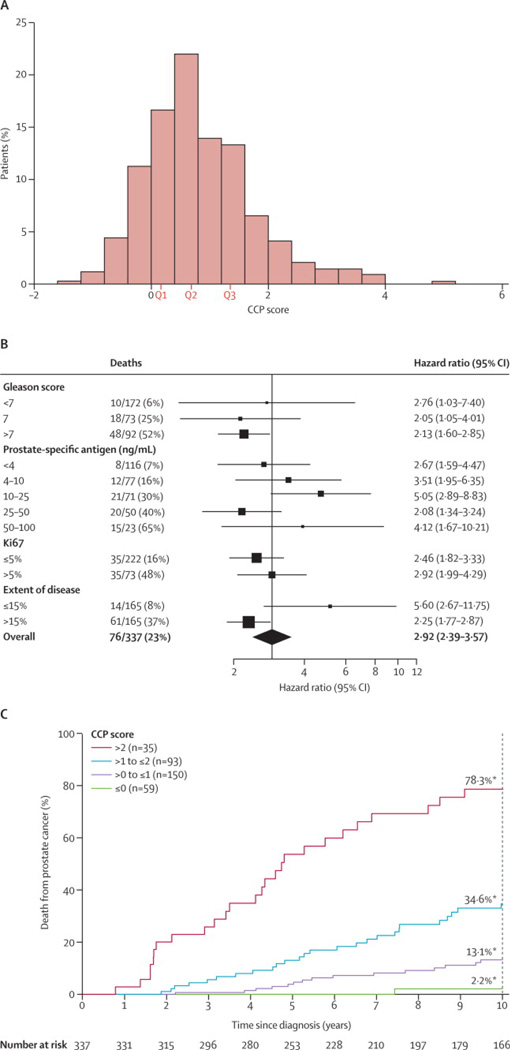
A histogram of the CCP score for these 337 patients is shown in Figure 5A. In a univariate analysis (Table 3), the increase in hazard ratio for a one unit change in CCP score was 2·92 (95% CI (2·38, 3·57)) leading to a χ2 of 92·7, p = 6.1 × 10−22. When compared to Gleason score, PSA, cancer extent and Ki67, the CCP score had the most prognostic information, being slightly greater than Gleason score (χ2= 92·7 vs. 80·0), and substantially larger than all other variables. As compared to the prostatectomy cohort, here the CCP score was somewhat more correlated with the other variables: Gleason, ρ = 0·61; log (1+ PSA), ρ = 0·27; Ki67, ρ = 0·50; extent of disease, ρ = 0·51. The prognostic value of the CCP score was similar in different Gleason categories as well as when stratified by PSA, extent of disease and Ki67 (Figure 5B). Addition of a quadratic term for CCP score was non-significant (p = 0·36). A Kaplan-Meier plot of the proportion dying of prostate cancer at different follow up times grouped by integer values of the CCP score is shown in Figure 5C.
Figure 5.
Analysis of the CCP score in the TURP cohort. A) Distribution of the CCP scores (N=337). The median value was 0·67 with an interquartile range of 0·16 to 1·35. B) Forest plot graphing the CCP score hazard ratio (HR) in different clinical subgroups. The prostate cancer death rate for each subgroup is also given (death /size), and the size of the each box is proportional to the number of deaths within that patient subgroup. The thin lines indicate the 95% CI for each HR. The diamond at the bottom is the 95% CI of the HR for the entire cohort. C) Kaplan-Meier plot of prostate cancer death versus time for the patients grouped by integer values of CCP score. Each bin corresponds to a 2-fold increase in CCP expression. The green line (59 patients) corresponds CCP score • 0, purple line (150 patients) corresponds to 0 < CCP • 1, pink line (93 patients) corresponds to 1 < CCP • 2, and red line (35 patients) corresponds to CCP score > 2. Also indicated are the 10-year prostate cancer death rates for each group. For these four groups, the 10-year prostate cancer death rates (%) are: 2.1, 13.2, 34.6 and 78.3, respectively.
Table 3.
Summary of the statistical analysis of TURP cohort
| Variable | Death (Prostate Cancer) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysisd | Final Model | |||||||||
| N | Hazard Ratio (95% CI) |
χ2 (1df) | p-value | Hazard Ratio (95% CI) |
χ2 (1df) | p-value | N | Hazard Ratio (95% CI) |
χ2 (1df) | p-value | |
| CCP Scorea | 337 | 2.92 (2.38, 3.57) | 92.7 | 6.1×10−22 | 2.56 (1.85, 3.53) | 32.3 | 1.3×10−8 | 337 | 2.57 (1.93, 3.43) | 42.2 | 8.2×10−11 |
| Gleason Score | |||||||||||
| <7 | 172 | 1(ref) | 1(ref) | 172 | 1(ref) | ||||||
| 7 | 73 | 5.20 (2.40, 11.29) | 80.0b | 3.7×10−19 | 2.45 (0.95, 6.30) | 4.2b | 0.04 | 73 | 2.45 (1.09, 5.48) | 5.4b | 0.02 |
| >7 | 92 | 13.67 (6.90, 27.11) | 2.91 (1.12, 7.55) | 92 | 2.72 (1.22, 6.08) | ||||||
| log(1+PSA) [ng/ml] | 337 | 2.30 (1.83, 2.88) | 57.5 | 3.4×10−14 | 1.87 (1.44, 2.44) | 24.9 | 6.0×10−7 | 337 | 1.84 (1.46, 3.32) | 29.3 | 6.2×10−8 |
| log(1+(10*Ki67)) [%] | 295 | 1.77 (1.43, 2.20) | 32.2 | 1.4×10−8 | 0.98 (0.80, 1.21) | 0.03 | 0.86 | - | - | - | |
The results of a full multivariate analysis are shown in Table 3. The prognostic value of CCP score was evaluated after adjusting for PSA, Gleason, Ki67 expression and extent of disease. In the resultant analysis, the CCP score was dominant (HR = 2.56, 95% CI (1.85, 3.53), χ2 = 32·3, p = 1·3 × 10−8), providing more information than any other variable. None of the variables exhibited a significant interaction with the CCP score and the hazard ratio for the CCP score was only slightly attenuated when other factors were added to the model (HR= 2·92, univariate; 2·56 with all variables). As in the post-surgical cohort, PSA was also highly predictive of death (HR= 1.87, 95% CI (1.44, 2.44), χ2 = 24·9, p = 6·0 × 10−7) and it also retained a significant amount of its univariate information. As seen in our previous paper13, hormone treatment was significant in a univariate model (HR = 3.81, 95% CI (2.40, 6.05), χ2= 27·7), but did not add significant predictive information in the multivariate model (HR = 1.12, 95% CI (0.68, 1.84), χ2 = 0·18), presumably because the decision to use this therapy was based on the known prognostic factors. Addition of a quadratic term for CCP was also non-significant in the multivariate model (χ2 = 2·33).
The final forward stepwise regression model consisted of the consisted of the following variables in the order they entered: CCP, PSA and Gleason score. The best predictor based on these variables was:
Combined Risk Score = 0·95 * CCP + 0·61* log (1 + PSA) { + 0·90*(Gleason = 7) + 1·00*(Gleason > 7) }.
The predicted 10-year death rate from prostate cancer as a function of Combined Score, along with the distribution of the score in different Gleason categories is shown in Figure 6.
Figure 6.
Ten-year predicted risk of prostate cancer death for different values of the Combined Risk Score for TURP cohort, and below, a histogram showing the distribution of the Combined Risk Score for different subgroups based on Gleason score. The probability of prostate cancer death was estimated from a Cox proportional hazard model using the Combined Risk Score. Gleason scores are shown in categories of <7, white bars; =7, grey bars; >7 black bars.
In terms of selecting patients for conservative management, it is informative to compare the Combined Score to clinical parameters only for identifying disease with a low fatality risk. For men with a tumour of Gleason score 6 or less, use of the Combined Score identified 15% of these patients with a 10-year risk of death from prostate cancer of less than 2% and 57% with a risk of less than 5%, but also identified 17% with a risk of greater that 10%, indicating not all Gleason 6 tumours are low risk. For Gleason score 7 (either 3+4 or 4+3) the corresponding percentages were 0·6%, 11% and 81% respectively. In the whole cohort, the percentages were 8%, 32% and 53% respectively.
We also looked at deaths from other causes. The only significant factor was age and neither the CCP score nor other major prognostic factors for prostate cancer death were significant, indicating that confounding by other deaths cannot explain these findings.
DISCUSSION
We developed a prognostic score for prostate cancer based on measuring mRNA expression levels of cell cycle progression (CCP) genes 31 and tested its ability to predict outcome in U.S. patients treated with radical prostatectomy, and in UK patients who were conservatively managed after being diagnosed with cancer by TURP. Both cohorts have their strengths and weaknesses. Both had long term follow up with a median approaching 10 years. The RP cohort was mainly screen detected and had lower baseline Gleason scores and PSA levels, but a very low death rate from prostate cancer. The TURP cohort had more aggressive disease so that prostate cancer mortality could be reliable modeled, but was all based on symptomatic patients and TURP samples, which is less common in contemporary practice. Unequivocally, the CCP score was predictive of outcome in both settings and provided substantially more prognostic information than clinical variables alone. CCP genes are expressed at higher levels in actively growing cells, and presumably by measuring the expression levels of CCP genes, we are indirectly measuring the growth rate and inherent aggressiveness of the tumour, which ultimately impacts outcome. . Since we are measuring fundamental tumour biology, we predict that the prognostic utility of CCP score to be highly robust. Three of the genes in our signature, TOP2A, RRM2 and BIRC5, have been previously associated with prostate cancer outcome 15, 16, but we feel use of a larger panel will ensure robustness of the score. It is also noted that these genes are putative targets of cytotoxic or radiation therapies: ribonucleotide reductase - pemetrexed and gemcitabine, topoisomerase 2A - anthracyclines, KIF11 and KIF20A - taxanes, TK1 - 5FU and RAD51 and RAD54L - radiation and alkylating agents as well as novel targeted agents: polokinase 1. Exploration of the value of the CCP score in clinical trials that have studied adjuvant radiation, androgen deprivation and other systemic therapies is warranted to determine whether this signature can help make therapeutic decisions in that setting as well.
Given the very different clinical settings in which we have tested the CCP score, it is remarkable to note that the resultant predictive models are quite similar. In both settings CCP and PSA were the dominant variables, while other clinical variables (including Gleason score) lost most of their univariate prognostic utility. These data suggest that CCP and PSA measure independent aspects of disease aggressiveness and are both required for an accurate outcome prediction. In contrast Ki67, which by itself can be predictive of outcome in this data set 33, was eliminated from models that include CCP score. This is not surprising because Ki67 is a CCP gene. It is dominated in the predictive models probably because measuring CCP through RNA levels is more quantitative and better approximates the underlying biological phenomenon.
The evidence presented here indicates that the CCP score adds significantly to the predictive power of the clinical parameters typically employed to predict disease outcome after surgery and at the time of disease diagnosis. Although potentially useful in both of these settings, the most pressing clinical need is to accurately separate indolent disease in men with newly diagnosed cancer, which is unlikely to be fatal, from the more aggressive cancers in need of radical treatment. Currently Gleason score and baseline PSA are the strongest predictors and extent of disease and Ki67 add only a small additional increment of discrimination33. Clinical stage, which was only available for a portion of the TURP cohort, was found to be only weakly predictive of outcome in previous analyses 14. Results from the TURP cohort have shown that the CCP score is a stronger univariate predictor of death from prostate cancer than any of these other variables, and multivariate analysis indicates that this score adds a substantial amount of prognostic information not captured by any other measure.. This is reflected by the hazard ratio of 1.74 in the RP cohort and 2.57 in the TURP cohort in the multivariate model for a one unit increase in the CCP score. When put on a standardized basis for the CCP score, this translates into hazard ratios of 1.68 and 3.08, respectively for a change from the 25th to 75th percentiles of the CCP score distributions in each cohort. Although it does not account for time-to-event information or censoring, the area under the ROC curve (AUC) for an event has become a common way to compare predictive scores. For an event in the first 5 yrs the AUC in the RP cohort for biochemical recurrence was 0.825 for the clinical score and 0.842 for the combined score (including the CCP score). For the TURP cohort the values were 0.806 and 0.878 for death from prostate cancer, respectively. In addition, the added value of the CCP score to clinical variables can be seen by comparing outcomes of patients in the lowest vs. highest quartile of the clinical score vs. the combined score. When comparing 10-year event proportions, in the highest quartile Kaplan-Meier estimates were 84.3% (clinical) vs. 83.6% (combined) for the RP cohort (biochemical recurrence), and 62.6% vs. 67.4%, respectively, for the TURP cohort (death from prostate cancer). For the lowest quartile they were 6.9% vs. 4.6% for the RP cohort and 2.7% vs. 1.3% for the TURP cohort. Thus, the combined score appears to be particularly helpful for the better prognosis patients in the lowest quartile, where the clinical need for better prediction is greatest. Therefore, addition of the CCP score to clinical variables allows one to predict more accurately which men can be safely managed by watchful waiting, and of equal importance, which men with apparently low-risk disease actually have a high risk of death from prostate cancer and might benefit from radical treatment. For example, 8% of the entire cohort and 15% of men with a Gleason score of 6 could be identified with a 10-year death rate from prostate cancer of less than 2%, and 17% of those with a Gleason score of 6 still had a 10-yr death rate from prostate cancer of more than 10%.
CCP genes have been shown to be prognostic in breast cancer 30. More recently, they have also proven prognostic in lung and brain cancers 34,35. These previous studies led us to evaluate whether CCP genes would be useful in prostate cancer. A major strength of the current study is that we that tested a single predefined score based on CCP genes, as opposed to evaluating a number of genes separately. As a result, the interpretation of this study is not complicated by the extensive multiple testing inherent in more comprehensive discovery strategies, and therefore, we can be highly confident about the main conclusions of this report. We do not claim, however, that the CCP score contains every gene that may be prognostic in prostate cancer, and additional studies may uncover biomarkers that significantly improve the overall performance of our predictive models.
We have shown that the CCP score is a robust predictor of prostate cancer outcome in two cohorts that were diverse in terms of their geographic origin and clinical setting. A total of 89% of the samples in the RP cohort and 85% in the TURP cohort generated valid CCP scores. This was based on old samples (>10 yrs in most cases) and experience with fresh FFPE material indicates a success rate of about 95%, indicating its widespread applicability. This is an important first-step in personalising treatment for prostate cancer. However, this score needs to be further validated in more contemporaneous cohorts diagnosed by needle biopsy. These studies are clearly warranted and are ongoing.
Research in Context
We performed Pubmed searches via NCBI search engine using key terms ‘prostate cancer’, ‘prognostic’, and ‘RNA expression’. These were reviewed for quality and relevance by JL and SS. The key publications are described and referenced in our introduction. We also downloaded and analyzed every large publicly available expression dataset that was associated with cancer outcome (GEO database). As a result, we concluded that CCP genes were the key component in every validated prognostic signature and this was supported in a review paper 30. However the previous studies were equivocal. For the most part, the authors did not show, or did not convincingly show, that their signatures added information beyond clinical parameters. Most studies also failed to demonstrate that their findings were robust to patient composition or clinical setting.
While the appropriate management of early prostate cancer is widely acknowledged to be a major clinical issue, none of the previously defined prognostic signatures have had a major impact on clinical care. Previous studies have indicated a need to better stratify these patients to guide the need for radical treatment. The current study addresses all these issues and indicates that the CCP score can be helpful in assessing prognosis in a range of settings, especially for relatively good prognosis patients such as those with Gleason score 6. The score is a robust measure of the proliferative activity in the tumour and may be useful in determining which prostates can be safely managed with a conservative strategy. Further validation studies are needed, especially for screened detected patients diagnosed by needle biopsy where conservative management is an option.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully recognize the patients who participated in this study and the support from Cancer Research UK, The Orchid Appeal, National Institutes of Health (SPORE CA92629), the Koch Foundation and Myriad Genetics. We also thank investigators and staff in the cancer registries and participating hospitals (Appendix) for their support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All authors approved the final manuscript and contributed to the study: overall concept, data analysis and finalising paper JC; experimental design, writing of manuscript and data analysis JC, SS, GS, study plan, design and data interpretation, JL, AG, GF, statistical analysis, JC, DM, JR, generation of CCP data and development of CCP score, SW, JP, AY, DF; sample preparation and assay, JW. For the TURP cohort study: experimental design, writing of manuscript and data analysis JC, GF, pathology review and interpretation, DB, CF; cancer registry clinical data coordination, HM, specimen preparation, ES; statistical analysis, JC, DM, study plan and design, PS, HM. For the RP cohort study: experimental design, writing of manuscript and data analysis, GS, AB; pathology review and interpretation VS, statistical analysis, JR, AG, DF.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Evans HS, Moller H. Recent trends in prostate cancer incidence and mortality in southeast England. Eur Urol. 2003;43(4):337–341. doi: 10.1016/s0302-2838(03)00085-x. [DOI] [PubMed] [Google Scholar]
- 3.Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360(13):1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 4.Breslow N, Chan CW, Dhom G, Drury RA, Franks LM, Gellei B, et al. Latent carcinoma of prostate at autopsy in seven areas. The International Agency for Research on Cancer, Lyons, France. Int J Cancer. 1977;20(5):680–688. doi: 10.1002/ijc.2910200506. [DOI] [PubMed] [Google Scholar]
- 5.Sakr WA, Grignon DJ, Crissman JD, Heilbrun LK, Cassin BJ, Pontes JJ, et al. High grade prostatic intraepithelial neoplasia (HGPIN) and prostatic adenocarcinoma between the ages of 20–69: an autopsy study of 249 cases. In Vivo. 1994;8(3):439–443. [PubMed] [Google Scholar]
- 6.Kattan MW, Eastham J. Algorithms for prostate-specific antigen recurrence after treatment of localized prostate cancer. Clin Prostate Cancer. 2003;1(4):221–226. doi: 10.3816/cgc.2003.n.003. [DOI] [PubMed] [Google Scholar]
- 7.Stephenson AJ, Scardino PT, Eastham JA, Bianco FJ, Jr, Dotan ZA, DiBlasio CJ, et al. Postoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Clin Oncol. 2005;23(28):7005–7012. doi: 10.1200/JCO.2005.01.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kattan MW, Vickers AJ, Yu C, Bianco FJ, Cronin AM, Eastham JA, et al. Preoperative and postoperative nomograms incorporating surgeon experience for clinically localized prostate cancer. Cancer. 2009;115(5):1005–1010. doi: 10.1002/cncr.24083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shariat SF, Karakiewicz PI, Roehrborn CG, Kattan MW. An updated catalog of prostate cancer predictive tools. Cancer. 2008;113(11):3075–3099. doi: 10.1002/cncr.23908. [DOI] [PubMed] [Google Scholar]
- 10.Han M, Partin AW, Zahurak M, Piantadosi S, Epstein JI, Walsh PC. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J Urol. 2003;169(2):517–523. doi: 10.1097/01.ju.0000045749.90353.c7. [DOI] [PubMed] [Google Scholar]
- 11.Capitanio U, Briganti A, Gallina A, Suardi N, Karakiewicz PI, Montorsi F, et al. Predictive models before and after radical prostatectomy. Prostate. 2010;70(12):1371–1378. doi: 10.1002/pros.21159. [DOI] [PubMed] [Google Scholar]
- 12.Tewari A, Johnson CC, Divine G, Crawford ED, Gamito EJ, Demers R, et al. Long-term survival probability in men with clinically localized prostate cancer: a case-control, propensity modeling study stratified by race, age, treatment and comorbidities. J Urol. 2004;171(4):1513–1519. doi: 10.1097/01.ju.0000117975.40782.95. [DOI] [PubMed] [Google Scholar]
- 13.Kattan MW, Cuzick J, Fisher G, Berney DM, Oliver T, Foster CS, et al. Nomogram incorporating PSA level to predict cancer-specific survival for men with clinically localized prostate cancer managed without curative intent. Cancer. 2008;112(1):69–74. doi: 10.1002/cncr.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuzick J, Fisher G, Kattan MW, Berney D, Oliver T, Foster CS, et al. Long-term outcome among men with conservatively treated localised prostate cancer. Br J Cancer. 2006;95(9):1186–1194. doi: 10.1038/sj.bjc.6603411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakagawa T, Kollmeyer TM, Morlan BW, Anderson SK, Bergstralh EJ, Davis BJ, et al. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS ONE. 2008;3(5):e2318. doi: 10.1371/journal.pone.0002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosari F, Munz JM, Savci-Heijink CD, Spiro C, Klee EW, Kube DM, et al. Identification of prognostic biomarkers for prostate cancer. Clin Cancer Res. 2008;14(6):1734–1743. doi: 10.1158/1078-0432.CCR-07-1494. [DOI] [PubMed] [Google Scholar]
- 17.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113(6):913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115(6):1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1(2):203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 20.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101(3):811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22(14):2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 22.Cheang MC, van de Rijn M, Nielsen TO. Gene expression profiling of breast cancer. Annu Rev Pathol. 2008;3:67–97. doi: 10.1146/annurev.pathmechdis.3.121806.151505. [DOI] [PubMed] [Google Scholar]
- 23.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 25.Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102(10):3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sotiriou C, Desmedt C. Gene expression profiling in breast cancer. Ann Oncol. 2006;17 Suppl 10:x259–x262. doi: 10.1093/annonc/mdl270. [DOI] [PubMed] [Google Scholar]
- 27.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 28.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 29.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel AB, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355(6):560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 30.Mosley JD, Keri RA. Cell cycle correlated genes dictate the prognostic power of breast cancer gene lists. BMC Med Genomics. 2008;1:11. doi: 10.1186/1755-8794-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13(6):1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanson GP, Riggs M, Hermans M. Pathologic findings at radical prostatectomy: risk factors for failure and death. Urol Oncol. 2007;25(2):110–114. doi: 10.1016/j.urolonc.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Berney DM, Gopalan A, Kudahetti S, Fisher G, Ambroisine L, Foster CS, et al. Ki-67 and outcome in clinically localised prostate cancer: analysis of conservatively treated prostate cancer patients from the Trans-Atlantic Prostate Group study. Br J Cancer. 2009;100(6):888–893. doi: 10.1038/sj.bjc.6604951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14(8):822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Liu B, Jiang X, Zhao H, Fan M, Fan Z, et al. A systems biology-based gene expression classifier of glioblastoma predicts survival with solid tumors. PLoS ONE. 2009;4(7):e6274. doi: 10.1371/journal.pone.0006274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.