Abstract
Full-length cDNA (FLcDNA) libraries consisting of 172,000 clones were constructed from a two-row malting barley cultivar (Hordeum vulgare ‘Haruna Nijo’) under normal and stressed conditions. After sequencing the clones from both ends and clustering the sequences, a total of 24,783 complete sequences were produced. By removing duplicates between these and publicly available sequences, 22,651 representative sequences were obtained: 17,773 were novel barley FLcDNAs, and 1,699 were barley specific. Highly conserved genes were found in the barley FLcDNA sequences for 721 of 881 rice (Oryza sativa) trait genes with 50% or greater identity. These FLcDNA resources from our Haruna Nijo cDNA libraries and the full-length sequences of representative clones will improve our understanding of the biological functions of genes in barley, which is the cereal crop with the fourth highest production in the world, and will provide a powerful tool for annotating the barley genome sequences that will become available in the near future.
In 2009, approximately 54 million hectares of barley (Hordeum vulgare) were cultivated, and harvests of this species produced approximately 150 million tons of grain worldwide, according to the Food and Agriculture Organization of the United Nations (http://faostat.fao.org/). Barley belongs to the Poaceae family (i.e. grasses), which also includes rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum), and rye (Secale cereale; cross-pollinated). These members of the Poaceae, including barley, are the major cereal crops cultivated throughout the world. Barley is utilized for animal feed, malting, and as a human food source. Because barley is self-pollinated and has a diploid (2n = 14) genome, it is recognized as a genetic model of the Triticeae tribe within the Poaceae. Studies of the barley genome, transcriptome, and proteome are currently advancing our understanding of the molecular functions of agriculturally important barley genes (Sreenivasulu et al., 2008a, 2008b).
Although barley has been extensively studied in terms of its genetics and breeding, molecular biology and genomics studies have been limited until recently because of the large genome size of this species (greater than 5 Gb, approximately 12 times that of rice; Varshney et al., 2007). To further promote barley genomics, a multinational collaboration, the International Barley Sequencing Consortium (IBSC), has been developed with the objective of obtaining the whole genome sequence of barley (http://barleygenome.org; Schulte et al., 2009).
The study of an organism's transcriptome (i.e. all transcribed sequences) is one of the most effective ways to investigate the structure and function of its active genes. In many plant species, transcript contigs have been constructed by assembling all the EST data available in PlantGDB, with the aim of identifying a data set of unique mRNA sequences and maximizing the information obtained for both protein-coding and noncoding regions in these sequences (Duvick et al., 2008). A large set of ESTs (501,620 from the vulgare subspecies and 24,161 from the spontaneum subspecies in the National Center for Biotechnology Information-dbEST release-100110 [www.ncbi.nlm.nih.gov/dbEST/] and 522,561 sequences in PlantGDB [www.plantgdb.org/]) has been accumulated in the public domain. These ESTs have been assembled into 23,595 clusters in National Center for Biotechnology Information’s UniGene data set (http://www.ncbi.nlm.nih.gov/unigene) and into 134,482 PlantGDB-assembled unique transcripts (http://www.plantgdb.org/prj/ESTCluster/progress.php). It is estimated that approximately 75% of the genes in the barley genome have been captured (Sreenivasulu et al., 2008a). Although these assemblies provide researchers with a tremendous amount of information for understanding partial barley gene structures, the assembly of sequences from different barley varieties might result in erroneous contigs or lead to the fusion of unrelated transcripts via common domains or motif sequences. Moreover, these data might lead to an overestimation of the number of genes in barley, because these assemblies do not consider virtual connections between 5′ and 3′ end sequences where both sequences are derived from the same cDNA clone.
Capturing the transcripts of all active genes under defined (temporal, spatial, and stressed) conditions can provide a “snapshot” of living cells. The resultant data can be used to obtain quantitative (expression frequency) and qualitative (predicted protein function) information about these genes. For barley transcriptome analysis, a 22K Barley1 GeneChip probe array based on an EST database containing 350,000 sequences from 84 different RNA sources has been established (Close et al., 2004). The results of high-throughput transcript profiling of barley using this chip can be retrieved from PLEXdb (http://www.plexdb.org/) and ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/). Using several platforms for transcript profiling, researchers have described the responses of barley to conditions such as high salinity (Ueda et al., 2006; Walia et al., 2006), low temperature or freezing (Koo et al., 2008), and drought (Talamè et al., 2007; Tommasini et al., 2008) as well as during various stages, such as grain development (Sreenivasulu et al., 2006), germination (Sreenivasulu et al., 2008b), and growth (Druka et al., 2006), by means of coordinated up- or down-regulation of sets of genes.
Moreover, the construction of a comprehensive gene set for barley by genome sequencing is under way. However, following sequencing, genome annotation in the absence of information on exon-intron junctions is not an easy task, even for organisms for which complete genome sequences have been revealed. Although several gene prediction programs have been developed, the predicted gene structures might not always be correct, because these programs may select and connect incorrectly predicted exons (Bennetzen et al., 2004; Cruveiller et al., 2004; Jabbari et al., 2004). Hence, most gene annotation projects refer to EST or mRNA sequences for the accurate structural annotation of genes (Imanishi et al., 2004; Itoh et al., 2007).
Full-length cDNA (FLcDNA) is defined as the DNA complementary to an mRNA sequence that extends from the region near the 5′ cap structure to the poly(A) tail, which is a structure specific to eukaryotic mRNAs (Maruyama and Sugano, 1994; Suzuki et al., 1997; Carninci et al., 2000). FLcDNA technology has been applied to the genomes of humans (Ota et al., 2004), mice (Kawai et al., 2001), and several plant species, including Arabidopsis (Arabidopsis thaliana; Seki et al., 2002), O. sativa (Kikuchi et al., 2003), T. aestivum (Ogihara et al., 2004; Kawaura et al., 2009), soybean (Glycine max; Umezawa et al., 2008), Z. mays (Soderlund et al., 2009), and tomato (Solanum lycopersicum; Aoki et al., 2010). Recently, an FLcDNA library has been constructed for the Japanese malting barley variety Haruna Nijo (Sato et al., 2009b). From more than 45,000 cDNA clones, 5,006 nonoverlapping sequences were obtained. These sequences and clones have been made publicly available through the BarleyDB (http://www.shigen.nig.ac.jp/barley/), and they have proven effective in the identification of functional genes. However, this information is insufficient, because the number of obtained FLcDNAs is much lower than all of the expressed genes in the barley genome.
In this study, we constructed 12 FLcDNA libraries for Haruna Nijo from various organs or under different conditions to produce a comprehensive data set for this barley variety. After 5′ and 3′ end sequencing and clustering of approximately 172,000 cDNA clones, 24,783 complete FLcDNA sequences were retrieved. In conjunction with publicly available barley FLcDNAs, a total of 22,651 nonredundant barley FLcDNAs were obtained. These sequenced clones should provide comprehensive information about the barley gene repertory.
RESULTS
Barley FLcDNA Data Set
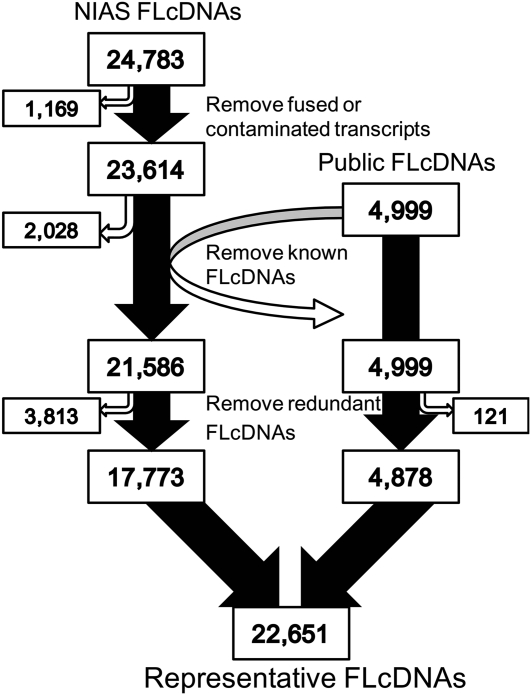
A total of 24,783 FLcDNAs were completely sequenced from the EST library (see Supplemental Information S1; Fig. 1). After eliminating contaminated sequences and possible fusions of unrelated transcripts, we obtained 23,614 barley FLcDNA sequences (henceforth, the National Institute of Agrobiological Sciences FLcDNAs, or NIAS FLcDNAs). These FLcDNAs were derived from the 12 cDNA libraries distinguished by library-specific tag sequences and an unknown group lacking the tag sequences (Supplemental Table S1). The number of FLcDNAs in the various libraries ranged from 857 in the library derived from flowers in the juvenile stage to 2,643 in the library derived from the plants treated with jasmonic acid. To characterize the data set, we compared the sequences with 4,999 Haruna Nijo FLcDNA sequences that had been published previously (henceforth, Public FLcDNAs; Sato et al., 2009b). We found that the NIAS FLcDNA inserts were clearly longer (1,701 ± 846 bp) than the Public FLcDNA inserts (1,455 ± 742 bp; Supplemental Fig. S1). For comparison, the average lengths of rice FLcDNAs in sets of 37,139 O. sativa var japonica FLcDNAs and 10,083 O. sativa var indica FLcDNAs (DNA Data Bank of Japan [DDBJ]/EMBL/GenBank) were 1,746 ± 945 bp and 1,042 ± 535 bp, respectively. Moreover, the estimated proportion of clones that might contain complete predicted open reading frames (ORFs; from Met to the stop codon) was similar between the NIAS and Public FLcDNAs (84.9% and 85.6%, respectively; Table I). These results suggest that the qualities of the two FLcDNA data sets are quite similar in terms of capturing complete ORFs.
Figure 1.
Determination of the representative barley FLcDNAs obtained in this study.
Table I.
Comparison of the completeness of the ORFs in the barley FLcDNAs
| ORF | NIAS | Public | Representative |
| Complete ORF | 20,055 | 4,279 | 19,335 |
| 5′ Truncated ORF | 3,404 | 685 | 3,181 |
| 3′ Truncated ORF | 100 | 11 | 79 |
| 5′ and 3′ Truncated ORFa | 29 | 4 | 28 |
| Nonprotein codingb | 26 | 20 | 28 |
| Total | 23,614 | 4,999 | 22,651 |
ORF had neither start nor stop codon.
FLcDNAs had no ORFs that were similar to known proteins or longest ORFs whose lengths were 70 or more amino acids.
To evaluate the novel barley FLcDNAs revealed by our data, we compared the sequence similarity between the NIAS and Public FLcDNAs. We found that 21,586 of the 23,614 NIAS FLcDNAs showed no significant hits for any of the Public FLcDNAs. After redundant clones were removed, the remaining 17,773 FLcDNAs represented novel barley sequences (Fig. 1). Combined with the 4,878 nonredundant Public FLcDNAs, 22,651 representative barley FLcDNAs were constructed. We have designated these FLcDNAs “Uni-FLcDNAs” in this study and used this set for further comparative analyses. The average length of the Uni-FLcDNAs was 1,711 ± 863 bp.
Recently, many paired sense/antisense transcripts, referred to as natural antisense transcripts (NATs; Osato et al., 2003; Wang et al., 2005), have been identified. In Arabidopsis, 958 NAT pairs have been confirmed (Alexandrov et al., 2006), and in rice, 687 bidirectional transcription units have been discovered by means of FLcDNA mapping onto genome sequences (Osato et al., 2003). To test for the existence of NATs in the obtained FLcDNAs, we used BLASTN to screen for paired sense/antisense transcripts among all of the FLcDNAs, with a positive match criterion of greater than 95% identity and an E-value of less than 1 × 10−5. We determined that 2,051 FLcDNAs could form sense/antisense pairs. An example of a sense/antisense pair presented by ClustalX alignment software is shown in Supplemental Figure S2. NIASHv1002F20 and NIASHv1022F01 exhibit complete reverse homology, despite the fact that they both contain predicted ORFs. This suggests that our data set contains NATs, even for sequences that encode predicted proteins.
Protein-Coding Genes
Based on homology searches and predictions of the longest ORF, 22,623 of 22,651 representative ORFs were identified; 19,212 of these ORFs (84.9%) were homologous to known functional genes according to the results of BLASTX searches against the RefSeq (Pruitt et al., 2009) and UniProtKB (UniProt Consortium, 2009) databases. The proportion of representative FLcDNAs that were deemed to contain complete ORFs was 85.4% (Table I), indicating that most of the FLcDNAs were associated with protein-coding genes.
To identify sequences in the Uni-FLcDNAs that were homologous to previously cloned rice trait genes, we employed 881 genes from Oryzabase (http://www.shigen.nig.ac.jp/rice/oryzabase/; Kurata and Yamazaki, 2006). Barley homologs of these genes could play important roles in barley traits (phenotypes). We searched for homologs of these sequences among the Uni-FLcDNAs using BLASTP (Supplemental Table S2). We found that 721 of the 881 genes (81.8%) had barley homologs with a high similarity (50% or greater identity). Although we currently do not know whether these homologs are orthologs, this result indicates that these trait genes are highly conserved between barley and rice. The detection of homologous sequences among the Uni-FLcDNAs could accelerate the comparative mapping of barley genes and the functional prediction of these homologous cDNAs to promote future barley genomics research.
We found 75 representative FLcDNAs that were longer than 5,000 bp in length, and some of these had the capacity to encode relatively long ORFs. For example, four of the 10 longest FLcDNAs encoded ORFs longer than 1,000 amino acids. In contrast, three of their ORFs were shorter than 100 amino acids (Table II).
Table II.
The 10 longest FLcDNAs in the Uni-FLcDNA library
| Clone Name | Nucleotide Length | Amino Acid Length | Gene Function |
| bp | |||
| NIASHv2010H17 | 7,384 | 460 | Mitogen-activated protein kinase |
| NIASHv3115C23 | 7,155 | 2,230 | Acetyl-CoA carboxylase |
| NIASHv2090B16 | 7,012 | 90 | Cycloeucalenol cycloisomerase |
| FLbaf76j08 | 6,765 | 98 | Hypothetical protein |
| NIASHv2019K03 | 6,637 | 1,731 | Chromatin-remodeling complex subunit |
| NIASHv2035I12 | 6,311 | 227 | Conserved hypothetical protein |
| NIASHv2017F18 | 6,183 | 152 | KN1-type homeobox transcription factor |
| NIASHv1031N03 | 6,072 | 33 | Hypothetical protein |
| NIASHv2125A18 | 6,037 | 1,859 | DNA-directed RNA polymerase |
| NIASHv3085A02 | 6,036 | 1,792 | Callose synthase 1 catalytic subunit |
We estimated that there were 28 noncoding FLcDNAs among the Uni-FLcDNAs (see “Materials and Methods”). Although these noncoding FLcDNAs could have been derived from microRNAs, a BLASTN search against rice microRNAs revealed no significant homologies. Moreover, as 26 of the 28 noncoding FLcDNAs exhibited no homologies with four fully sequenced grass genomes (O. sativa, Z. mays, Sorghum bicolor, and Brachypodium distachyon), they might represent poorly conserved noncoding RNAs.
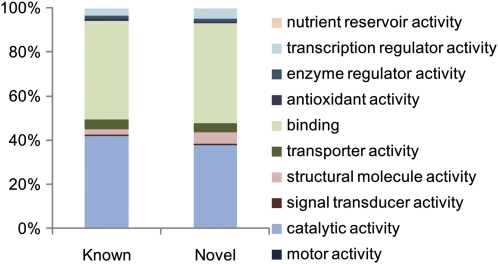
An analysis using InterProScan revealed that 16,859 of the 22,623 representative ORFs (74.5%) contained conserved domains, and 12,595 ORFs (55.7%) were assigned Gene Ontology (GO) terms (Barrell et al., 2009). Using GO2slim, we found that “binding” (GO:0005488) and “catalytic activity” (GO:0003824) were the most common second-level GO terms found in the data (Supplemental Fig. S3). The distribution of GO terms related to the barley Uni-FLcDNAs was quite similar to that seen in representative Rice Annotation Project (RAP) data (Tanaka et al., 2008).
Comparison of Barley FLcDNAs with the Triticeae Transcriptome
We described 17,773 novel barley nonredundant FLcDNAs (Fig. 1). To evaluate the amount of novel gene information in these sequences, homology searches of the FLcDNAs were conducted against sequences from publicly available transcripts. For this purpose, 6,625 mRNAs and 525,559 ESTs from barley and 3,433 mRNAs and 1,107,168 ESTs from wheat were downloaded from DDBJ/EMBL/GenBank.
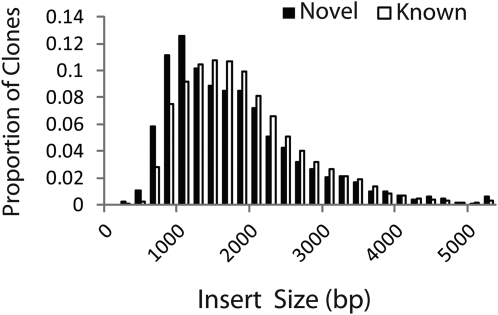
A BLASTN search showed that 3,278 of the representative FLcDNAs had no homologous sequences. The average insert length of these novel FLcDNAs (1,602 ± 921 bp) was slightly shorter than that of the known FLcDNAs (1,729 ± 851 bp; Fig. 2). Characterization of protein functions using the second-level GO terms showed a similar distribution of terms among the novel and known FLcDNAs, except for “structural molecule activity” (GO:0005198; Fig. 3; 2.4% in the “known” gene set and 5.1% in the “novel” gene set), indicating that the molecular functions of the novel FLcDNAs were distributed similarly to those of the known FLcDNAs. The novel transcripts were predicted to code for many different kinds of essential proteins, such as proteins involved in protein synthesis, including ribosomal proteins and elongation factors, transcription factors, cytochromes, and stress-related proteins, including jasmonic acid-induced protein, CBFII-5.1, heat shock protein, and nucleotide-binding site-Leu-rich repeat disease resistance protein homolog (Supplemental Table S3). Hence, we anticipate that these novel transcripts will support the construction of an essential gene network for barley.
Figure 2.
Distribution of nucleotide lengths of known and novel FLcDNAs. Size distributions of FLcDNAs with high homology to known grass genes (white bars) and with no homology to known genes (black bars) are shown.
Figure 3.
GO categorization of known and novel FLcDNAs. Functional motifs/domains were detected and categorized using GO criteria. Results for the representative FLcDNAs with high homology to known grass genes (Known) and with no homology to known genes (Novel) are shown.
Of the 3,278 novel FLcDNAs, 1,974 were conserved in all four sequenced grass genomes (i.e. O. sativa, Z. mays, S. bicolor, and B. distachyon; International Rice Genome Sequencing Project, 2005; Paterson et al., 2009; Schnable et al., 2009; International Brachypodium Initiative, 2010) according to the results of BLASTN searches, but 942 had no grass homolog (Supplemental Table S4). We conducted BLASTP searches of the amino acid sequences derived from the novel ORFs against predicted protein sequences from the four species and found that 1,731 ORFs were homologous to genes annotated in at least one species. These results suggest that the molecular functions of approximately two-thirds of the novel barley FLcDNAs could be predicted by comparison with annotated genes in major grass species.
Detection of Barley-Specific FLcDNAs
To elucidate whether the Uni-FLcDNAs contained gene structures that are conserved among crop plants, BLASTN searches were conducted against the four complete genome sequences. The results showed that 1,699 FLcDNAs (specific FLcDNAs) were not associated with homologous regions in the four genomes, whereas 20,952 FLcDNAs (common FLcDNAs) had a homolog in at least one of the four plant species (Supplemental Table S5). Most FLcDNAs (19,778) were conserved in all four species, but more barley ORFs were homologous to ORFs in O. sativa than in B. distachyon, which shares a more recent common ancestor with barley than rice does. This result might have been caused by differences in the methodology used to sequence O. sativa and B. distachyon and the resulting quality of the reference genome sequences. The number of conserved genes detected in Z. mays was less than half the number detected in S. bicolor, even though the evolutionary distances of barley from Z. mays and S. bicolor are similar. The length of the inserts in the barley-specific FLcDNAs was shorter (1,301 ± 897 bp) than in the other FLcDNAs (1,745 ± 851 bp).
Of the specific FLcDNAs, 263 contained known functional domains based on InterProScan analysis and 25 were noncoding transcripts; thus, 85% of the specific FLcDNAs were not assigned to any putative function. GO data suggested that the relative proportions of the genes related to “signal transducer activity” (GO:0004871) and “enzyme regulator activity” (GO:0030234) were greater in the specific FLcDNAs than in the total representative FLcDNAs (data not shown). This suggests that barley exhibits species-specific regulatory networks involved in signal transduction, transcription, and metabolism.
Comparison of FLcDNA Sequences with Published BAC Sequences of Barley
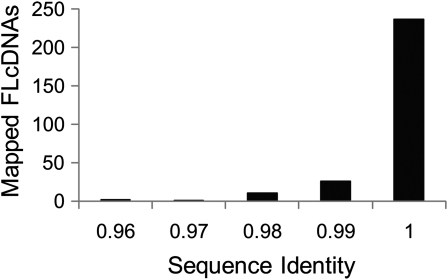
To evaluate the barley FLcDNA data set in terms of its ability to capture a large number of active FLcDNAs, we mapped all of the FLcDNAs on publicly available barley bacterial artificial chromosome (BAC) sequences. Taketa et al. (2008) reported a Haruna BAC contig (AP009567) in which two genes were predicted. BAG12385.1 encodes a putative iron deficiency-specific 4 protein, and BAG12386.1 is a putative ethylene-responsive transcription factor responsible for the nud (hull-less) phenotype. Even though the public FLcDNAs could not be mapped to the predicted loci, multiple FLcDNAs determined in this study could be mapped at each locus, which suggests that the structures of these genes are accurate (Supplemental Fig. S4). We also mapped the FLcDNAs on BACs of a different cultivar, Morex. Haruna Nijo is an established Japanese two-row malting cultivar, whereas Morex is a six-row cultivar that is used as the American malting industry’s standard cultivar. Because Morex will provide the first reference sequence for the barley genome, there have been increasing numbers of submissions of Morex sequences in the DDBJ (www.ddbj.nig.ac.jp/). To estimate the conservation of gene sequences between these cultivars, we mapped the FLcDNAs to publicly available Morex BAC sequences using EST2genome (Mott, 1997). We found that 373 FLcDNAs mapped to genomic sequences from 113 of 181 BACs. Forty-five FLcDNAs (31 representatives) mapped to more than one locus. For example, NIASHv1123K08 was mapped to four loci in three BACs (one locus each of AY758233.1 and DQ249273.1 and two loci of AC239053.1). These genes were highly conserved (99.4% identity on average; Fig. 4), and 61 of them were completely identical. The results of this analysis indicate that the Haruna Nijo FLcDNAs can be utilized by IBSC to annotate the Morex genome.
Figure 4.
Distribution of sequence identities of mapped Haruna Nijo FLcDNAs to published Morex BAC sequences.
DISCUSSION
Scientists have recently recognized that FLcDNAs are indispensable resources for gene identification and annotation. As the barley genome sequencing project progresses, obtaining information about barley FLcDNAs has become increasingly important. There are a considerable amount of Triticeae (barley and wheat) transcript data in the public domain; however, the number of barley FLcDNAs is insufficient compared with the number available for other crops, such as O. sativa, Z. mays, and G. max (Kikuchi et al., 2003; Umezawa et al., 2008; Soderlund et al., 2009).
The sequences developed in this study represent the largest collection of Triticeae FLcDNA data produced to date. To comprehensively cover the entire barley transcriptome with FLcDNA clones, we constructed novel libraries from Haruna Nijo from 12 different developmental stages, organs, and/or stressed conditions and isolated more than 170,000 clones (Supplemental Information S1). To identify the source of clones in the pooled library constructions, tagged sublibraries were used.
The ESTs we identified were clustered, and representative cDNA clones were selected for full sequencing. The quality of the FLcDNAs obtained in this study was evaluated by comparing their average sequence length (1,701 bp) with that of publicly available barley and rice FLcDNAs. A recent report in which the average length of 27,455 maize FLcDNAs was found to be 1,442 bp (Soderlund et al., 2009) also supports the length quality of the FLcDNAs in this study. The method of FLcDNA construction used here was the cap-trapper method (Carninci et al., 1996), which has been used in FLcDNA projects in mice, Arabidopsis (Seki et al., 2002, 2004), rice (Kikuchi et al., 2003), and soybean (Umezawa et al., 2008). Because this methodology is based on the selection of mRNAs with an existing cap structure, it provides high coverage of mRNA structures.
The 22,651 representative barley FLcDNAs described here were constructed using the clones produced in this study and publicly available barley FLcDNAs. Functional categorization of the Haruna Nijo FLcDNAs based on ORF prediction and GO assignments produced results similar to those found for rice FLcDNAs. This could indicate conservation of the entire gene sets between barley and rice. We also detected 28 noncoding genes. Only four of the 28 noncoding transcripts were included in the NAT pairs, suggesting that NATs are not the sole reason underlying the observed the noncoding transcripts.
Comparative analysis of the Uni-FLcDNAs against public data indicated the importance of this data set. First, 17,773 FLcDNAs were found to be novel in barley, and 3,278 cDNAs from the Uni-FLcDNAs exhibited no homology to public EST or mRNA data from barley or wheat. We consider it unlikely that this paucity of homologous sequences resulted from natural variation among Haruna Nijo and other barley varieties, because 1,974 of the 3,278 novel FLcDNAs had common homologs in all four grass species examined (i.e. O. sativa, Z. mays, S. bicolor, and B. distachyon). As full-length sequences are available for these four grass species, these 1,974 genes might be structurally conserved and functionally active in grasses in general. These findings should help us to assign gene functions to the novel barley FLcDNAs.
Second, we detected 1,699 FLcDNAs that showed no homology to any of the four grass genomes. Even though the mean insert length of these FLcDNAs was shorter than that of the other cDNAs, these 1,699 FLcDNAs still have the capacity to encode functional proteins. Therefore, we concluded that these genes are Triticeae-specific (or at least Hordeum-specific) sequences. Unfortunately, only 263 specific FLcDNAs could be assigned putative functions based on InterPro domains, and the gene functions of the other genes are unknown. The complete gene structure information and the FLcDNA clones of the barley-specific genes can be used in future experimental studies, such as for overexpression of recombinant proteins or microarray analyses, to reveal the functions of these genes. We note that the 20,952 FLcDNAs with homologs in all four grass genomes might still include some barley-specific genes, because the coding potential of their mapped regions has not been verified; additionally, there were cases where FLcDNAs mapped to nongenic regions in the latest annotated genome data.
The barley cv Haruna Nijo, which was used here as the source of the cDNA libraries generated, was released as a malting barley variety in 1981 and has been intensively used in the pedigree of Japanese malting barleys because of its excellent quality profiles for brewing. Additionally, several genetic/genomic resources have been established for this cultivar. More than 140,000 ESTs have been sequenced, and the positions of more than 2,890 of these have been located in genetic maps (Sato et al., 2009a). A BAC library has been constructed (Saisho et al., 2007), and some of these BAC clones have been beneficial for map-based cloning of trait genes in barley (Taketa et al., 2008). These resources have made Haruna Nijo a useful variety for investigating barley genetics and genomics (http://www.shigen.nig.ac.jp/barley/index.html).
The IBSC is currently sequencing the complete genome of the Morex cultivar. Mapping of all of the Haruna Nijo FLcDNAs to genome sequences from Morex BACs clearly showed that the Haruna Nijo FLcDNAs can serve as good resources for the genome annotation of Morex. Based on the density of the mapped FLcDNAs (i.e. 48.8 kb per gene; 277 loci mapped to 181 BAC sequences, covering 13.52 Mb), we estimated that the total gene number in the barley genome is approximately 100,000. This estimation is much larger than an estimation presented in a previous report (Mayer et al., 2009) of 38,000 to 48,000 genes in the barley genome (i.e. 104 to 132 kb per gene on average). It is likely that we have overestimated the number of genes, because the BAC clones used for genome sequencing come from gene-rich regions targeted for the purpose of map-based cloning or the investigation of genic regions. Based on the previously estimated gene number (Mayer et al., 2009), our Haruna Nijo data set of 22,651 nonredundant FLcDNAs could represent 47% to 59% of the total number of genes present in barley. The fact that 54% to 70% of the rice genes predicted by RAP have been validated by rice FLcDNAs indicates that our data set is similarly comprehensive. Not all genes present in barley are expected to have been expressed in the 12 conditions examined in this study, so we suggest that the proportion of active genes captured could be much greater than 47% to 59%.
CONCLUSION
We cloned more than 170,000 Haruna Nijo barley FLcDNAs and identified more than 24,000 complete FLcDNAs. The final set of 22,651 representative FLcDNAs obtained will be a very useful resource for future studies of barley as well as for the annotation of barley genomic sequences in the ongoing IBSC genome sequencing project. These data will also support the future wheat genome sequencing project currently being organized by the International Wheat Genome Sequencing Consortium (http://www.wheatgenome.org/).
All FLcDNA data obtained in this study are available from our full-length Barley cDNA Database (http://barleyflc.dna.affrc.go.jp/hvdb/index.html).
MATERIALS AND METHODS
Full-Sequencing of Representative Clones
For each representative clone determined after clustering the end sequences of clones, both the 5′ and 3′ ends were resequenced using the Big Dye Terminator version 3.1 Cycle Sequencing Kit and then analyzed with an ABI 3730xl DNA sequencer (Applied Biosystems) to confirm the clone sequence identity. Next, the internal regions were sequenced using a primer-walking method until the sequences from opposite directions overlapped. We designed the sequencing strategy so that at least two reads covered every part of the insert region of each cDNA clone to ensure sequence quality. Finally, we assembled these reads to create a consensus sequence for each clone.
Removing Redundancy
All-against-all BLASTN searches (Altschul et al., 1990) were conducted among the barley (Hordeum vulgare) FLcDNA sequences to detect redundant sequences. When an FLcDNA was similar to another FLcDNA with 90% or greater identity and 95% or greater coverage, the FLcDNAs were clustered. If two FLcDNAs that did not overlap were in the same cluster, we considered that a fused transcript that bridged the two FLcDNAs was contained in this cluster. In this case, the cDNA was discarded and the cluster was separated into two clusters.
Prediction of ORFs and Functional Annotation
Representative ORFs were predicted by BLASTX searches against the RefSeq and UniProtKB databases with a positive match cutoff set at an E value of less than 1 × 10−20. If there were no homologous proteins in the databases, the longest ORFs (more than 70 amino acids in length) were assigned as predicted ORFs. If no ORFs were predicted for an FLcDNA, it was defined as a noncoding FLcDNA. Noncoding FLcDNAs were compared with rice microRNAs downloaded from miRBase (http://www.mirbase.org/; Griffiths-Jones et al., 2008) using BLASTN. To assign a gene function on the basis of conserved domains or motifs, InterProScan searches (Zdobnov and Apweiler, 2001; Hunter et al., 2009) were conducted using the predicted ORFs. Based on the results of the InterProScan searches, GO categories were assigned to each predicted ORF (Barrell et al., 2009). The GO assignments were categorized using map2slim software (http://www.geneontology.org/GO.slims.shtml#whatIs).
Comparison of FLcDNA Sequences with Complete Genome Sequences from Other Crop Species
To conduct genome-wide comparisons of the FLcDNA sequences, genome sequences and annotation data from four completely sequenced crop plants were obtained at the following sites: the Rice Annotation Project Database (RAP-DB; http://rapdb.dna.affrc.go.jp/) for Oryza sativa; the MaizeSequence database (http://www.maizesequence.org/index.html) for Zea mays; the JGI (http://genome.jgi-psf.org/Sorbi1/Sorbi1.home.html) for Sorghum bicolor; and BrachyBase (http://www.brachybase.org/) for Brachypodium distachyon. The FLcDNA sequences were mapped onto the genome sequences using BLASTN search at an E value of less than 1 × 10−5.
Detection of Novel Barley FLcDNAs
mRNAs and ESTs from barley and wheat (Triticum aestivum) were downloaded from the DDBJ. The poly(A) sequences were then trimmed, and repetitive regions were masked using RepeatMasker software (http://repeatmasker.org). We conducted BLASTN searches of the sequences against all barley FLcDNAs with the threshold of a positive match set at an identity of 90% or greater and a coverage of 90% or greater. Because the data set of barley FLcDNAs contained redundant sequences, multiple hits for the query mRNA or EST sequences were accepted.
Comparison with Rice Trait Genes
A list of rice trait genes was downloaded from Oryzabase (http://www.shigen.nig.ac.jp/rice/oryzabase/genes/genesTop.jsp). Of 4,124 trait genes, 881 genes had RAP-DB transcript information. The amino acid sequences of these genes were used as queries for BLASTP analyses with the predicted ORFs of the Uni-FLcDNAs.
Comparison with Barley BAC Sequences
To compare the FLcDNAs with BAC sequences, one Haruna Nijo BAC sequence (Taketa et al., 2008) and 181 Morex BAC sequences were downloaded from the DDBJ/EMBL/GenBank. The Morex BACs were downloaded using the keywords “Morex” and “BAC.” After masking repeat sequences using RepeatMasker and the libraries of MIPS Repeat Element Database 4.3 (http://mips.helmholtz-muenchen.de/plant/genomes.jsp; Spannagl et al., 2007) and Triticeae Repeat Sequence Database release 10 (http://wheat.pw.usda.gov/ITMI/Repeats/), the FLcDNAs were mapped onto the BACs using the same method used for RAP annotation (Tanaka et al., 2008). The FLcDNAs were first mapped using BLASTN to determine the possible mapping regions in the genomic sequences and further mapped using EST2genome (Mott, 1997) with thresholds of 95% or greater identity and 90% or greater coverage. When two or more FLcDNAs were mapped to the same locus, the longest FLcDNA was used for further analysis. All of the sequence data for the representative FLcDNAs have been submitted to the DDBJ (accession nos. AK353559–AK377172).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Distribution of insert lengths of FLcDNAs.
Supplemental Figure S2. Alignment of sense/antisense FLcDNAs.
Supplemental Figure S3. GO categorization of Uni-FLcDNAs.
Supplemental Figure S4. Annotation of a BAC sequence using barley FLcDNAs.
Supplemental Table S1. Conditions used for sample preparation for library construction.
Supplemental Table S2. Homologous sequences for rice trait genes in the Uni-FLcDNA library.
Supplemental Table S3. Automated annotation of barley FLcDNAs.
Supplemental Table S4. Distribution of homologs of novel barley FLcDNAs in four grass species.
Supplemental Table S5. Distribution of homologs of representative barley FLcDNAs in four grass species.
Supplemental Information S1. Construction and evaluation of the barley FLcDNA libraries.
Acknowledgments
We are grateful to Sapporo Breweries Co., Ltd., for providing the germinating barley seeds and to Dr. J.F. Ma from Okayama University for experimental assistance. Published barley FLcDNA information was provided by the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- Alexandrov NN, Troukhan ME, Brover VV, Tatarinova T, Flavell RB, Feldmann KA. (2006) Features of Arabidopsis genes and genome discovered using full-length cDNAs. Plant Mol Biol 60: 69–85 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Aoki K, Yano K, Suzuki A, Kawamura S, Sakurai N, Suda K, Kurabayashi A, Suzuki T, Tsugane T, Watanabe M, et al. (2010) Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genomics 11: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R. (2009) The GOA database in 2009: an integrated Gene Ontology Annotation resource. Nucleic Acids Res 37: D396–D403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Coleman C, Liu R, Ma J, Ramakrishna W. (2004) Consistent over-estimation of gene number in complex plant genomes. Curr Opin Plant Biol 7: 732–736 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kvam C, Kitamura A, Ohsumi T, Okazaki Y, Itoh M, Kamiya M, Shibata K, Sasaki N, Izawa M, et al. (1996) High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics 37: 327–336 [DOI] [PubMed] [Google Scholar]
- Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y. (2000) Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res 10: 1617–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ, Wanamaker SI, Caldo RA, Turner SM, Ashlock DA, Dickerson JA, Wing RA, Muehlbauer GJ, Kleinhofs A, Wise RP. (2004) A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiol 134: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruveiller S, Jabbari K, Clay O, Bernardi G. (2004) Incorrectly predicted genes in rice? Gene 333: 187–188 [DOI] [PubMed] [Google Scholar]
- Druka A, Muehlbauer G, Druka I, Caldo R, Baumann U, Rostoks N, Schreiber A, Wise R, Close T, Kleinhofs A, et al. (2006) An atlas of gene expression from seed to seed through barley development. Funct Integr Genomics 6: 202–211 [DOI] [PubMed] [Google Scholar]
- Duvick J, Fu A, Muppirala U, Sabharwal M, Wilkerson MD, Lawrence CJ, Lushbough C, Brendel V. (2008) PlantGDB: a resource for comparative plant genomics. Nucleic Acids Res 36: D959–D965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37: D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi T, Itoh T, Suzuki Y, O’Donovan C, Fukuchi S, Koyanagi KO, Barrero R A, Tamura T, Yamaguchi-Kabata Y, Tanino M, et al. (2004) Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol 2: 0859–0875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Itoh T, Tanaka T, Barrero RA, Yamasaki C, Fujii Y, Hilton PB, Antonio BA, Aono H, Apweiler R, Bruskiewich R, et al. (2007) Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res 17: 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari K, Cruveiller S, Clay O, Le Saux J, Bernardi G. (2004) The new genes of rice: a closer look. Trends Plant Sci 9: 281–285 [DOI] [PubMed] [Google Scholar]
- Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, et al. (2001) Functional annotation of a full-length mouse cDNA collection. Nature 409: 685–690 [DOI] [PubMed] [Google Scholar]
- Kawaura K, Mochida K, Enju A, Totoki Y, Toyoda A, Sakaki Y, Kai C, Kawai J, Hayashizaki Y, Seki M, et al. (2009) Assessment of adaptive evolution between wheat and rice as deduced from full-length common wheat cDNA sequence data and expression patterns. BMC Genomics 10: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al. (2003) Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301: 376–379 [DOI] [PubMed] [Google Scholar]
- Koo BC, Bushman BS, Mott IW. (2008) Transcripts associated with non-acclimated freezing response in two barley cultivars. Plant Genome 1: 21–32 [Google Scholar]
- Kurata N, Yamazaki Y. (2006) Oryzabase: an integrated biological and genome information database for rice. Plant Physiol 140: 12–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Sugano S. (1994) Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138: 171–174 [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Taudien S, Martis M, Simková H, Suchánková P, Gundlach H, Wicker T, Petzold A, Felder M, Steuernagel B, et al. (2009) Gene content and virtual gene order of barley chromosome 1H. Plant Physiol 151: 496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R. (1997) EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA. Comput Appl Biosci 13: 477–478 [DOI] [PubMed] [Google Scholar]
- Ogihara Y, Mochida K, Kawaura K, Murai K, Seki M, Kamiya A, Shinozaki K, Carninci P, Hayashizaki Y, Shin-I T, et al. (2004) Construction of a full-length cDNA library from young spikelets of hexaploid wheat and its characterization by large-scale sequencing of expressed sequence tags. Genes Genet Syst 79: 227–232 [DOI] [PubMed] [Google Scholar]
- Osato N, Yamada H, Satoh K, Ooka H, Yamamoto M, Suzuki K, Kawai J, Carninci P, Ohtomo Y, Murakami K, et al. (2003) Antisense transcripts with rice full-length cDNAs. Genome Biol 5: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. (2004) Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet 36: 40–45 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR. (2009) NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res 37: D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisho D, Myoraku E, Kawasaki S, Sato K, Takeda K. (2007) Construction and characterization of a bacterial artificial chromosome (BAC) library from the Japanese malting barley variety Haruna Nijo. Breed Sci 57: 29–38 [Google Scholar]
- Sato K, Nankaku N, Takeda K. (2009a) A high-density transcript linkage map of barley derived from a single population. Heredity 103: 110–117 [DOI] [PubMed] [Google Scholar]
- Sato K, Shin I T, Seki M, Shinozaki K, Yoshida H, Takeda K, Yamazaki Y, Conte M, Kohara Y. (2009b) Development of 5006 full-length CDNAs in barley: a tool for accessing cereal genomics resources. DNA Res 16: 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schulte D, Close TJ, Graner A, Langridge P, Matsumoto T, Muehlbauer G, Sato K, Schulman AH, Waugh R, Wise RP, et al. (2009) The international barley sequencing consortium: at the threshold of efficient access to the barley genome. Plant Physiol 149: 142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Seki M, Satou M, Sakurai T, Akiyama K, Iida K, Ishida J, Nakajima M, Enju A, Narusaka M, Fujita M, et al. (2004) RIKEN Arabidopsis full-length (RAFL) cDNA and its applications for expression profiling under abiotic stress conditions. J Exp Bot 55: 213–223 [DOI] [PubMed] [Google Scholar]
- Soderlund C, Descour A, Kudrna D, Bomhoff M, Boyd L, Currie J, Angelova A, Collura K, Wissotski M, Ashley E, et al. (2009) Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet 5: e1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spannagl M, Noubibou O, Haase D, Yang L, Gundlach H, Hindemitt T, Klee K, Haberer G, Schoof H, Mayer KF. (2007) MIPSPlantsDB: plant database resource for integrative and comparative plant genome research. Nucleic Acids Res 35: D834–D840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Graner A, Wobus U. (2008a) Barley genomics: an overview. Int J Plant Genomics 2008: 486258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Radchuk V, Strickert M, Miersch O, Weschke W, Wobus U. (2006) Gene expression patterns reveal tissue-specific signaling networks controlling programmed cell death and ABA-regulated maturation in developing barley seeds. Plant J 47: 310–327 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Usadel B, Winter A, Radchuk V, Scholz U, Stein N, Weschke W, Strickert M, Close TJ, Stitt M, et al. (2008b) Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol 146: 1738–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S. (1997) Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene 200: 149–156 [DOI] [PubMed] [Google Scholar]
- Taketa S, Amano S, Tsujino Y, Sato T, Saisho D, Kakeda K, Nomura M, Suzuki T, Matsumoto T, Sato K, et al. (2008) Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc Natl Acad Sci USA 105: 4062–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamè V, Ozturk NZ, Bohnert HJ, Tuberosa R. (2007) Barley transcript profiles under dehydration shock and drought stress treatments: a comparative analysis. J Exp Bot 58: 229–240 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y, Numa H, Sakai H, Wu J, Itoh T, Sasaki T, et al. (2008) The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res 36: D1028–D1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasini L, Svensson JT, Rodriguez EM, Wahid A, Malatrasi M, Kato K, Wanamaker S, Resnik J, Close TJ. (2008) Dehydrin gene expression provides an indicator of low temperature and drought stress: transcriptome-based analysis of barley (Hordeum vulgare L.). Funct Integr Genomics 8: 387–405 [DOI] [PubMed] [Google Scholar]
- Ueda A, Kathiresan A, Bennett J, Takabe T. (2006) Comparative transcriptome analyses of barley and rice under salt stress. Theor Appl Genet 112: 1286–1294 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sakurai T, Totoki Y, Toyoda A, Seki M, Ishiwata A, Akiyama K, Kurotani A, Yoshida T, Mochida K, et al. (2008) Sequencing and analysis of approximately 40,000 soybean cDNA clones from a full-length-enriched cDNA library. DNA Res 15: 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2009) The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res 37: D169–D174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Langridge P, Graner A. (2007) Application of genomics to molecular breeding of wheat and barley. Adv Genet 58: 121–155 [DOI] [PubMed] [Google Scholar]
- Walia H, Wilson C, Wahid A, Condamine P, Cui X, Close TJ. (2006) Expression analysis of barley (Hordeum vulgare L.) during salinity stress. Funct Integr Genomics 6: 143–156 [DOI] [PubMed] [Google Scholar]
- Wang XJ, Gaasterland T, Chua NH. (2005) Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol 6: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. (2001) InterProScan: an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848 [DOI] [PubMed] [Google Scholar]