Abstract
The plant hormone abscisic acid (ABA) plays a crucial role in the control of the stress response and the regulation of plant growth and development. ABA binding to PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS intracellular receptors leads to inhibition of key negative regulators of ABA signaling, i.e. clade A protein phosphatases type 2C (PP2Cs) such as ABA-INSENSITIVE1 and HYPERSENSITIVE TO ABA1 (HAB1), causing the activation of the ABA signaling pathway. To gain further understanding on the mechanism of hormone perception, PP2C inhibition, and its implications for ABA signaling, we have performed a structural and functional analysis of the PYR1-ABA-HAB1 complex. Based on structural data, we generated a gain-of-function mutation in a critical residue of the phosphatase, hab1W385A, which abolished ABA-dependent receptor-mediated PP2C inhibition without impairing basal PP2C activity. As a result, hab1W385A caused constitutive inactivation of the protein kinase OST1 even in the presence of ABA and PYR/PYL proteins, in contrast to the receptor-sensitive HAB1, and therefore hab1W385A qualifies as a hypermorphic mutation. Expression of hab1W385A in Arabidopsis (Arabidopsis thaliana) plants leads to a strong, dominant ABA insensitivity, which demonstrates that this conserved tryptophan residue can be targeted for the generation of dominant clade A PP2C alleles. Moreover, our data highlight the critical role of molecular interactions mediated by tryptophan-385 equivalent residues for clade A PP2C function in vivo and the mechanism of ABA perception and signaling.
Abscisic acid (ABA) is required for biotic and abiotic stress responses as well as the control of plant growth and development. Plant growth can be severely impaired by adverse environmental conditions like drought, salinity, cold, or high temperature, which can reduce average productivity of crops by 50% to 80% (Bray et al., 2000). ABA plays a key role in orchestrating the adaptive response of the plant to cope with these forms of abiotic stress (Verslues et al., 2006; Cutler et al., 2010). Under drought stress, cleavage of ABA from ABA conjugates stored in the vacuole or apoplastic space (Lee et al., 2006) as well as de novo ABA biosynthesis (Nambara and Marion-Poll, 2005) are stimulated, leading to a sharp increase in the cellular concentration of the hormone. This elicits a plant response that limits water loss and, under prolonged stress, the hormone response adapts plant metabolism to the low water potential of the cellular environment.
A large number of cellular components have been implicated in the ABA signaling pathway (Hirayama and Shinozaki, 2007). However, recently it has become clear that just three types of proteins constitute the so-called core ABA pathway (Cutler et al., 2010). These include the family of PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR) ABA receptors, the clade A of protein phosphatases type 2Cs (PP2Cs), and three ABA-activated protein kinases from the Suc nonfermenting1-related subfamily2 (SnRK2; Cutler et al., 2010). Under nonstress conditions clade A PP2Cs can interact with and dephosphorylate three SnRK2s, i.e. 2.2, 2.3, and 2.6/OST1, reducing their catalytic activity (Umezawa et al., 2009; Vlad et al., 2009). The increase of ABA levels in the plant cell leads to the PYR/PYL/RCAR receptor-mediated inhibition of the PP2C activity that results in the activation of the three SnRK2s and ultimately of the ABA signaling pathway (Ma et al., 2009; Park et al., 2009; Umezawa et al., 2009; Vlad et al., 2009). Upon activation, the SnRK2s directly phosphorylate transcription factors that bind to ABA-responsive promoter elements, named ABFs/AREBs for ABA-responsive promoter element-binding factors, and components of the machinery regulating stomatal aperture like the anion channel SLAC1 (Fujii et al., 2009; Fujita et al., 2009; Geiger et al., 2009; Lee et al., 2009).
To date, three receptors, i.e. PYR1, PYL1, and PYL2, and two receptor-ABA-phosphatase complexes, i.e. PYL1-ABA-INSENSITIVE1 (ABI1) and PYL2-HYPERSENSITIVE TO ABA1 (HAB1), have been studied at a structural level, which has contributed to the understanding of the molecular interactions between receptor, hormone, and phosphatase (Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Santiago et al., 2009a; Yin et al., 2009). The PYR/PYL/RCAR proteins belong to the superfamily of START/Bet v proteins, whose members are widespread in eukaryotes and are characterized by the presence of a cavity able to accommodate hydrophobic ligands (Iyer et al., 2001; Radauer et al., 2008). This cavity represents the hormone-binding pocket and is flanked by two flexible loops (b3-b4 and b5-b6), the so-called gating loops, which close over the hormone once inside the binding pocket. In the two structures available from ternary complexes, the ABA-bound receptor contacts the PP2C through the gating loops that cover the ABA-binding pocket. Thus, the side chains of Ser-112 of PYL1 and the Ser-89 of PYL2, located in the b3-b4 loop, insert into the PP2C active site and presumably occlude the access of the substrates (Melcher et al., 2009; Miyazono et al., 2009; Yin et al., 2009). These conserved Ser residues establish contacts with Gly-180 of ABI1 or Gly-246 of HAB1, next to the PP2C active site, and the metal-coordinating residue Glu-142 of ABI1 or Glu-203 of HAB1, respectively. Another important feature of the ternary complex, involves a key water-mediated interaction between the ABA’s ketone group and the Trp-300 or Trp-385 residue of ABI1 or HAB1, respectively. Indeed, this is the only residue of the PP2C approaching the ABA molecule and accordingly, this interaction has been postulated to play a key role in the stabilization of the whole ternary complex, contributing to the higher ABA affinity measured for PYR/PYL/RCAR receptors in the presence of the PP2Cs (Ma et al., 2009; Santiago et al., 2009b). However, beyond the structural data, no in planta evidence has been provided for its direct role in ABA signaling. Moreover, the ternary complexes analyzed at a structural level have not included PYR1, which plays a predominant role in germination (Park et al., 2009).
Plants harboring abi1G180D, abi2G180D, and hab1G246D dominant mutations have represented valuable tools to dissect ABA signaling (Leung et al., 1994, 1997; Meyer et al., 1994; Rodriguez et al., 1998; Robert et al., 2006). Their ABA-insensitive phenotypes are in agreement with a reduced capacity of the mutant PP2Cs to interact with PYR/PYL/RCAR receptors (Park et al., 2009; Santiago et al., 2009b; Umezawa et al., 2009). In spite of their utility, these alleles bear mutations close to the phosphatase catalytic site and have reduced basal PP2C activity (Bertauche et al., 1996; Leung et al., 1997; Leube et al., 1998; Rodriguez et al., 1998; Robert et al., 2006), which has complicated the interpretation of their in vivo phenotypes. Mutations in the conserved Trp residue described above have not been isolated by forward genetic screens, or engineered in Arabidopsis (Arabidopsis thaliana) plants, and the functional relevance of this residue has been documented uniquely on in vitro studies for the case of ABI1 (Miyazono et al., 2009). Since mutations in the Trp residue are expected to affect the stability of the ternary complex without compromising the phosphatase catalytic activity, they represent an ideal tool for studying in planta the effect of decoupling the receptor and phosphatase interaction.
Here we present a combined structural and functional analysis of the ternary complex formed by PYR1-ABA-HAB1. We analyzed the effect of PYR1-HAB1 mutations on OST1 kinase activity in vitro, since this SnRK2 is a key target of HAB1 (Vlad et al., 2009). We also performed in planta analysis of a hab1W385A mutation that decouples receptor and phosphatase interaction without impairing PP2C activity. These transgenic plants show an acute ABA insensitivity, demonstrating the importance of ABA-mediated PYR/PYL/RCAR-PP2C contacts for receptor function in vivo, and enabling a new method for probing PP2C function with dominant receptor-insensitive mutations.
RESULTS
Architecture of the PYR1-ABA-ΔNHAB1 Ternary Complex
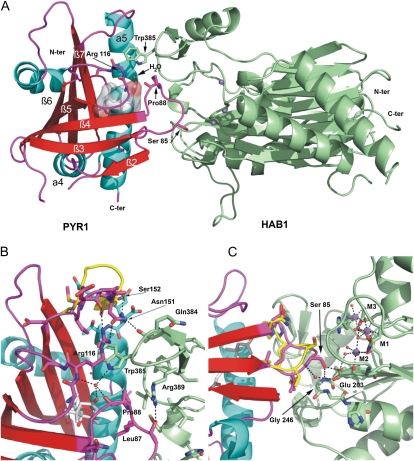
The PYR1 receptor and the catalytic domain of the HAB1 phosphatase (residues 179–511, ΔΝHAB1) were separately overexpressed in Escherichia coli, purified, and mixed in equimolar amounts in the presence of 1 mm (+)-ABA. The resulting complex was assayed for crystallization at the high-throughput crystallization facility of the EMBL Grenoble Outstation (https://embl.fr/htxlab; Dimasi et al., 2007). X-ray diffraction data were collected from orthorhombic crystals at the ID14-4 beam line of the European Synchrotron Radiation Facility (ESRF) to 1.8 Å resolution. Initial phases were obtained by the molecular replacement method using the two central β-sheets of the catalytic domain of the human PP2Cα protein (1A6Q; Das et al., 1996) as a search model. The initial phases provided an easily interpretable electron density map extending outside the search model region. Successive rounds of automatic refinement and manual building resulted in a refined model with a Rwork and Rfree of 17.4% and 21.8%, respectively. In the refined model, the crystallographic asymmetric unit contains one molecule of PYR1, one molecule of ΔNHAB1, one molecule of ABA, and three manganese ions (Fig. 1; Table I).
Figure 1.
Structure of the PYR1-ABA-HAB1 complex. A, The PYR1 receptor is shown with strands in red, loops in magenta, and helices in cyan. The HAB1 catalytic domain is shown in green. The (+)-ABA molecule is shown as stick model with semitransparent surface. The three metal ions at the phosphatase catalytic site are depicted (blue spheres). The gating loops containing Pro-88, Ser-85, and Arg-116 are indicated. The flap subdomain containing Trp-385 can be easily appreciated. The water molecule (red sphere) at the narrow channel between the gating loops is hydrogen bonded to the ketone group of the hormone, the backbone atoms of PYR1 Pro-88 and Arg-116, and the side chain of HAB1 Trp-385. B, Detail of the interaction between HAB1 Trp-385 region and the PYR1 gating loops. C, Detail of the interaction between the β3-β4 loop containing Pro-88 and Ser-85 and the phosphatase catalytic site. Relevant amino acids are shown as sticks, and hydrogen bonds are indicated by dotted lines. The conformation rearrangements in the β7/β5 and β3/β4 loops of PYR1 upon binding to the phosphatase (magenta) as compared to the ABA-bound subunit of the PYR1 dimer (yellow) can be appreciated.
Table I. Crystallographic data collection and refinement statistics.
| Data Collection and Refinement | |
| Data collection | |
| Space group | P212121 |
| Unit cell a, b, c, | 45.849, 65.857, 170.867 |
| α, β, γ | 90, 90, 90 |
| Resolution | 30.0–1.80 |
| Highest resolution shell | (1.9–1.8) |
| Rsym | 6.2% (19.6%) |
| Completeness | 97% (91%) |
| I/σ (I) | 22.6 (5.2) |
| Refinement | |
| Resolution range (A) | 28.24–1.8 |
| No. reflections | 340.949 |
| No. unique reflections | 47.524 |
| Rwork (%) | 17.386 |
| Rfree (%) | 21.760 |
| No. atoms | 4,170 |
| Protein | 3,720 |
| Ligand | 21 |
| Solvent | 475 |
| R.m.s. deviations | |
| Bond length | 0.02 |
| Angles | 1.655 |
The structure of PYR1 in the complex is very similar to that of the ABA-bound subunit in dimeric PYR1 (Nishimura et al., 2009; Santiago et al., 2009a). The ABA molecule is located in the receptor cavity stabilized by both polar and hydrophobic interactions and the gating loops are in the closed conformation, as described previously (Nishimura et al., 2009; Santiago et al., 2009a; Fig. 1). Subtle differences between the two PYR1 structures likely induced by interaction with HAB1 are found around Ser-85 in one of the gating loops, and the loop β7/α5, adjacent to the gating loops (Fig. 1, B and C). The structure of the HAB1 catalytic domain is similar to those of Arabidopsis ABI1 (Melcher et al., 2009; Miyazono et al., 2009; Yin et al., 2009) and the human PP2Cα protein phosphatase (Das et al., 1996). It is formed by a central 10-strand antiparallel β-sandwich flanked by two long α-helices at each side. A 55-amino acid α/β domain, which has been named the flap subdomain in some bacterial PP2Cs (Schlicker et al., 2008) is inserted between strands β8 and β12 of HAB1. This subdomain contains the HAB1 Trp-385 (Fig. 1A), which is highly conserved in plant clade A PP2Cs. Small conformational differences between the three phosphatases are found at the β2-β3 and α1-α2 loop regions of HAB1. In addition to this, HAB1 displays a 16-amino acid insertion at the α3/β4 loop not found in ABI1 and the human PP2Cα (Supplemental Fig. S1).
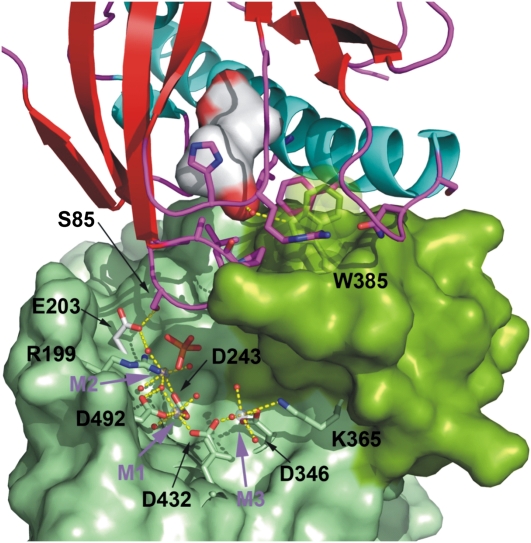
The catalytic site of HAB1 is located inside a deep channel formed at the top of the β-sandwich and flanked by the flap subdomain (Fig. 2; Supplemental Fig. S2). In our structure, the catalytic site of HAB1 contains three metal ions designated here as M1, M2, and M3 according to Alzari and coworkers (Wehenkel et al., 2007; Fig. 2). While some protein phosphatases contain two metal ions at the catalytic site, a few bacterial phosphatases have been shown to display a third conserved metal ion site, M3 (Pullen et al., 2004; Wehenkel et al., 2007; Schlicker et al., 2008). The M3 site, is located at the exit of the catalytic channel and is typically coordinated by one conserved aspartic residue also involved in coordination of the metal at M1 (Asp-432 for HAB1), and one residue from the flap domain. In some bacterial PP2Cs coordination of the third metal ion at M3 has been correlated with a change in position of the flap subdomain (Wehenkel et al., 2007), however, this site displays low metal-binding affinity and has been shown to be dispensable for catalysis (Wehenkel et al., 2007). To our knowledge, HAB1 is the first eukaryotic PP2C with three metal sites.
Figure 2.
The PYR1 β3/β4 loop docks at the catalytic site of HAB1. The ABA-bound PYR1 receptor is shown as in Figure 1. The accessible surface of the HAB1 phosphatase is depicted in light green with the flap subdomain containing Trp-385 in dark green. Residues coordinating the three metal ions at the catalytic site were excluded in the calculation of the molecular surface and are depicted as stick models. The water molecules involved in metal coordination are depicted as red spheres. The human PP2Cα structure (not shown), which contains a phosphate ion (shown as stick model) in the active site, was superposed on HAB1 to transfer the position of the phosphate ion into the catalytic site of HAB1.
Molecular Interactions Stabilizing the PYR1-ABA-HAB1 Complex
The PYR1-HAB1 interface comprises a total protein buried surface area of 1,691 Å2. As in the case of the PYL2-HAB1 and PYL1-ABI1 structures (Melcher et al., 2009; Miyazono et al., 2009; Yin et al., 2009), HAB1 docks into the ABA-bound receptor establishing interactions with the gating loops (loops β3/β4 and β5/β6), the N-terminal part of the α5 helix, and the α4/β2 loop of PYR1 (Fig. 1, A–C). The HAB1 residues involved in those interactions are located both in the flap subdomain including Trp-385 and the phosphatase active site including the β1/β2, β3/α1, and α2/β4 loops (Figs. 1, A–C and 2; Supplemental Fig. S3). The HAB1 Trp-385 residue is inserted between the PYR1 gating loops with the nitrogen in the indole group establishing a hydrogen bond with the water located at the channel between the gating loops (Fig. 1B). This water molecule represents a critical point in the ternary complex, establishing hydrogen bonds not only with HAB1 Trp-385 but also with the receptor gating loops (with the backbone carbonyl and amine groups of Pro-88 and Arg-116, respectively) and with the hormone itself, through its ketone group. Comparison of the present structure with the previously reported structures of isolated PYR1 reveals a conformational rearrangement in the β7/α5 loop of PYR1 upon binding to HAB1. This loop moves forward toward the flap domain of HAB1 (Fig. 1B), establishing new interactions that stabilize both the closed conformation of the gating loops and the receptor-phosphatase complex. These interactions involve Asn-151 of PYR1, which is hydrogen bonded to both the carbonyl group of HAB1 Gln-384 in the flap domain and PYR1 Arg-116, located in one of the gating loops. At the same time, in the present structure the side chain of PYR1 Ser-152 is involved in a helix-capping interaction (Presta and Rose, 1988) that stabilizes the forward movement of the β7/α5 loop.
Another important interaction region involves the PYR1 β3/β4 loop containing Ser-85 and the catalytic site of the phosphatase (Fig. 1C). PYR1 Ser-85 takes part in a hydrogen bond network with the backbone amine of Gly-246 and the carboxylic group of Glu-203 at the catalytic site of HAB1. This interaction is likely to be responsible for the inhibition of the phosphatase activity, as the β3/β4 loop containing Ser-85 seems to block access to the phosphatase catalytic site (Fig. 2). The structure of the human PP2Cα contains a phosphate ion at the catalytic site, which is likely mimicking the position of the phosphorylated amino acid substrate (Das et al., 1996). Interestingly, when PP2Cα and HAB1 catalytic cores are superimposed the phosphate ion of human PP2Cα is 2.9 Å away from the Cβ carbon of Ser 85 of PYR1 (Fig. 2; Supplemental Fig. S4), which suggests that a phospho-Ser substrate might enter the catalytic site in a similar manner.
Mutational Analysis of the PYR1-HAB1 Interaction and Effect on the HAB1-Dependent Inhibition of OST1 Activity
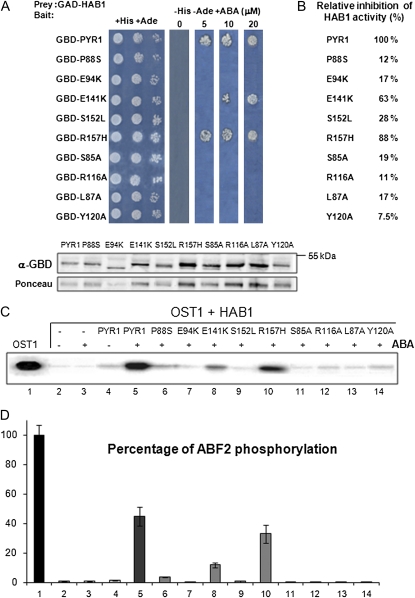
To test the biological relevance of the interactions observed in the PYR1-HAB1 complex, we analyzed the effect of a number of single-point mutations on both proteins. In the case of PYR1, we mutated key amino acid residues involved in either direct ABA binding (Glu-94Lys, Glu-141Lys, and Tyr-120Ala) or both ABA binding and PP2C interaction, particularly residues located in the gating loops (Ser-85Ala, Leu-87Ala, Pro-88Ser, Arg-116Ala) and the loop β7-α5 (Ser-152Leu). For HAB1 we chose the Gly-246Asp mutation, equivalent to abi1-1D and abi2-1D mutations, since expression of hab1G246D in planta leads to a dominant ABA-insensitive phenotype (Robert et al., 2006) and Trp-385Ala, due to its critical interactions with the PYR1 gating loops and ABA. For each PYR1 mutant we first tested both its capacity to interact with HAB1 and inhibit its activity through yeast (Saccharomyces cerevisiae) two-hybrid (Y2H) interaction and in vitro phosphatase activity assays, respectively (Fig. 3, A and B; Supplemental Fig. S5). In general, the PYR1 mutations abolished or severely reduced the ABA-mediated interaction and the inhibition of HAB1 phosphatase activity as compared to the wild type. An exception is the PYR1R157H variant. Although this mutation confers resistance to pyrabactin, a seed ABA agonist (Park et al., 2009), it shows very limited effect in both the Y2H and phosphatase activity assays.
Figure 3.
Analysis of the PYR1 mutations and their effect on the HAB1-dependent inhibition of OST1 activity. A, Interaction between HAB1 and PYR1 variants was analyzed by the Y2H growth assay on medium lacking His and Ade in the presence of 5, 10, or 20 μm (+)-ABA. Immunoblot analysis using antibody against the Gal4 binding domain (GBD) verifies the expression of the different fusion proteins in the Y2H assay. Ponceau staining from a representative yeast protein is shown as loading control. GAD, Gal4 activation domain. B, Relative inhibition of HAB1 activity by the different PYR1 variants in the presence of 8 μm ABA with respect to wild-type PYR1 (100%; sd was below 7%). C, OST1 in vitro kinase activity assay in the presence of HAB1, PYR1 wild type, and mutated versions, ΔCABF2 and 10 μm ABA, when indicated. The autoradiography shows the levels of autophosphorylation of OST1. D, Quantification of ΔCABF2 phosphorylation levels in the previous assay using the phosphoimager Image Gauge V.4.0. se measurements are shown (n = 3). [See online article for color version of this figure.]
In vitro reconstitution of an ABA signaling cascade can be achieved by combining PYR1, PP2C, SnRK2.6/OST1, and ABF2 in a test tube (Fujii et al., 2009). In this system, OST1 activity is measured as autophosphorylation as well as transphosphorylation of its natural substrate ABF2. We used this assay to determine how the different mutations affect the control of the OST1 activity. Figure 3 shows that HAB1 dephosphorylates OST1 and inhibits its kinase activity (lanes 1 and 2, Fig. 3, C and D). However, if ABA and PYR1 are added, HAB1 is inactivated, and consequently a significant recovery of OST1 activity is observed (lane 5, Fig. 3, C and D). All the PYR1 mutants assayed, except R157H, showed a strongly decreased capacity to antagonize the HAB1-mediated dephosphorylation of OST1 and were unable to promote ABA-dependent recovery of the OST1 protein kinase activity.
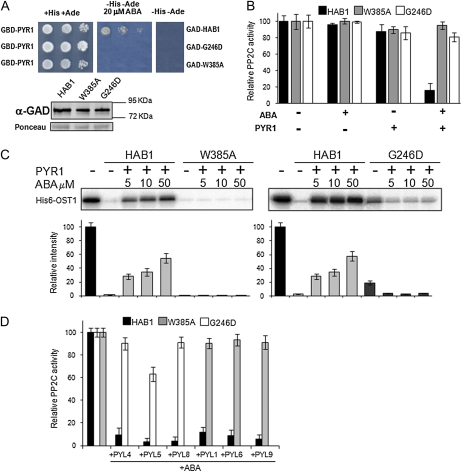
Both HAB1 Trp-385Ala and Gly-246Asp mutations abolished the ABA-dependent interaction between HAB1 and PYR1, as revealed by the Y2H and in vitro phosphatase activity assays (Fig. 4, A and B). In agreement with these results and in contrast to wild-type HAB1, both mutant PP2Cs were able to dephosphorylate OST1 in the presence of ABA and PYR1 (Fig. 4C). Thus, both mutant PP2Cs were refractory to inhibition by PYR1 under these experimental conditions. This result indicates that both hab1W385A and hab1G246D qualify as hypermorphic mutants compared to wild-type HAB1 in the presence of ABA and PYR1 (Wilkie, 1994). However, the basal dephosphorylation of OST1 by hab1G246D was less effective than wild type in the absence of ABA and PYR1 (Vlad et al., 2009; this work), which can be explained because this mutation is located close to the PP2C active site. Indeed, using p-nitrophenol as substrate, hab1G246D showed 4-times-lower specific activity as compared to wild-type HAB1 (4.86 ± 0.43 and 18.76 ± 2.13 nmol inorganic phosphate/min mg, respectively). Instead, the activity of hab1W385A was similar to wild type both in the p-nitrophenyl phosphate (pNPP; 20.52 ± 2.53 nmol inorganic phosphate/min mg) and the OST1 dephosphorylation assays (Fig. 4C).
Figure 4.
The hab1W385A and hab1G246D PP2Cs are refractory to inhibition by PYR1 and dephosphorylate OST1 in the presence of ABA and PYR1. A, The HAB1 mutations Trp-385Ala and Gly-246Asp abolish the interaction of the PP2C and PYR1 in a Y2H assay. Immunoblot analysis using antibody against the Gal4 activation domain (GAD) is shown to verify the expression of the different fusion proteins. Ponceau staining from a representative yeast protein is shown as loading control. GBD, Gal4 binding domain. B, Phosphatase activity of HAB1, hab1W385A, and hab1G246D proteins was measured in vitro using pNPP as substrate in the absence or presence of PYR1 and ABA, as indicated. Assays were performed in a 100 μL reaction volume containing 2 μm phosphatase and, when indicated, 4 μm HIS6-PYR1 and 1 μm (+)-ABA. Data are averages ± sd from three independent experiments. C, In vitro OST1 kinase activity in the presence of wild-type and mutated versions of HAB1, PYR1, and ABA, as indicated. The autoradiography shows the level of autophosphorylation of OST1 in each reaction. The graphs show the quantitative analysis of the autoradiogram. D, hab1W385A and hab1G246D proteins are resistant to ABA-mediated inhibition by different PYR/PYLs. The assay was performed as described in B. [See online article for color version of this figure.]
In summary, the mutational analysis of both PYR1 and HAB1 confirms that the interactions revealed by the structural analysis of the ternary complex are crucial for the inhibition of HAB1 activity. Additionally, these results illustrate that certain mutations in the PP2C lead to escape of the inhibitory ABA-mediated PYR/PYL mechanism. The results obtained for hab1G246D provide additional support to the model proposed by Merlot and coworkers (Vlad et al., 2009) to explain the negative regulation of OST1 activity by HAB1 and the strong ABA-insensitive phenotype of 35S:hab1G246D plants (Robert et al., 2006), assuming that a general escape from PYR/PYL receptors occurs in these plants. Indeed, we have demonstrated in vitro that hab1G246D phosphatase, as well as hab1W385A, are refractory to inhibition by different PYR/PYL proteins (Fig. 4D).
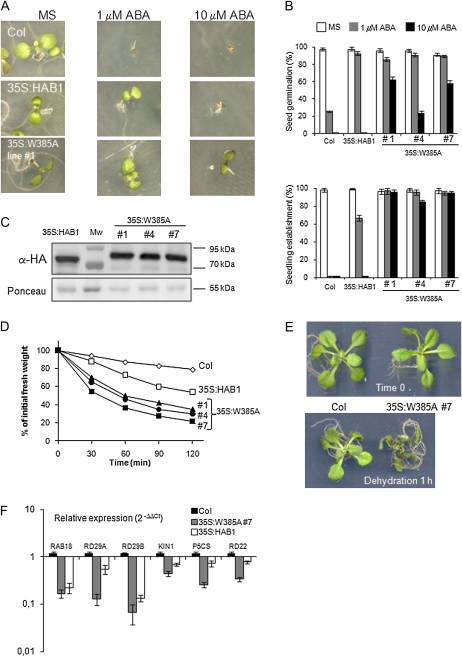
Expression of hab1W385A in Arabidopsis Plants Leads to Reduced ABA Sensitivity
To test the biological relevance of the PYR1-ABA-HAB1 interaction mediated by the residue Trp-385 of HAB1, we generated 35S:hab1W385A transgenic lines and examined their ABA response compared to 35S:HAB1 plants (Fig. 5). For this analysis, we selected three 35S:hab1W385A transgenic lines that showed expression levels of the recombinant protein similar to those of the previously described 35S:HAB1 plants (Saez et al., 2004), as determined by immunoblot analysis against the hemagglutinin (HA) epitope added to each protein (Fig. 5C). Germination and early seedling establishment of 35S:HAB1 and 35S: hab1W385A seeds were less sensitive to ABA-mediated inhibition than wild-type seeds (Fig. 5, A and B). Moreover, 35S:hab1W385A seeds were able to germinate and establish seedlings at 10 μm ABA, which is an inhibitory concentration for establishment of 35S:HAB1 seeds (Fig. 5, A and B).
Figure 5.
Constitutive expression of hab1W385A leads to reduced ABA sensitivity. A, Seed germination and seedling establishment of representative Columbia (Col) wild-type, 35S:HAB1, and 35S:hab1W385A plants in medium lacking or supplemented with ABA. Photographs were taken 7 d after sowing. B, Inhibition of seed germination and seedling establishment by ABA in Columbia wild-type, 35S:HAB1, and 35S:hab1W385A plants. C, Immunoblot analysis using antibody against HA tag to quantify phosphatase expression in transgenic lines. Ponceau staining from the large subunit of Rubisco is shown as loading control. D, Enhanced water loss measured in detached leaves of 35S:HAB1 and 35S:hab1W385A plants as compared to Columbia wild type. Values are averages from two independent experiments (n = 10), and sd (not shown) was below 7%. E, The photograph illustrates the severe phenotype observed in 35S:hab1W385A plants after 60 min of water loss. F, Reduced expression of ABA-inducible genes in 35S:hab1W385A (line 4) and 35S:HAB1 plants compared with Columbia wild type. Values are expression levels reached in the transgenic lines with respect to wild type (value 1) as determined by reverse transcription-quantitative PCR analysis. Expression of gene markers was analyzed in 10-d-old seedlings treated with 10 μm ABA for 3 h. Values are averages ± sd for two independent experiments (n = 30–40 seedlings per experiment). MS, Murashige and Skoog medium. [See online article for color version of this figure.]
Stomatal closing is a key ABA-controlled process that preserves water under drought conditions. We mimicked drought by exposing plants to the drying atmosphere of a flow laminar hood and under these conditions we measured water loss in 2-week-old seedlings (Fig. 5, D and E). Both 35S:HAB1 and 35S:hab1W385A plants showed a higher transpiration rate than wild type, and water loss in plants overexpressing the mutated phosphatase was higher than in the wild-type PP2C. The increased insensitivity to ABA of the 35S:hab1W385A plants as compared to 35S:HAB1, is consistent with the inability of the PYRL/PYL/RCAR receptors to inhibit in vitro the activity of hab1W385A (Fig. 4D). Finally, the expression of ABA-inducible genes was severely reduced in 35S:hab1W385A plants as compared to the wild type (Fig. 5F). The accumulation of these transcripts was also impaired in 35S:HAB1 plants; in some cases, RAB18 and RD29B, the effect was similar to 35S:hab1W385A plants, however, ABA induction of other transcripts, KIN1, RD29A, P5CS, and RD22, was less affected (Fig. 5F).
DISCUSSION
The structure of the PYR1-ABA-HAB1 complex presented here and those of the ternary complexes studied previously (Melcher et al., 2009; Miyazono et al., 2009; Yin et al., 2009) contribute to explain how ABA binding induces the interaction between receptor and phosphatase and its inhibitory nature on phosphatase activity. Interestingly, these complexes show a 1:1 receptor:phosphatase stoichiometry. Since it has been shown that PYR1 forms a dimer in vivo (Nishimura et al., 2009), evidence that is not yet available for PYL1 and PYL2, our data confirm that PYR1 dimer dissociation is required for the formation of the ternary complex, as Yan and coworkers have suggested (Yin et al., 2009). However, a detailed understanding of the dimer dissociation process is not available yet.
Once the hormone enters the receptor cavity, the cyclic moiety of the ABA molecule establishes interactions with the receptor gating loops, which favors their closed conformation. This closed conformation offers an optimal surface for the docking of the phosphatase, which contributes in turn to the stability of the ternary complex by locking the gating loops in their closed conformation and trapping the hormone inside the binding cavity. For instance, PYL9 and PYL5 bind to ABA with a Kd of 0.70 and 1.1 μm, respectively, whereas inclusion of ABI2 and HAB1 in the binding assay leads to a Kd of 64 and 38 nm, respectively (Ma et al., 2009; Santiago et al., 2009b). The HAB1 Trp-385 residue plays a major role in this stabilization process by inserting between the gating loops, and additionally via an indirect contact with the ABA’s ketone group through a hydrogen bond network mediated by a critical water molecule. This water molecule establishes hydrogen bonds not only with HAB1 Trp-385 and the hormone, but also with key residues (Pro-88 and Arg-116) of the receptor gating loops. This complex network of interactions provides a mechanism through which the phosphatase is able to monitor hormone occupancy of the ABA binding cavity, and therefore ensuring that the conserved Trp residue will only contribute to the stabilization of the receptor-phosphatase complex if the hormone is present. The in vitro data presented here for hab1W385A and by Miyazono et al. (2009) for abi1W300A support this conclusion. Moreover, our results show that this hormone-sensing mechanism is critical for ABA response in planta. Thus, expression of hab1W385A in Arabidopsis plants leads to a strong ABA-insensitive phenotype, which can’t be explained solely by enhanced PP2C gene dosage, since 35S:HAB1 plants, although less sensitive to ABA than wild type, show milder phenotypes. The reduced sensitivity to ABA-mediated inhibition of seed germination and seedling establishment, enhanced water loss, and reduced expression of ABA-responsive genes in 35S:hab1W385A plants support the relevance of this locking interaction, postulated by structural studies. Additionally, these plants represent a valuable tool to dissect the ABA pathway by using dominant receptor-insensitive PP2C mutants that do not compromise the intrinsic phosphatase activity. Taking into account the large number of screenings performed to identify ABA-insensitive plants, the failure to isolate mutants harboring missense mutations in this Trp residue is somehow surprising. However, since ethylmethanesulfonate mutagenesis usually leads to G → A transitions, such mutation in the Trp codon (UGG) would lead to stop codons and presumably loss-of-function alleles. The locking mechanism provided by the Trp residue appears to be a particular evolution of the plant clade A PP2Cs, since with the exception of AHG1, they are the unique plant PP2Cs that present this residue in the appropriated position of the flap PP2C subdomain. Interestingly, AHG1 was less sensitive to ABA-dependent PYL8-mediated inhibition than other clade A PP2Cs, such as PP2CA and At5g59220 (Supplemental Fig. S6).
This work and previous structural analyses indicate that the insertion of the PYR1 Ser-85-containing β3-β4 loop (Ser-112 of PYL1 and Ser-89 of PYL2) into the phosphatase catalytic site could account for the inhibition of PP2C catalytic activity by blocking access of potential substrates to the phosphatase catalytic site in a competitive manner. However, although this mechanism looks plausible, the phosphatase catalytic channel remains open in its lower part in the ternary complexes formed by both HAB1 and ABI1 (Supplemental Fig. S2). This lower part of the phosphatase catalytic groove might represent an alternative entry site for substrates and indeed initial studies based on biochemical assays with a nonpeptidic substrate, suggested that inhibition of the PP2C activity by PYR/PYL/RCAR proteins occurs by a noncompetitive, rather than competitive mechanism (Ma et al., 2009). In contrast, in other studies the inhibition of HAB1 by ABA-bound PYL2 was overcome by increasing concentrations of an OST1 phosphopeptide containing residues of the kinase activation loop (Melcher et al., 2009). Unfortunately the structure of a PP2C in complex with a natural peptide substrate is lacking, which could contribute to resolve this issue. However, one striking observation arising from this structural analysis is the proximity of Ser-85 in the gating loop of the PYR1 receptor to the position expected to be occupied by the phosphoryl group of the substrate of the phosphatase reaction. Superposition of this structure and the catalytic domain of human PP2Cα shows that the β-carbon of PYR1 Ser-85 is next to the phosphate ion oxygen atom that Barford and coworkers have proposed as the seryl/threonyl oxygen in their analysis of the PP2Cα catalytic site (Das et al., 1996). This would suggest that the PYR1 Ser-85, and its equivalent in other PYR/PYL proteins, might act as a product mimic and occupy a similar position as the phosphorylated Ser residues in SnRK2s and other PP2C targets. In our view, this important observation lends weight to the interpretation that the formation of the receptor-phosphatase complex prevents access of natural PP2C substrates to the catalytic site, supporting the competitive nature of the inhibition mechanisms. At the same time it would support the catalytic mechanism proposed by Barford (Das et al., 1996), where the water molecule linked to the metal at the M2 site and Glu-37 of human PP2Cα (Glu-203 in HAB1) would contribute to catalysis by facilitating the protonation of the oxygen atom in the P-O scissile bond.
Since Ser-85 of PYR1, Ser-112 of PYL1, and Ser-89 of PYL2 insert into the PP2C active site and establish contacts with Gly-180 of ABI1 or Gly-246 of HAB1, the structural data provide a framework to explain the effect of abi1G180D and hab1G246D mutations. However, no direct biochemical evidence had been previously provided in the case of hab1G246D. This analysis shows that hab1G246D is insensitive to inhibition by various PYR/PYL proteins, which leads to the escape from the ABA-dependent PYR/PYL inhibitory mechanism and the subsequent constitutive inhibition of OST1 activity. Therefore, these data are in agreement with the notion that hab1G246D behaves as a hypermorphic mutation in the presence of ABA, as noted by Schroeder and coworkers (Robert et al., 2006). Paradoxically, in the absence of ABA, hab1G246D shows lower intrinsic phosphatase activity than wild-type HAB1, probably because this mutation perturbs the PP2C active site to some extent.
Even though other ABA receptors have been identified (Pandey et al., 2009; Shang et al., 2010) and therefore other input sources exist for ABA signaling, the phenotypes of both 35S:hab1G246D and 35S:hab1W385A plants indicate that constitutive activation of the PP2Cs (and the consequent inactivation of the SnRK2s) leads to a severe blockade of ABA signaling. Therefore, the action of the SnRK2s is likely localized downstream of the other putative inputs and could represent a core ABA signaling component shared by all ABA receptors. This would be in agreement with the extreme ABA insensitivity of triple snrk2.2/2.3/2.6 mutant plants (Fujii and Zhu, 2009).
MATERIALS AND METHODS
Construction of Plasmids
Plasmids pETM11 or pET28a were used to generate N-terminal His6-tagged recombinant proteins. The cloning of 6xHis-ΔNHAB1 (lacking residues 1–178), PYR1, PYL4, PYL5, and PYL8 constructs was previously described (Santiago et al., 2009b). Using a similar approach, PYL1 and PYL6 were cloned in pETM11, whereas PYL9 was cloned in pET28a. HAB1(W385A), HAB1(G246D), PYR1(S85A), PYR1(R116A), PYR1(L87A), and PYR1(Y120A) mutants were produced using the overlap extension procedure (Ho et al., 1989) and cloned into pETM11. PYR1(S152L), PYR1(P88S), PYR1(R157H), PYR1(E141K), and PYR1(E94K) mutants were obtained from the pyr1-2, pyr1-3, pyr1-4, pyr1-5, and pyr1-6 alleles, respectively (Park et al., 2009), and cloned into pET28a. The coding sequence of OST1 and a C-terminal deletion of ABF2 (ΔCABF2, amino acids 1–173) were cloned into pET28a.
Protein Expression and Purification
BL21(DE3) cells transformed with the corresponding constructs in pETM11 or pET28a vectors were grown in Luria-Bertani medium to an OD600 of 0.6 to 0.8. At this point 1 mm isopropylthio-β-galactoside was added and the cells were harvested after overnight incubation at 20°C. Proteins used for crystallization were purified as described (Santiago et al., 2009a). For small-scale protein preparations, the following protocol was used. Pellets were resuspended in lysis buffer (50 mm Tris pH 7.5, 250 mm KCl, 10% glycerol, 1 mm β-mercaptoethanol) and lysed by sonication with a Branson Sonifier 250. The clear lysate obtained after centrifugation was purified by nickel affinity. A washing step was performed using 50 mm Tris, 250 mm KCl, 20% glycerol, 30 mm imidazole, and 1 mm β-mercaptoethanol washing buffer, and finally the protein was eluted using 50 mm Tris, 250 mm KCl, 20% glycerol, 250 mm imidazole, and 1 mm β-mercaptoethanol elution buffer.
Crystallization and Structure Solution
The PYR1-ABA-HAB1 ternary complex was prepared by mixing PYR1, ΔNHAB1, and 1 mm ABA to a final concentration of 3 mg/mL, 5 mg/mL, and 1 mm, respectively, in 20 mm Tris pH 7.5, 150 mm NaCl, 1 mm MnCl2, and 1 mm β-mercaptoethanol. Crystallization conditions for the complex were identified at the High Throughput Crystallization Laboratory of EMBL Grenoble Outstation (https://htxlab.embl.fr) as described in Marquez et al. (2007). The crystals used for data collection were obtained by vapor diffusion method in 0.25 m NaCl, 19% Peg 3350 at 20°C. X-ray diffraction data were collected at the ID14-4 beam line of the ESRF to 1.8 Å resolution. Initial phases were obtained by the molecular replacement method using the two central β-sheets of the catalytic domain of the human PP2Cα protein (1A6Q; Das et al., 1996) as a search model and the program Phaser (McCoy et al., 2007). Successive rounds of automatic refinement and manual building were carried out with RefMac5 (Murshudov et al., 1997) and Coot (Emsley and Cowtan, 2004). Atomic coordinates from the final model have been deposited in the Protein Data Bank under accession code 3QN1.
PP2C and OST1 in Vitro Activity Assays
Phosphatase activity was measured using either the Ser/Thr phosphatase assay system (Promega) using the RRA(phosphoT)VA peptide as substrate or pNPP. In the first case assays were performed in a 100-μL reaction volume containing 25 mm Tris-HCl pH 7.5, 10 mm MgCl2, 1 mm dithiothreitol, 25 μm peptide substrate, and the PP2C. When indicated, PYR-PYL recombinant proteins and ABA were included in the PP2C activity assay. After incubation for 60 min at 30°C, the reaction was stopped by addition of 30 μL molybdate dye (Baykov et al., 1988) and the absorbance was read at 630 nm with a 96-well plate reader. For the pNPP phosphatase activity assays a 100 μL solution containing 25 mm Tris-HCl pH 7.5, 2 mm MnCl2, and 5 mm pNPP substrate and the indicated amount of the PP2Cs was used. Measurements were taken with a ViktorX5 reader at 405 nm every 60 s over 30 min.
Phosphorylation assays were done basically as described previously (Belin et al., 2006; Vlad et al., 2009). Assays to test recovery of OST1 activity were done by previous incubation for 10 min of the protein phosphatase HAB1 together with the PYR1 wild type or PYR1 mutant proteins in the presence of the indicated concentration of (+)-ABA. Next, the reaction mixture was incubated for 50 min at room temperature in 30 μL of kinase buffer: 20 mm Tris-HCl pH 7.8, 20 mm MgCl2, 2 mm MnCl2, and 3.5 μCi of γ-32ATP (3,000 Ci/mmol). The reaction was stopped by adding Laemmli buffer. When indicated, ΔCABF2 recombinant protein (100 ng) was added as substrate of OST1. After the reaction proteins were separated by SDS-PAGE using an 8% acrylamide gel and transferred to an Immobilon-P membrane (Millipore). Radioactivity was detected using a phosphorimage system (FLA5100, Fujifilm). After scanning, the same membrane was used for Ponceau staining. The data presented are averages of at least three independent experiments.
Y2H Assays
Protocols were similar to those described previously (Saez et al., 2006).
Generation of 35S:hab1W385A Transgenic Lines
The mutated hab1W385A was cloned into pCR8/GW/TOPO entry vector (Invitrogen) and recombined by LR reaction into the gateway-compatible ALLIGATOR2 vector. This construct drives expression of hab1W385A under control of the 35S cauliflower mosaic virus promoter and introduces a triple HA epitope at the N terminus of the protein. Selection of transgenic lines is based on the visualization of GFP in seeds, whose expression is driven by the specific seed promoter At2S3. The ALLIGATOR2-35S:3HA-hab1W385A construct was transferred to Agrobacterium tumefaciens C58C1 (pGV2260; Deblaere et al., 1985) by electroporation and used to transform Columbia wild-type plants by the floral-dip method. T1 transgenic seeds were selected based on GFP visualization and sowed in soil to obtain the T2 generation. Homozygous T3 progeny was used for further studies and hab1W385A protein level was verified by immunoblot analysis using anti-HA-peroxidase (Roche). The generation of 35S:HAB1-dHA lines was described previously (Saez et al., 2004).
Seed Germination and Seedling Establishment Assays
After surface sterilization of the seeds, stratification was conducted in the dark at 4°C for 3 d. Next, approximately 200 seeds per experiment were sowed on solid medium composed of Murashige and Skoog basal salts, 1% Suc, and supplemented with different ABA concentrations. To score seed germination, radical emergence was analyzed at 72 h after sowing. Seedling establishment was scored as the percentage of seeds that developed green expanded cotyledons and the first pair of true leaves at 7 d.
Water Loss Assays
Two- to 3-week-old seedlings growing in Murashige and Skoog plates were used. Three seedlings per genotype with similar growth were submitted to the drying atmosphere of a flow laminar hood. Kinetic analysis of water loss was performed and represented as the percentage of initial fresh weight loss at each scored time point. Data are averages ± se from two independent experiments.
RNA Analyses
ABA treatment, RNA extraction, and reverse transcription-quantitative PCR amplifications were performed as previously described (Saez et al., 2004).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structural superposition of ternary receptor complexes.
Supplemental Figure S2. Detail of the catalytic groove of HAB1.
Supplemental Figure S3. Amino acid sequence and secondary structure alignment of plant PP2Cs and the catalytic core of human PP2C.
Supplemental Figure S4. Detail of the HAB1 catalytic site around the PYR1 Ser-85.
Supplemental Figure S5. Comparison of the ABA-dependent inhibitory effect of PYR1 wild-type and mutant proteins on HAB1 activity.
Supplemental Figure S6. Amino acid sequence alignment of Arabidopsis clade A PP2Cs and representative PP2Cs from other groups.
Acknowledgments
We are grateful to the ESRF and the EMBL for access to macromolecular crystallography beam lines ID14eh4 and BM14 and Andrew McCarthy and Hassan Belrhali for support with data collection.
References
- Baykov AA, Evtushenko OA, Avaeva SM. (1988) A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem 171: 266–270 [DOI] [PubMed] [Google Scholar]
- Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier-Brygoo H, Thomine S. (2006) Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol 141: 1316–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertauche N, Leung J, Giraudat J. (1996) Protein phosphatase activity of abscisic acid insensitive 1 (ABI1) protein from Arabidopsis thaliana. Eur J Biochem 241: 193–200 [DOI] [PubMed] [Google Scholar]
- Bray EA, Bailey-Serres J, Weretilnyk E. (2000) Responses to abiotic stresses. Buchanan BB, Gruissem W, Jones RL, , Biochemistry & Molecular Biology of Plants. John Wiley & Sons, Somerset, UK, pp 1158–1203 [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Das AK, Helps NR, Cohen PT, Barford D. (1996) Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J 15: 6798–6809 [PMC free article] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De GH, Deboeck F, Schell J, Van MM, Leemans J. (1985) Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res 13: 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimasi N, Flot D, Dupeux F, Marquez JA. (2007) Expression, crystallization and X-ray data collection from microcrystals of the extracellular domain of the human inhibitory receptor expressed on myeloid cells IREM-1. Acta Crystallogr Sect F Struct Biol Cryst Commun 63: 204–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK. (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, et al. (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12: 343–351 [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. (2001) Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins 43: 134–144 [DOI] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I. (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S. (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leube MP, Grill E, Amrhein N. (1998) ABI1 of Arabidopsis is a protein serine/threonine phosphatase highly regulated by the proton and magnesium ion concentration. FEBS Lett 424: 100–104 [DOI] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Marquez JA, Galfre E, Dupeux F, Flot D, Moran O, Dimasi N. (2007) The crystal structure of the extracellular domain of the inhibitor receptor expressed on myeloid cells IREM-1. J Mol Biol 367: 310–318 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. (2007) Phaser crystallographic software. J Appl Cryst 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. (2009) Structural basis of abscisic acid signalling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. (2009) Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Nelson DC, Assmann SM. (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TFF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presta LG, Rose GD. (1988) Helix signals in proteins. Science 240: 1632–1641 [DOI] [PubMed] [Google Scholar]
- Pullen KE, Ng HL, Sung PY, Good MC, Smith SM, Alber T. (2004) An alternate conformation and a third metal in PstP/Ppp, the M. tuberculosis PP2C-family Ser/Thr protein phosphatase. Structure 12: 1947–1954 [DOI] [PubMed] [Google Scholar]
- Radauer C, Lackner P, Breiteneder H. (2008) The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol 8: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert N, Merlot S, N'guyen V, Boisson-Dernier A, Schroeder JI. (2006) A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett 580: 4691–4696 [DOI] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E. (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421: 185–190 [DOI] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL. (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Saez A, Robert N, Maktabi MH, Schroeder JI, Serrano R, Rodriguez PL. (2006) Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol 141: 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA. (2009a) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Marquez JA, Cutler SR, Rodriguez PL. (2009b) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60: 575–588 [DOI] [PubMed] [Google Scholar]
- Schlicker C, Fokina O, Kloft N, Grune T, Becker S, Sheldrick GM, Forchhammer K. (2008) Structural analysis of the PP2C phosphatase tPphA from Thermosynechococcus elongatus: a flexible flap subdomain controls access to the catalytic site. J Mol Biol 376: 570–581 [DOI] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, et al. (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK. (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Lauriere C, Merlot S. (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehenkel A, Bellinzoni M, Schaeffer F, Villarino A, Alzari PM. (2007) Structural and binding studies of the three-metal center in two mycobacterial PPM Ser/Thr protein phosphatases. J Mol Biol 374: 890–898 [DOI] [PubMed] [Google Scholar]
- Wilkie AO. (1994) The molecular basis of genetic dominance. J Med Genet 31: 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. (2009) Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol 16: 1230–1236 [DOI] [PubMed] [Google Scholar]